Deep Transfer Learning Enables Robust Prediction of Antimicrobial Resistance for Novel Antibiotics
Abstract
:1. Introduction
2. Results
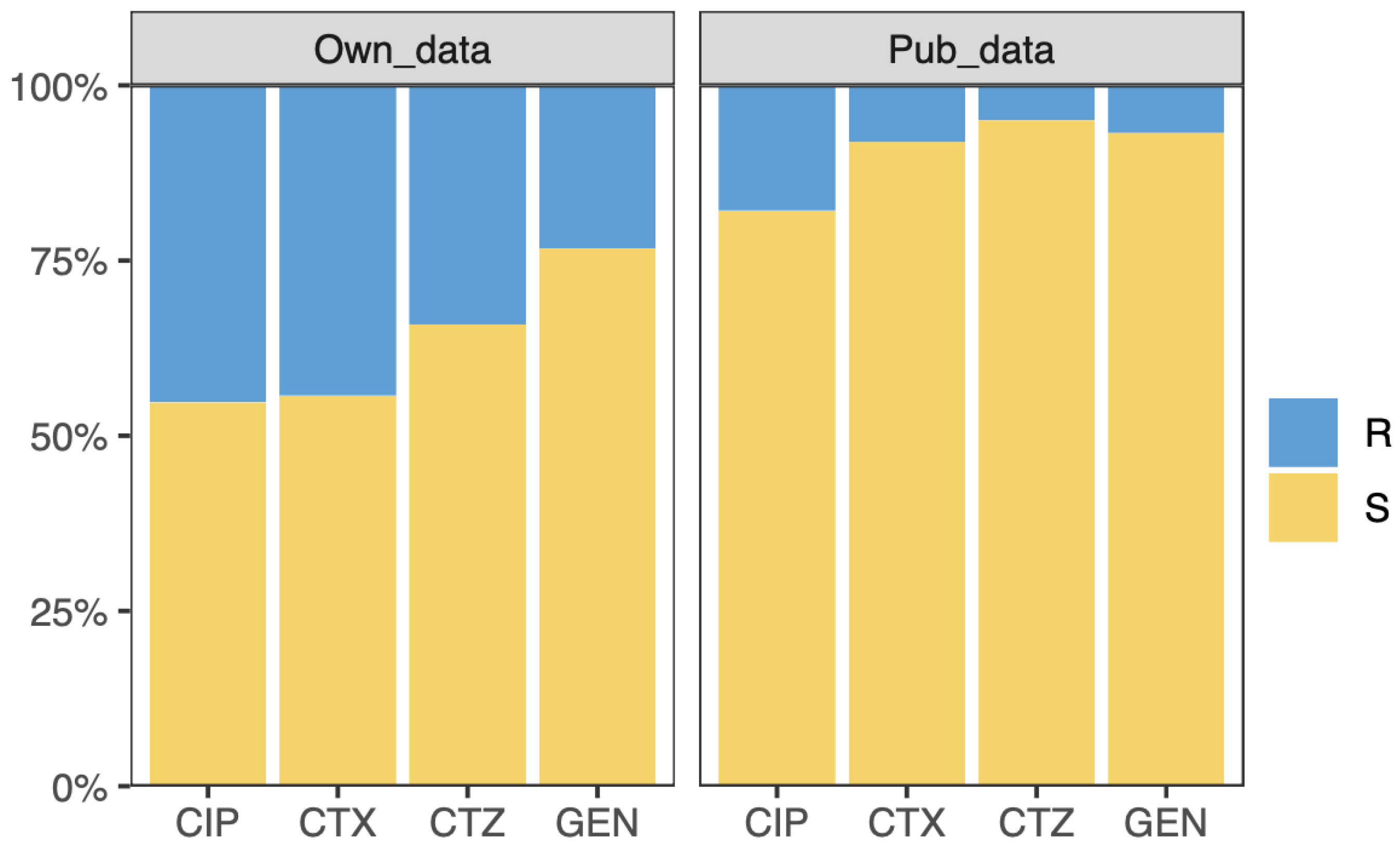
2.1. Datasets
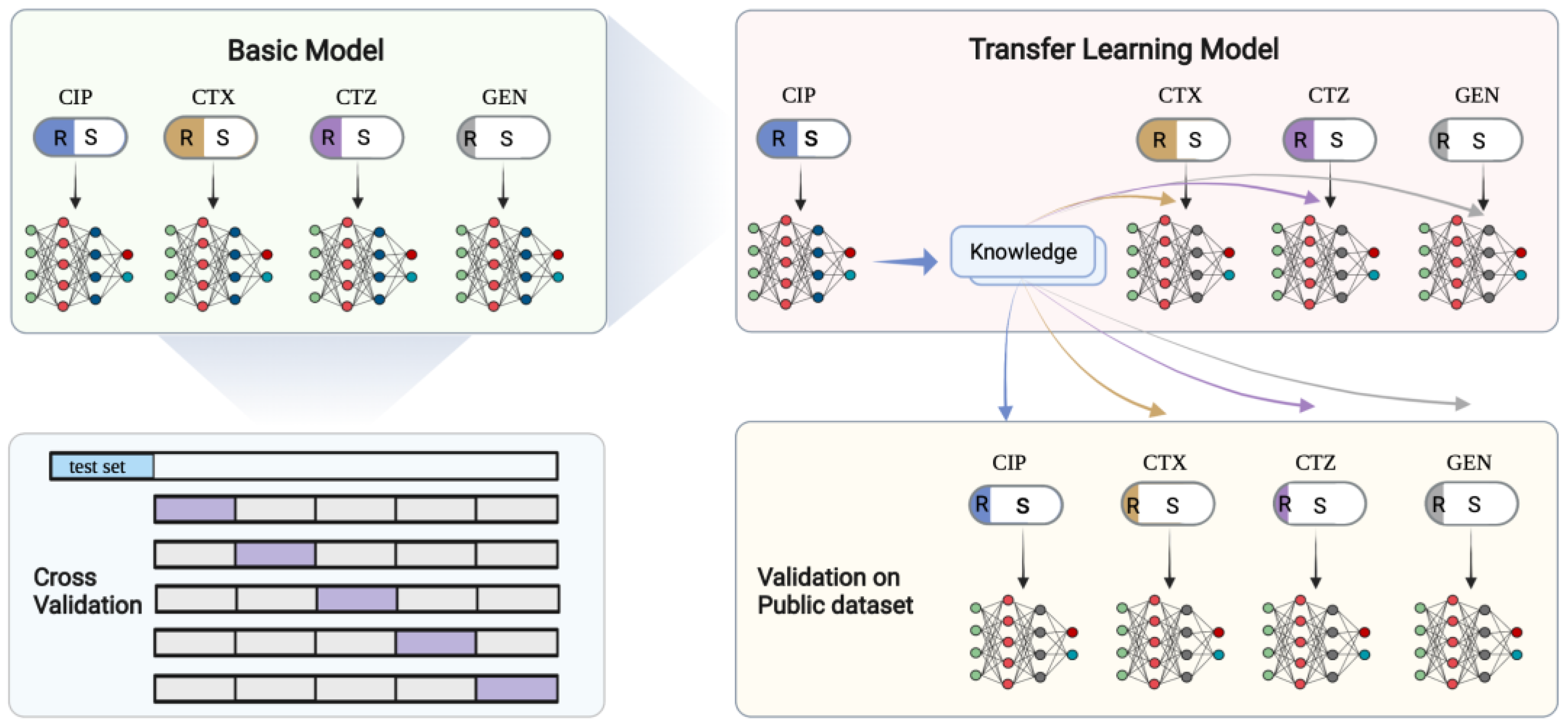
2.2. Study Design
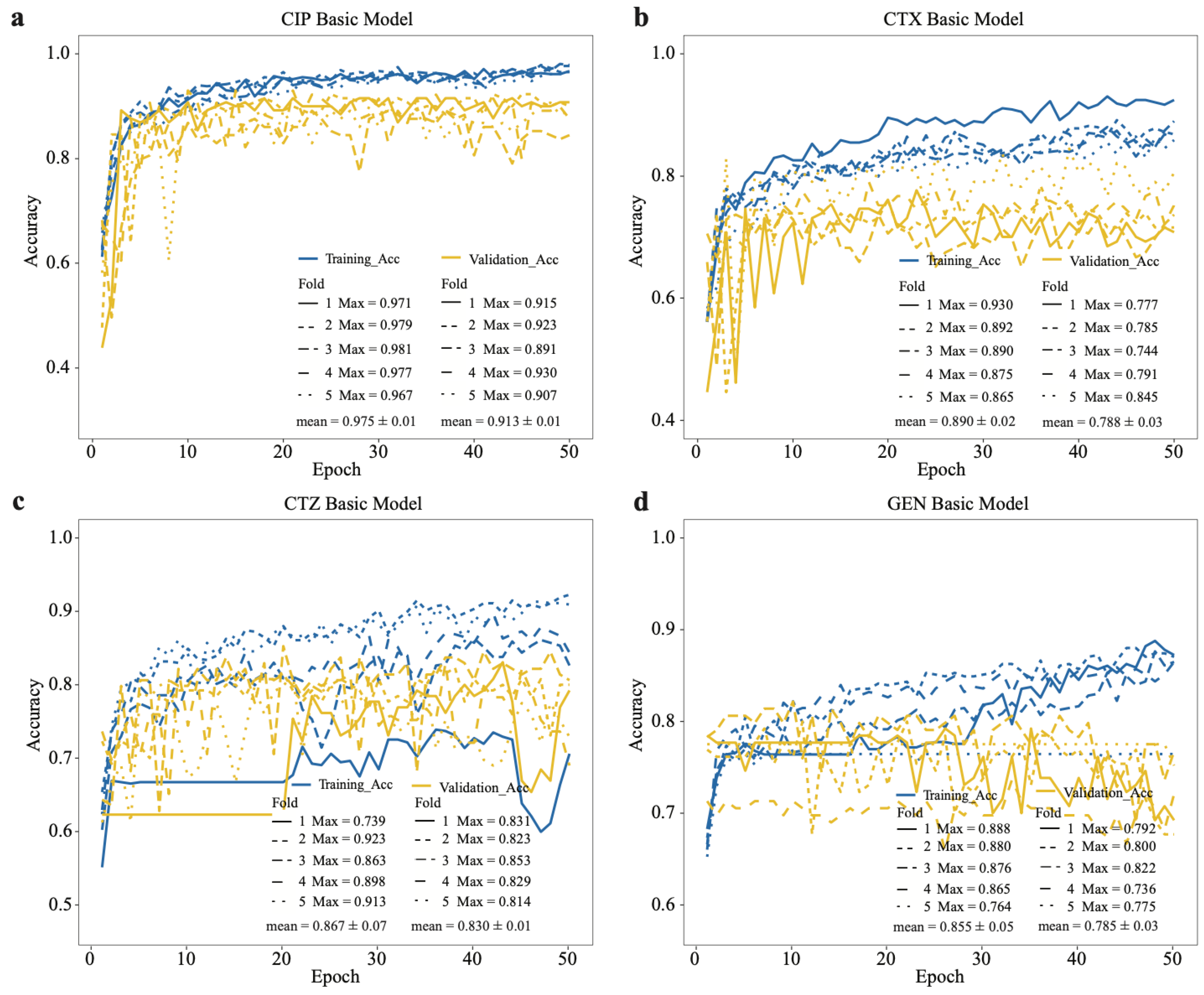
2.3. Performance of the Basic CNN Models
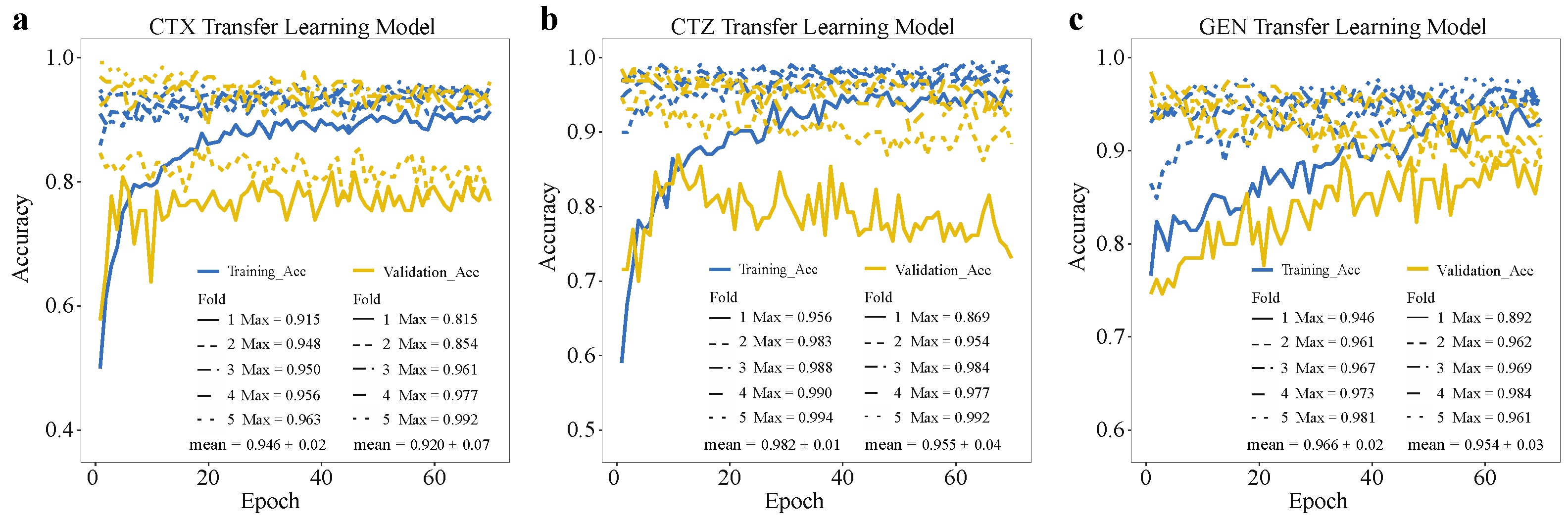
2.4. Deep Transfer Learning Improves the Model Performance on the Minority Group
2.5. Model Evaluation on Independent Public Data
3. Discussion
4. Materials and Methods
4.1. Data Pre-processing
4.2. Basic CNN Model
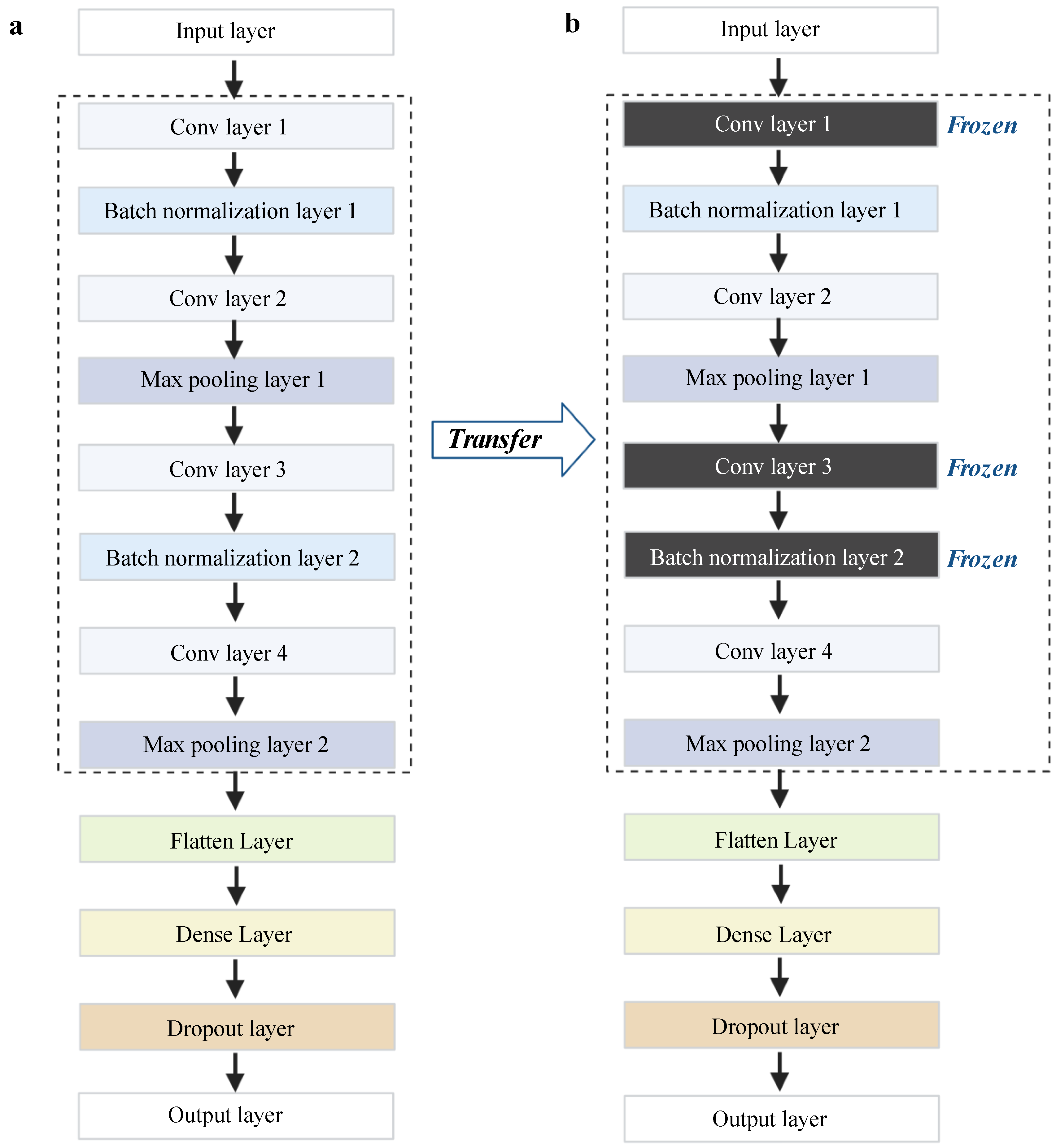
4.3. Deep Transfer Learning Architecture
4.4. Model Evaluation Metrics
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial Resistance: A Global Multifaceted Phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO-Antimicrobial_Resistance_Whitepaper. Available online: https://www.who.int/docs/default-source/documents/no-time-to-wait-securing-the-future-from-drug-resistant-infections-en.pdf (accessed on 15 October 2021).
- Boolchandani, M.; D’Souza, A.W.; Dantas, G. Sequencing-Based Methods and Resources to Study Antimicrobial Resistance. Nat. Rev. Genet. 2019, 20, 356–370. [Google Scholar] [CrossRef]
- Macesic, N.; Polubriaginof, F.; Tatonetti, N.P. Machine Learning: Novel Bioinformatics Approaches for Combating Antimicrobial Resistance. Curr. Opin. Infect. Dis. 2017, 30, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.H.; Wright, S.N.; Hamblin, M.; McCloskey, D.; Alcantar, M.A.; Schrübbers, L.; Lopatkin, A.J.; Satish, S.; Nili, A.; Palsson, B.O.; et al. A White-Box Machine Learning Approach for Revealing Antibiotic Mechanisms of Action. Cell 2019, 177, 1649–1661.e9. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Chakraborty, T.; Doijad, S.; Falgenhauer, L.; Falgenhauer, J.; Goesmann, A.; Hauschild, A.-C.; Schwengers, O.; Heider, D. Prediction of Antimicrobial Resistance Based on Whole-Genome Sequencing and Machine Learning. Bioinformatics 2021, 38, 325–334. [Google Scholar] [CrossRef]
- Arango-Argoty, G.A.; Garner, E.; Pruden, A.; Heath, L.S.; Vikesland, P.; Zhang, L. DeepARG: A Deep Learning Approach for Predicting Antibiotic Resistance Genes from Metagenomic Data. Microbiome 2018, 6, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.; Bhadra, P.; Li, A.; Sethiya, P.; Qin, L.; Tai, H.K.; Wong, K.H.; Siu, S.W.I. Deep-AmPEP30: Improve Short Antimicrobial Peptides Prediction with Deep Learning. Mol. Ther. Nucleic Acids 2020, 20, 882–894. [Google Scholar] [CrossRef]
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.M.; MacNair, C.R.; French, S.; Carfrae, L.A.; Bloom-Ackerman, Z.; et al. A Deep Learning Approach to Antibiotic Discovery. Cell 2020, 180, 688–702.e13. [Google Scholar] [CrossRef] [Green Version]
- Al-Stouhi, S.; Reddy, C.K. Transfer Learning for Class Imbalance Problems with Inadequate Data. Knowl. Inf. Syst. 2016, 48, 201–228. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, F.; Qi, Z.; Duan, K.; Xi, D.; Zhu, Y.; Zhu, H.; Xiong, H.; He, Q. A Comprehensive Survey on Transfer Learning. Proc. IEEE 2020, 109, 43–76. [Google Scholar] [CrossRef]
- Chen, Y. A Transfer Learning Model with Multi-Source Domains for Biomedical Event Trigger Extraction. BMC Genom. 2021, 22, 31. [Google Scholar] [CrossRef]
- Yu, J.; Deng, Y.; Liu, T.; Zhou, J.; Jia, X.; Xiao, T.; Zhou, S.; Li, J.; Guo, Y.; Wang, Y.; et al. Lymph Node Metastasis Prediction of Papillary Thyroid Carcinoma Based on Transfer Learning Radiomics. Nat. Commun. 2020, 11, 4807. [Google Scholar] [CrossRef]
- Mahbod, A.; Schaefer, G.; Wang, C.; Dorffner, G.; Ecker, R.; Ellinger, I. Transfer Learning Using a Multi-Scale and Multi-Network Ensemble for Skin Lesion Classification. Comput. Methods Programs Biomed. 2020, 193, 105475. [Google Scholar] [CrossRef]
- Farahani, A.; Pourshojae, B.; Rasheed, K.; Arabnia, H.R. A Concise Review of Transfer Learning. In Proceedings of the 2020 International Conference on Computational Science and Computational Intelligence (CSCI), Las Vegas, NV, USA, 16–18 December 2020. [Google Scholar]
- Radha, M.; Fonseca, P.; Moreau, A.; Ross, M.; Cerny, A.; Anderer, P.; Long, X.; Aarts, R.M. A Deep Transfer Learning Approach for Wearable Sleep Stage Classification with Photoplethysmography. NPJ Digit. Med. 2021, 4, 135. [Google Scholar] [CrossRef]
- Mallesh, N.; Zhao, M.; Meintker, L.; Höllein, A.; Elsner, F.; Lüling, H.; Haferlach, T.; Kern, W.; Westermann, J.; Brossart, P.; et al. Knowledge Transfer to Enhance the Performance of Deep Learning Models for Automated Classification of B Cell Neoplasms. Patterns 2021, 2, 100351. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Weiss, K.; Khoshgoftaar, T.M.; Wang, D. A Survey of Transfer Learning. J. Big Data 2016, 3, 9. [Google Scholar] [CrossRef] [Green Version]
- Kopas, L.; Kusalik, A.; Schneider, D. Antimicrobial Resistance Prediction from Whole-Genome Sequence Data Using Transfer Learning. F1000Research 2019, 8, 1333. [Google Scholar] [CrossRef]
- Ebbehoj, A.; Thunbo, M.Ø.; Andersen, O.E.; Glindtvad, M.V.; Hulman, A. Transfer Learning for Non-Image Data in Clinical Research: A Scoping Review. PLoS Digit. Health 2022, 1, e0000014. [Google Scholar] [CrossRef]
- Liu, Z.; Jiang, M.; Luo, T. Leverage Electron Properties to Predict Phonon Properties via Transfer Learning for Semiconductors. Sci. Adv. 2020, 6, eabd1356. [Google Scholar] [CrossRef]
- Plested, J.; Gedeon, T. Deep Transfer Learning for Image Classification: A Survey. arXiv 2022, arXiv:2205.09904. [Google Scholar]
- Li, X.; Grandvalet, Y.; Davoine, F.; Cheng, J.; Cui, Y.; Zhang, H.; Belongie, S.; Tsai, Y.-H.; Yang, M.-H. Transfer Learning in Computer Vision Tasks: Remember Where You Come From. Image Vis. Comput. 2020, 93, 103853. [Google Scholar] [CrossRef]
- Gao, Y.; Mosalam, K.M. Deep Transfer Learning for Image-Based Structural Damage Recognition: Deep Transfer Learning for Image-Based Structural Damage Recognition. Comput.-Aided Civ. Infrastruct. Eng. 2018, 33, 748–768. [Google Scholar] [CrossRef]
- Shao, L.; Zhu, F.; Li, X. Transfer Learning for Visual Categorization: A Survey. IEEE Trans. Neural Netw. Learn. Syst. 2015, 26, 1019–1034. [Google Scholar] [CrossRef] [PubMed]
- Schwessinger, R.; Gosden, M.; Downes, D.; Brown, R.C.; Oudelaar, A.M.; Telenius, J.; Teh, Y.W.; Lunter, G.; Hughes, J.R. DeepC: Predicting 3D Genome Folding Using Megabase-Scale Transfer Learning. Nat. Methods 2020, 17, 1118–1124. [Google Scholar] [CrossRef]
- Mikolov, T.; Chen, K.; Corrado, G.; Dean, J. Efficient Estimation of Word Representations in Vector Space. arXiv 2013, arXiv:1301.3781. [Google Scholar]
- Gupta, V.; Choudhary, K.; Tavazza, F.; Campbell, C.; Liao, W.; Choudhary, A.; Agrawal, A. Cross-Property Deep Transfer Learning Framework for Enhanced Predictive Analytics on Small Materials Data. Nat. Commun. 2021, 12, 6595. [Google Scholar] [CrossRef]
- Park, Y.; Hauschild, A.-C.; Heider, D. Transfer Learning Compensates Limited Data, Batch-Effects, And Technical Heterogeneity In Single-Cell Sequencing. bioRxiv 2021. [Google Scholar] [CrossRef]
- Okerinde, A.; Shamir, L.; Hsu, W.; Theis, T.; Nafi, N. EGAN: Unsupervised Approach to Class Imbalance Using Transfer Learning. arXiv 2021, arXiv:2104.04162. [Google Scholar]
- Weiss, K.R.; Khoshgoftaar, T.M. Investigating Transfer Learners for Robustness to Domain Class Imbalance. In Proceedings of the 2016 15th IEEE International Conference on Machine Learning and Applications (ICMLA), Anaheim, CA, USA, 18–20 December 2016; pp. 207–213. [Google Scholar]
- Minvielle, L.; Atiq, M.; Peignier, S.; Mougeot, M. Transfer Learning on Decision Tree with Class Imbalance. In Proceedings of the 2019 IEEE 31st International Conference on Tools with Artificial Intelligence (ICTAI), Portland, OR, USA, 4–6 November 2019; pp. 1003–1010. [Google Scholar]
- Krawczyk, B. Learning from Imbalanced Data: Open Challenges and Future Directions. Prog. Artif. Intell. 2016, 5, 221–232. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Cui, Y. Deep Transfer Learning for Reducing Health Care Disparities Arising from Biomedical Data Inequality. Nat. Commun. 2020, 11, 5131. [Google Scholar] [CrossRef] [PubMed]
- Falgenhauer, L.; Nordmann, P.; Imirzalioglu, C.; Yao, Y.; Falgenhauer, J.; Hauri, A.M.; Heinmüller, P.; Chakraborty, T. Cross-Border Emergence of Clonal Lineages of ST38 Escherichia Coli Producing the OXA-48-like Carbapenemase OXA-244 in Germany and Switzerland. Int. J. Antimicrob. Agents 2020, 56, 106157. [Google Scholar] [CrossRef] [PubMed]
- Moradigaravand, D.; Palm, M.; Farewell, A.; Mustonen, V.; Warringer, J.; Parts, L. Prediction of Antibiotic Resistance in Escherichia Coli from Large-Scale Pan-Genome Data. PLoS Comput. Biol. 2018, 14, e1006258. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Wang, S.; Xu, Y.; Zhang, W.; Tang, K.; Ouyang, Q.; Lai, L.; Pei, J. Transfer Learning for Drug Discovery. J. Med. Chem. 2020, 63, 8683–8694. [Google Scholar] [CrossRef]
- Chicco, D.; Jurman, G. The Advantages of the Matthews Correlation Coefficient (MCC) over F1 Score and Accuracy in Binary Classification Evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef] [Green Version]
- Chicco, D.; Starovoitov, V.; Jurman, G. The Benefits of the Matthews Correlation Coefficient (MCC) Over the Diagnostic Odds Ratio (DOR) in Binary Classification Assessment. IEEE Access 2021, 9, 47112–47124. [Google Scholar] [CrossRef]
- Boughorbel, S.; Jarray, F.; El-Anbari, M. Optimal Classifier for Imbalanced Data Using Matthews Correlation Coefficient Metric. PLoS ONE 2017, 12, e0177678. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Vakili, M.; Ghamsari, M.; Rezaei, M. Performance Analysis and Comparison of Machine and Deep Learning Algorithms for IoT Data Classification. arXiv 2020, arXiv:2001.09636. [Google Scholar]


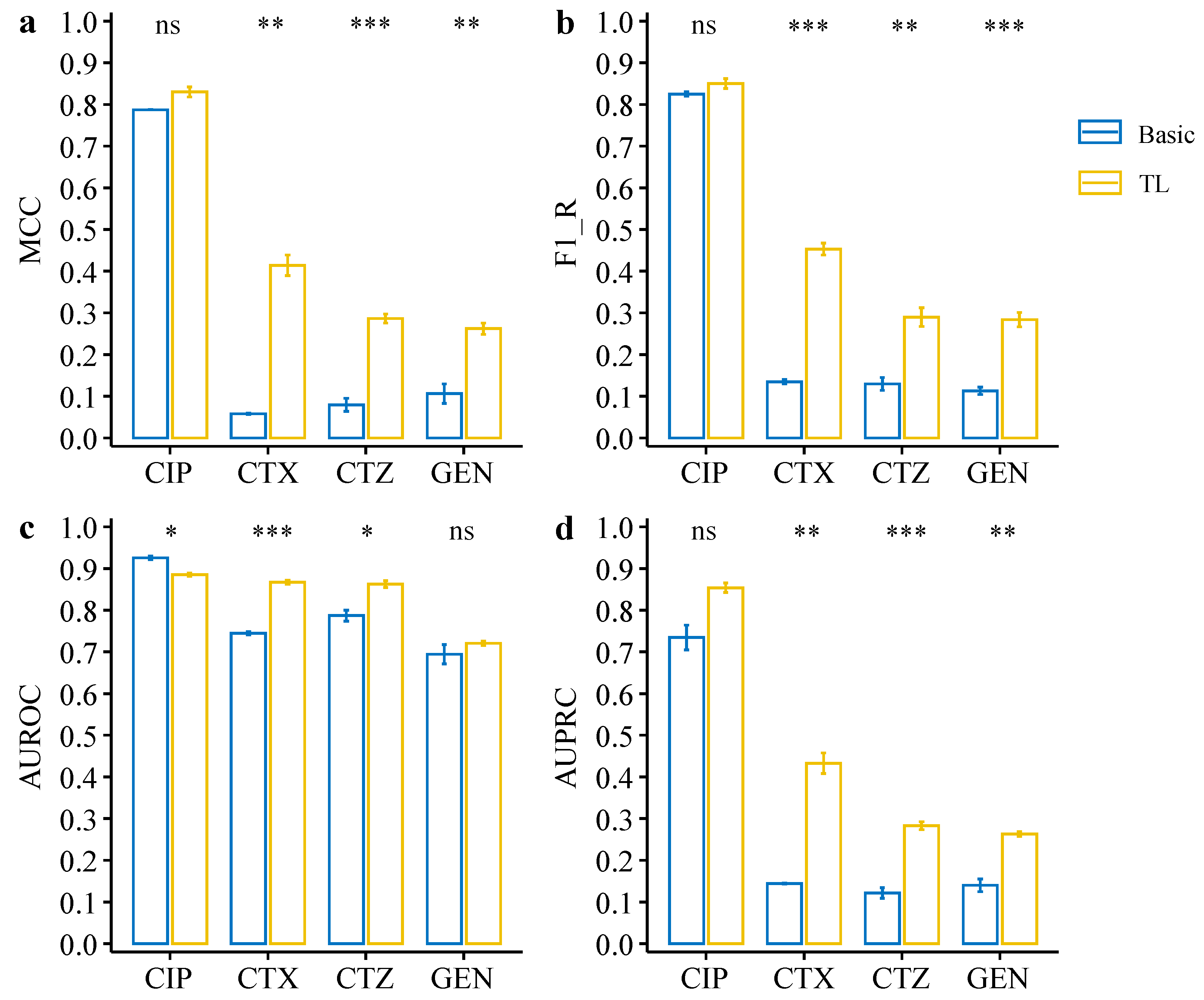
| Drugs | CTX | CTZ | GEN | |||
|---|---|---|---|---|---|---|
| Metrics | MCC | F1-R | MCC | F1-R | MCC | F1-R |
| Basic | 0.47 ± 0.03 | 0.70 ± 0.02 | 0.46 ± 0.03 | 0.65 ± 0.02 | 0.33 ± 0.01 | 0.41 ± 0.02 |
| TL | 0.56 ± 0.03 | 0.76 ± 0.02 | 0.55 ± 0.03 | 0.71 ± 0.02 | 0.53 ± 0.03 | 0.63 ± 0.02 |
| Drugs | CIP | CTX | CTZ | GEN | ||||
|---|---|---|---|---|---|---|---|---|
| Model | Basic | TL | Basic | TL | Basic | TL | Basic | TL |
| MCC | 0.79 ± 0.00 | 0.83 ± 0.02 | 0.06 ± 0.00 | 0.41 ± 0.04 | 0.08 ± 0.03 | 0.29 ± 0.02 | 0.11 ± 0.04 | 0.26 + 0.03 |
| F1-R | 0.83 ± 0.01 | 0.85 ± 0.02 | 0.14 ± 0.01 | 0.45 ± 0.03 | 0.13 ± 0.03 | 0.29 ± 0.05 | 0.11 ± 0.02 | 0.28 + 0.04 |
| AUROC | 0.93 ± 0.01 | 0.89 ± 0.01 | 0.74 ± 0.00 | 0.87 ± 0.01 | 0.79 ± 0.02 | 0.86 ± 0.02 | 0.69 ± 0.04 | 0.72 + 0.01 |
| AUPRC | 0.73 ± 0.04 | 0.85 ± 0.02 | 0.14 ± 0.00 | 0.43 ± 0.04 | 0.12 ± 0.02 | 0.28 ± 0.02 | 0.14 ± 0.03 | 0.26 + 0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, Y.; Chakraborty, T.; Doijad, S.; Falgenhauer, L.; Falgenhauer, J.; Goesmann, A.; Schwengers, O.; Heider, D. Deep Transfer Learning Enables Robust Prediction of Antimicrobial Resistance for Novel Antibiotics. Antibiotics 2022, 11, 1611. https://doi.org/10.3390/antibiotics11111611
Ren Y, Chakraborty T, Doijad S, Falgenhauer L, Falgenhauer J, Goesmann A, Schwengers O, Heider D. Deep Transfer Learning Enables Robust Prediction of Antimicrobial Resistance for Novel Antibiotics. Antibiotics. 2022; 11(11):1611. https://doi.org/10.3390/antibiotics11111611
Chicago/Turabian StyleRen, Yunxiao, Trinad Chakraborty, Swapnil Doijad, Linda Falgenhauer, Jane Falgenhauer, Alexander Goesmann, Oliver Schwengers, and Dominik Heider. 2022. "Deep Transfer Learning Enables Robust Prediction of Antimicrobial Resistance for Novel Antibiotics" Antibiotics 11, no. 11: 1611. https://doi.org/10.3390/antibiotics11111611
APA StyleRen, Y., Chakraborty, T., Doijad, S., Falgenhauer, L., Falgenhauer, J., Goesmann, A., Schwengers, O., & Heider, D. (2022). Deep Transfer Learning Enables Robust Prediction of Antimicrobial Resistance for Novel Antibiotics. Antibiotics, 11(11), 1611. https://doi.org/10.3390/antibiotics11111611







