Isolation and Identification of Bioactive Compounds from Streptomyces actinomycinicus PJ85 and Their In Vitro Antimicrobial Activities against Methicillin-Resistant Staphylococcus aureus
Abstract
:1. Introduction
2. Results
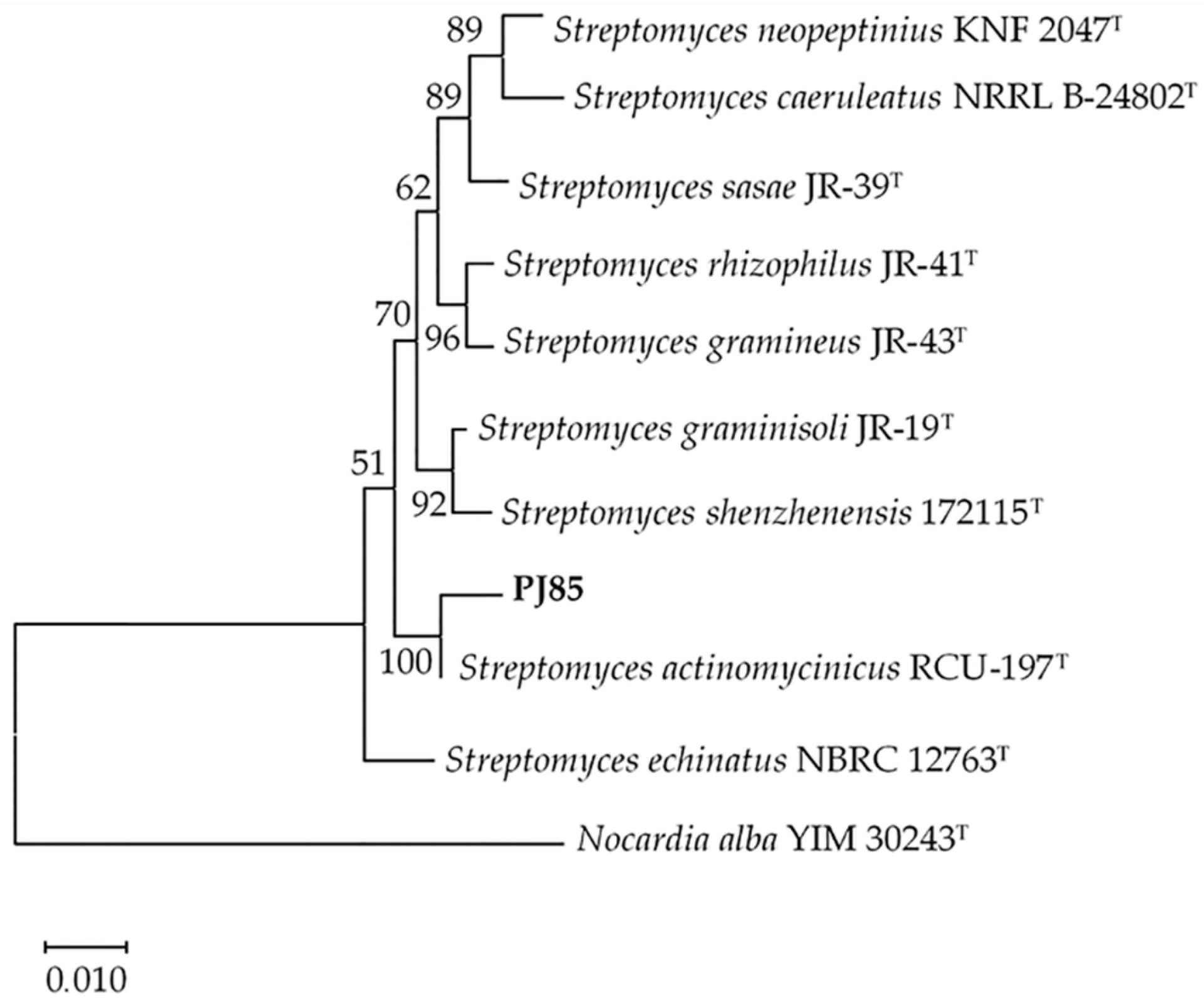
2.1. Identification and Characterization of PJ85 Strain
2.2. Antibacterial Activity of PJ85
2.3. Incubation Temperature, Period Affect Growth, and Antibacterial Activity of PJ85
2.4. Crude Compound Preparation and MIC Values
2.5. Purification of the Active Compounds of Streptomyces sp. PJ85 with Thin-Layer Chromatography, Column Chromatography, and Bioautography Analysis
2.6. Liquid Chromatography–Mass Spectrometry (LC–MS) Analysis
3. Discussion
4. Materials and Methods
4.1. Isolation, Cultivation, and Maintenance of PJ85
4.2. Cultural and Morphological Characteristics
4.3. Amplification and Sequencing of the 16S rRNA Gene
4.4. Phylogenetic Tree Analysis
4.5. Antibacterial Assay
4.5.1. Bacterial Strains
4.5.2. Perpendicular Cross Streak Method
4.5.3. Preparation of Crude Compounds of PJ85
4.5.4. Disc Diffusion Method
4.5.5. Determination of the Minimum Inhibitory Concentration (MIC) of Crude Compounds of PJ85
4.6. The Study of Incubation Temperature and Incubation Period on Growth and Activity of Antibacterial Agents
4.7. Purification of Active Compounds
4.7.1. Purification of Antibacterial Compounds with Thin-Layer Chromatography
4.7.2. Purification of Antibacterial Compounds with Column Chromatography
4.7.3. Contact Bioautography Analysis
4.8. Identification of Active Compounds with Liquid Chromatography–Mass Spectrometry (LC–MS)
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zaffiri, L.; Gardner, J.; Toledo-Pereyra, L.H. History of Antibiotics. From Salvarsan to Cephalosporins. J. Investig. Surg. 2012, 25, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Clardy, J.; Fischbach, M.A.; Currie, C.R. The natural history of antibiotics. Curr. Biol. 2009, 19, R437–R441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, F.; Qin, Y.H.; Zheng, X.; Zhao, H.W.; Chai, D.Y.; Li, W.; Pu, M.X.; Zuo, X.S.; Qian, W.; Ni, P. Biogeography and adaptive evolution of Streptomyces strains from saline environments. Sci. Rep. 2016, 6, 32718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baltz, R.H. Antimicrobials from actinomycetes: Back to the future. Microbe 2007, 2, 125–133. [Google Scholar]
- Aminov, R.I. A brief history of the antibiotic era: Lessons learned and challenges for the future. Front. Microbiol. 2010, 1, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharmacol. Ther. 2015, 40, 277–283. [Google Scholar]
- Fair, R.J.; Tor, Y. Antibiotics and Bacterial Resistance in the 21st Century. Perspect. Medicinal. Chem. 2014, 6, 25–64. [Google Scholar] [CrossRef] [Green Version]
- Enright, M.C.; Robinson, D.A.; Randle, G.; Feil, E.J.; Grundmann, H.; Spratt, B.G. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 2002, 99, 7687–7692. [Google Scholar] [CrossRef] [Green Version]
- Darabpour, E.; Ardakani, M.R.; Motamedi, H.; Ronagh, M.T. Isolation of a potent antibiotic producer bacterium, especially against MRSA, from northern region of the Persian Gulf. Bosn. J. Basic Med. Sci. 2012, 12, 108–121. [Google Scholar] [CrossRef] [Green Version]
- Wangai, F.K.; Masika, M.M.; Maritim, M.C.; Seaton, R.A. Methicillin-resistant Staphylococcus aureus (MRSA) in East Africa: Red alert or red herring? BMC Infect. Dis. 2019, 19, 596. [Google Scholar] [CrossRef]
- Chinnambedu, R.S.; Marimuthu, R.R.; Sunil, S.S.; Amrose, P.; Ramachandran, V.; Pachamuthu, B. Changing antibiotic resistance profile of Staphylococcus aureus isolated from HIV patients (2012–2017) in Southern India. J. Infect. Public Health 2020, 13, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Kemung, H.M.; Tan, L.T.H.; Khan, T.M.; Chan, K.G.; Pusparajah, P.; Goh, B.H.; Lee, L.H. Streptomyces as a prominent resource of future anti-MRSA drugs. Front. Microbiol. 2018, 9, 2221. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, A.; Dick, J.D.; PERL, T.M. Vancomycin resistance in staphylococci. Clin. Microbiol. Rev. 2002, 15, 430–438. [Google Scholar] [CrossRef] [Green Version]
- Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance. Microbiol. Spectr. 2016, 4, 481–511. [Google Scholar] [CrossRef] [Green Version]
- Balsalobre, L.C.; Dropa, M.; Matté, M.H. An overview of antimicrobial resistance and its public health significance. Braz. J. Microbiol. 2014, 45, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, L.H.; Zainal, N.; Azman, A.S.; Eng, S.-K.; Ab Mutalib, N.S.; Yin, W.-F.; Chan, K.-G. Streptomyces pluripotens sp. nov., a bacteriocin-producing streptomycete that inhibits meticillin-resistant Staphylococcus aureus. Int. J. Syst. Evol. Microbiol. 2014, 64, 3297–3306. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.M.; Kim, J. Streptomyces olivicoloratus sp. nov., an antibiotic-producing bacterium isolated from soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 3262–3270. [Google Scholar] [CrossRef]
- Moghannem, S.A.; El-Sherbiny, G.M.; Kalaba, M.H. Isolation and identification of Streptomyces baarnensis MH-133 produce bioactive metabolite inhibiting multidrug resistant bacteria (MDRB). World J. Pharm. Med. 2017, 3, 64–75. [Google Scholar]
- Sabaou, N.; Bijani, C.; Zitouni, A.; Pont, F.; Mathieu, F.; Badji, B. Streptomyces sp. AT37 isolated from a Saharan soil produces a furanone derivative active against multidrug-resistant Staphylococcus aureus. World J. Microbiol. Biotechnol. 2017, 33, 105. [Google Scholar] [CrossRef] [Green Version]
- Singh, L.S.; Sharma, H.; Talukdar, N.C. Production of potent antimicrobial agent by actinomycete, Streptomyces sannanensis strain SU118 isolated from phoomdi in Loktak Lake of Manipur, India. BMC Microbiol. 2014, 14, 278. [Google Scholar] [CrossRef]
- Ravi, R.; Vasantba, J.; Bhoomi, M.; Bonisha, T.; Bhumika, C. Antibacterial potentials of actinomycetes isolated from Gujarat. Int. J. Pharm. Sci. Rev. Res. 2015, 30, 78–83. [Google Scholar]
- El-Sherbini, A.; Khattab, A. Induction of novel mutants of Streptomyces lincolnensis with high Lincomycin production. J. Appl. Pharm. Sci. 2018, 8, 128–135. [Google Scholar] [CrossRef] [Green Version]
- Subramani, R.; Sipkema, D. Marine Rare Actinomycetes: A Promising Source of Structurally Diverse and Unique Novel Natural Products. Mar. Drugs 2019, 17, 249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shetty, P.R.; Buddana, S.K.; Tatipamula, V.B.; Naga, Y.V.V.; Ahmad, J. Production of polypeptide antibiotic from Streptomyces parvulus and its antibacterial activity. Braz. J. Microbiol. 2014, 45, 303–312. [Google Scholar] [CrossRef] [PubMed]
- De Jesus Sousa, J.A.; Olivares, F.L. Plant growth promotion by streptomycetes: Ecophysiology, mechanisms and applications. Chem. Biol. Technol. Agric. 2016, 3, 24. [Google Scholar] [CrossRef] [Green Version]
- Chanthasena, P.; Nantapong, N. Biodiversity of Antimicrobial-Producing Actinomycetes Strains Isolated from Dry Dipterocarp Forest soil in Northeast Thailand. Braz. Arch. Biol. Technol. 2016, 59, e16150674. [Google Scholar] [CrossRef] [Green Version]
- Rios, J.; Recio, M.; Villar, A. Screening methods for natural products with antimicrobial activity: A review of the literature. J. Ethnopharmacol. 1988, 23, 127–149. [Google Scholar] [CrossRef]
- Dewanjee, S.; Gangopadhyay, M.; Bhattacharya, N.; Khanra, R.; Dua, T.K. Bioautography and its scope in the field of natural product chemistry. J. Pharm. Anal. 2015, 5, 75–84. [Google Scholar] [CrossRef] [Green Version]
- Jumpathong, J.; Nuengchamnong, N.; Masin, K.; Nakaew, N.; Suphrom, N. Thin Layer Chromatography-Bioautography Assay for Antibacterial Compounds from Streptomyces sp. TBRC 8912, a Newly Isolated Actinomycin D Producer. Chiang Mai J. Sci. 2019, 46, 839–849. [Google Scholar]
- Tanasupawat, S.; Phongsopitanun, W.; Suwanborirux, K.; Ohkuma, M.; Kudo, T. Streptomyces actinomycinicus sp. nov., isolated from soil of a peat swamp forest. Int. J. Syst. Evol. Microbiol. 2016, 66, 290–295. [Google Scholar] [CrossRef]
- Kawashima, H.; Akimotoa, K.; Higashiyamaa, K.; Fujikawaa, S.; Shimizub, S. Industrial Production of Dihomo-γ-linolenic Acid by a ∆5 Desaturase-defective Mutant of Mortierella alpina 1S-4 Fungus. JAOCS 2000, 77, 1135. [Google Scholar] [CrossRef]
- Desbois, A.P.; Lawlor, K.C. Antibacterial Activity of Long-Chain Polyunsaturated Fatty Acids against Propionibacterium acnes and Staphylococcus aureus. Mar. Drugs 2013, 11, 4544–4557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grantina-levina, L.; Berzina, A.; Nikolajeva, V.; Mekss, P.; Muiznieks, I. Production of fatty acids by Mortierella and Umbelopsis species isolated from temperate climate soils. Environ. Exp. Biol. 2014, 12, 15–27. [Google Scholar]
- Singh, B.N.; Shankar, S.; Srivastava, R.K. Green tea catechin, epigallocatechin-3-gallate (EGCG): Mechanisms, perspectives and clinical applications. Biochem. Pharmacol. 2011, 82, 1807–1821. [Google Scholar] [CrossRef] [Green Version]
- Lewisa, D.A.; Shawb, G.P. A natural flavonoid and synthetic analogues protect the gastric mucosa from aspirin-induced erosions. J. Nutr. Biochem. 2001, 12, 95–100. [Google Scholar] [CrossRef]
- Cacan, M.; Moreau, S.; Tailliez, R. In vitro metabolism of Penicillium roqueforti toxin (PRT) and a structurally related compound, eremofortin A, by rat liver. Toxicology 1977, 8, 205–212. [Google Scholar] [CrossRef]
- Kumar, V.; Tewary, D.K.; Ravindranath, S.D.; Shanker, A. Investigation in tea on fate of fenazaquin residue and its transfer in brew. Food. Chem. Toxicol. 2006, 44, 596–600. [Google Scholar] [CrossRef]
- Jin, S.; Yoshida, M. Antifungal Compound, Feruloylagmatine, Induced in Winter Wheat Exposed to a Low Temperature. Biosci. Biotechnol. Biochem. 2000, 64, 1614–1617. [Google Scholar] [CrossRef]
- Aljamali, N.M. Synthesis of Antifungal Chemical Compounds from Fluconazole with (Pharma-Chemical) Studying. Res. J. Pharm. Biol. Chem. Sci. 2017, 8, 564–573. [Google Scholar]
- Xiong, B.J.; Xu, Y.; Jin, G.L.; Liu, M.; Yang, J.; Yu, C.X. Analgesic efects and pharmacologic mechanisms of the Gelsemium alkaloid koumine on a rat model of postoperative pain. Sci. Rep. 2017, 7, 14269. [Google Scholar] [CrossRef] [Green Version]
- Roca, I.; Akova, M.; Baquero, F.; Carlet, J.; Cavaleri, M.; Coenen, S.; Cohen, J.; Findlay, D.; Gyssens, I.; Heure, O.; et al. The global threat of antimicrobial resistance: Science for intervention. New Microbes New Infect. 2015, 6, 22–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benndorf, R.; Guo, H.; Sommerwerk, E.; Weigel, C.; Garcia-Altares, M.; Martin, K.; Hu, H.; Küfner, M.; de Beer, Z.W.; Poulsen, M.; et al. Natural Products from Actinobacteria Associated with Fungus-Growing Termites. Antibiotics 2018, 7, 83. [Google Scholar] [CrossRef] [Green Version]
- Lane, D. 16S/23S rRNA Sequencing. Nucleic Acid Techniques in Bacterial Systematics; John Wiley and Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Adzitey, F.; Huda, N.; Rusul, G.; Ali, R. Molecular techniques for detecting and typing of bacteria, advantages and application to foodborne pathogens isolated from ducks. 3 Biotech 2013, 3, 97–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Salaza, J.K. Culture-Independent Rapid Detection Methods for Bacterial Pathogens and Toxins in Food Matrices. Compr. Rev. Food. Sci. Food Saf. 2016, 15, 183–205. [Google Scholar] [CrossRef]
- Tan, L.T.-H.; Chan, K.-G.; Chan, C.K.; Khan, T.M.; Lee, L.-H.; Goh, B.-H. Antioxidative potential of a Streptomyces sp. MUM292 isolated from mangrove soil. BioMed Res. Int. 2018, 2018, 4823126. [Google Scholar] [CrossRef] [Green Version]
- Al-dhabi, N.A.; Esmail, G.A.; Duraipandiyan, V.; Arasu, M.V.; Salem-Bekhit, M.M. Isolation, identification and screening of antimicrobial thermophilic Streptomyces sp. Al-Dhabi-1 isolated from Tharban hot spring, Saudi Arabia. Extremophiles 2016, 20, 79–90. [Google Scholar] [CrossRef]
- Komaki, H. Resolution of housekeeping gene sequences used in MLSA for the genus Streptomyces and reclassification of Streptomyces anthocyanicus and Streptomyces tricolor as heterotypic synonyms of Streptomyces violaceoruber. Int. J. Syst. Evol. Microbiol. 2022, 72, 5. [Google Scholar] [CrossRef]
- Antony-Babu, S.; Stien, D.; Eparvier, V.; Parrot, D.; Tomasi, S.; Suzuki, M.T. Multiple Streptomyces species with distinct secondary metabolomes have identical 16S rRNA gene sequences. Sci. Rep. 2017, 7, 11089. [Google Scholar] [CrossRef]
- Liu, Y.; Lai, Q.; Shao, Z. A Multilocus Sequence Analysis Scheme for Phylogeny of Thioclava Bacteria and Proposal of Two Novel Species. Front. Microbiol. 2017, 8, 1321. [Google Scholar] [CrossRef] [Green Version]
- Ian, E.; Malko, D.B.; Sekurova, O.N.; Bredholt, H.; Ru, C.; Borisova, M.E.; Albersmeier, A.; Kalinowski, J.; Gelfand, M.S.; Zotchev, S.B. Genomics of Sponge-Associated Streptomyces spp. Closely Related to Streptomyces albus J1074: Insights into Marine Adaptation and Secondary Metabolite Biosynthesis Potential. PLoS ONE 2014, 9, e96719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arasu, M.V.; Rejiniemon, T.S.; Al-Dhabi, N.A.; Duraipandiyan, V.; Agastian, P.; Huxley, V.A.J.; Song, C.E.; Choi, K.C. In vitro antimicrobial potential of organic solvent extracts of novel actinomycetes isolated from forest soil. Afr. J. Biotechnol. 2014, 13, 1891–1897. [Google Scholar] [CrossRef]
- Naumann, D. Infrared spectroscopy in microbiology. In Encyclopedia of Analytical Chemistry; Wiley: Hoboken, NJ, USA, 2000. [Google Scholar] [CrossRef]
- Manimaran, M.; Gopal, J.V.; Kannabiran, K. Antibacterial activity of Streptomyces sp. VITMK1 isolated from mangrove soil of Pichavaram, Tamil Nadu, India. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2017, 87, 499–506. [Google Scholar] [CrossRef]
- Kumar, R.R.; Jadeja, V.J. Characterization and partial purification of an antibacterial agent from halophilic actinomycetes Kocuria sp. strain rsk4. BioImpacts 2018, 8, 253–261. [Google Scholar] [CrossRef] [Green Version]
- Sharma, M.; Manhas, R.K. Purification and characterization of actinomycins from Streptomyces strain M7 active against methicillin resistant Staphylococcus aureus and vancomycin resistant Enterococcus. BMC Microbiol. 2019, 19, 44. [Google Scholar] [CrossRef] [Green Version]
- Maiti, P.K.; Das, S.; Sahoo, P.; Mandal, S. Streptomyces sp. SM01 isolated from Indian soil produces a novel antibiotic picolinamycin effective against multi drug resistant bacterial strains. Sci. Rep. 2020, 10, 10092. [Google Scholar] [CrossRef]
- Awla, H.K.; Kadir, J.; Othman, R.; Rashid, T.S.; Wong, M.-Y. Bioactive Compounds Produced by Streptomyces sp. Isolate UPMRS4 and Antifungal Activity against Pyricularia oryzae. Am. J. Plant. Sci. 2016, 7, 1077–1085. [Google Scholar] [CrossRef] [Green Version]
- Bibi, F.; Naseer, M.I.; Yasir, M.; Al-Ghamdi, A.A.K.; Azhar, E.I. LC-MS based identification of secondary metabolites from marine antagonistic endophytic bacteria. Genet. Mol. Res. 2017, 17, gmr16039857. [Google Scholar] [CrossRef] [Green Version]
- Wei, Z.; Xu, C.; Wang, J.; Lu, F.; Bie, X.; Lu, Z. Identification and characterization of Streptomyces flavogriseus NJ-4 as a novel producer of actinomycin D and holomycin. PeerJ 2017, 5, e3601. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Jia, Y.; Xie, Y.; Zhang, C.; Ma, J.; Sun, C.; Ju, J. Identification of the Actinomycin D Biosynthetic Pathway from Marine-Derived Streptomyces costaricanus SCSIO ZS0073. Mar. Drugs 2019, 17, 240. [Google Scholar] [CrossRef] [Green Version]
- Khieu, T.N.; Liu, M.J.; Nimaichand, S.; Quach, N.T.; Chu-Ky, S.; Phi, Q.T.; Vu, T.T.; Nguyen, T.D.; Xiong, Z.; Prabhu, D.M.; et al. Characterization and evaluation of antimicrobial and cytotoxic effects of Streptomyces sp. HUST012 isolated from medicinal plant Dracaena cochinchinensis Lour. Front. Microbiol. 2015, 8, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Poosarla, A.; Krishna, R.M. Isolation of potent antibiotic producing Actinomycetes from marine sediments of Andaman and Nicobar Marine Islands. J. Microbiol. Antimicrob. 2013, 5, 6–12. [Google Scholar] [CrossRef] [Green Version]
- Charousová, I.; Medo, J.; Hleba, L.; Javoreková, S. Streptomyces globosus DK15 and Streptomyces ederensis ST13 as new producers of factumycin and tetrangomycin antibiotics. Braz. J. Microbiol. 2018, 49, 816–822. [Google Scholar] [CrossRef] [PubMed]
- Vu, H.-N.T.; Nguyen, D.T.; Nguyen, H.Q.; Chu, H.H.; Chu, S.K.; Van Chau, M.; Phi, Q.-T. Antimicrobial and cytotoxic properties of bioactive metabolites produced by Streptomyces cavourensis YBQ59 isolated from Cinnamomum cassia Prels in Yen Bai Province of Vietnam. Curr. Microbiol. 2018, 75, 1247–1255. [Google Scholar] [CrossRef] [PubMed]
- Al-dhabi, N.A.; Ghilan, A.-K.M.; Esmail, G.A.; Arasu, M.V.; Duraipandiyan, V.; Ponmurugan, K. Bioactivity assessment of the Saudi Arabian Marine Streptomyces sp. Al-Dhabi-90, metabolic profiling and its in vitro inhibitory property against multidrug resistant and extended-spectrum beta-lactamase clinical bacterial pathogens. J. Infect. Public Health 2019, 12, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Arasu, M.V.; Duraipandiyan, V.; Agastian, P.; Ignacimuthu, S. In vitro antimicrobial activity of Streptomyces spp. ERI-3 isolated from Western Ghats rock soil (India). J. Mycol. Med. 2009, 19, 22–28. [Google Scholar] [CrossRef]
- Singh, V.; Haque, S.; Singh, H.; Verma, J.; Vibha, K.; Singh, R.; Jawed, A.; Tripathi, C. Isolation, screening, and identification of novel isolates of actinomycetes from India for antimicrobial applications. Front. Microbiol. 2016, 7, 1921. [Google Scholar] [CrossRef] [Green Version]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [Green Version]
- Khatun, M.F.; Haque, M.U.; Islam, M.A.U. Antibacterial and Cytotoxic Activities of Crude Ethyl Acetate Extract of Streptomyces sp. FEAI-1 Isolated From Soil Samples of Rajshahi, Bangladesh. Bangladesh. J. Pharmacol. 2017, 20, 188–193. [Google Scholar] [CrossRef]
- Cockerill, F.R.; Wikler, M.A.; Alder, J.; Dudley, M.N.; Eliopoulos, G.M.; Ferraro, M.J.; Hardy, D.; Hecht, D.; Hindler, J.; Patel, J. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard; CLSI Document M07-A9; CLSI (Clinical and Laboratory Standard Institute): Pennsylvania, PA, USA, 2012. [Google Scholar]




| Test Pathogens | Zone of Inhibition (mm) |
|---|---|
| MRSA DMST20651 | 50.00 ± 0.00 a |
| S. aureus ATCC29213 | 46.67 ± 0.58 ab |
| S. epidermidis TISTR518 | 48.33 ± 2.89 ab |
| B. subtilis TISTR008 | 45.00 ± 3.00 b |
| B. cereus TISTR687 | 38.33 ± 1.15 c |
| Test Microorganisms | Zone of Inhibition (mm) | p-Value | |
|---|---|---|---|
| At 30 °C | At 37 °C | ||
| MRSA DMST20651 | 15.67 ± 0.58 | 16.67 ± 0.58 | 0.259 |
| S. aureus ATCC29213 | 19.33 ± 0.58 | 20.33 ± 1.15 | 0.188 |
| S. epidermidis TISTR518 | 21.33 ± 1.15 | 21.67 ± 0.58 | 0.757 |
| B. subtilis TISTR008 | 17.00 ± 1.00 | 22.33 ± 1.15 * | 0.001 |
| B. cereus TISTR687 | 18.33 ± 1.15 | 20.00 ± 0.00 * | 0.003 |
| Test Pathogens | Minimum Inhibitory Concentration (MIC) (μg/mL) | |||
|---|---|---|---|---|
| Crude Compounds | Oxacillin | Vancomycin | Tetracycline | |
| MRSA DMST20651 | 2 | 512 | 2 | 32 |
| S. aureus ATCC29213 | 2 | 0.125 | 1 | 0.5 |
| S. epidermidis TISTR518 | 16 | 0.125 | 2 | 0.0625 |
| B. subtilis TISTR008 | 2 | 0.25 | 0.5 | 4 |
| B. cereus TISTR687 | 1 | 64 | 2 | 0.0312 |
| Name of the Compound | Chemical Formula | Molecular Weight (g/mol) | Sources | Properties | References |
|---|---|---|---|---|---|
| Dihomo-γ-linolenic acid (DGLA) | C20H34O2 | 306.50 | Mortierella spp. | Antibacterial | [33] |
| Epigallocatechin | C15H14O7 | 306.27 | Camellia sinensis | Antioxidant | [34] |
| 2,3-trans-3,4-cis-leucocyanidin | C15H14O7 | 306.27 | Plantain banana | Antiulcerogenic | [35] |
| Eremofortin A | C17H22O5 | 306.35 | Penicillium roqueforti | Mycotoxin | [36] |
| Fenazaquin | C20H22N20 | 306.40 | Synthetic chemical | Acaricide/insecticide | [37] |
| Feruloyl agmatine | C15H22N403 | 306.36 | Triticum aestivum L. cv Chihokukomugi | Antifungal | [38] |
| Fluconazole | C13H12F2N60 | 306.10 | Synthetic chemical | Antifungal | [39] |
| Koumine | C20H22N20 | 306.40 | Gelsemium | Anti-inflammatory, analgesic and neurosteroid-modulating | [40] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chanthasena, P.; Hua, Y.; Rosyidah, A.; Pathom-Aree, W.; Limphirat, W.; Nantapong, N. Isolation and Identification of Bioactive Compounds from Streptomyces actinomycinicus PJ85 and Their In Vitro Antimicrobial Activities against Methicillin-Resistant Staphylococcus aureus. Antibiotics 2022, 11, 1797. https://doi.org/10.3390/antibiotics11121797
Chanthasena P, Hua Y, Rosyidah A, Pathom-Aree W, Limphirat W, Nantapong N. Isolation and Identification of Bioactive Compounds from Streptomyces actinomycinicus PJ85 and Their In Vitro Antimicrobial Activities against Methicillin-Resistant Staphylococcus aureus. Antibiotics. 2022; 11(12):1797. https://doi.org/10.3390/antibiotics11121797
Chicago/Turabian StyleChanthasena, Panjamaphon, Yanling Hua, A’liyatur Rosyidah, Wasu Pathom-Aree, Wanwisa Limphirat, and Nawarat Nantapong. 2022. "Isolation and Identification of Bioactive Compounds from Streptomyces actinomycinicus PJ85 and Their In Vitro Antimicrobial Activities against Methicillin-Resistant Staphylococcus aureus" Antibiotics 11, no. 12: 1797. https://doi.org/10.3390/antibiotics11121797






