Persistence of Antibiotic-Resistant Escherichia coli Strains Belonging to the B2 Phylogroup in Municipal Wastewater under Aerobic Conditions
Abstract
1. Introduction
2. Results
2.1. Changes to TOC Concentrations and Number of E. coli Colonies under Aerobic Conditions
2.2. Identification of E. coli Isolates
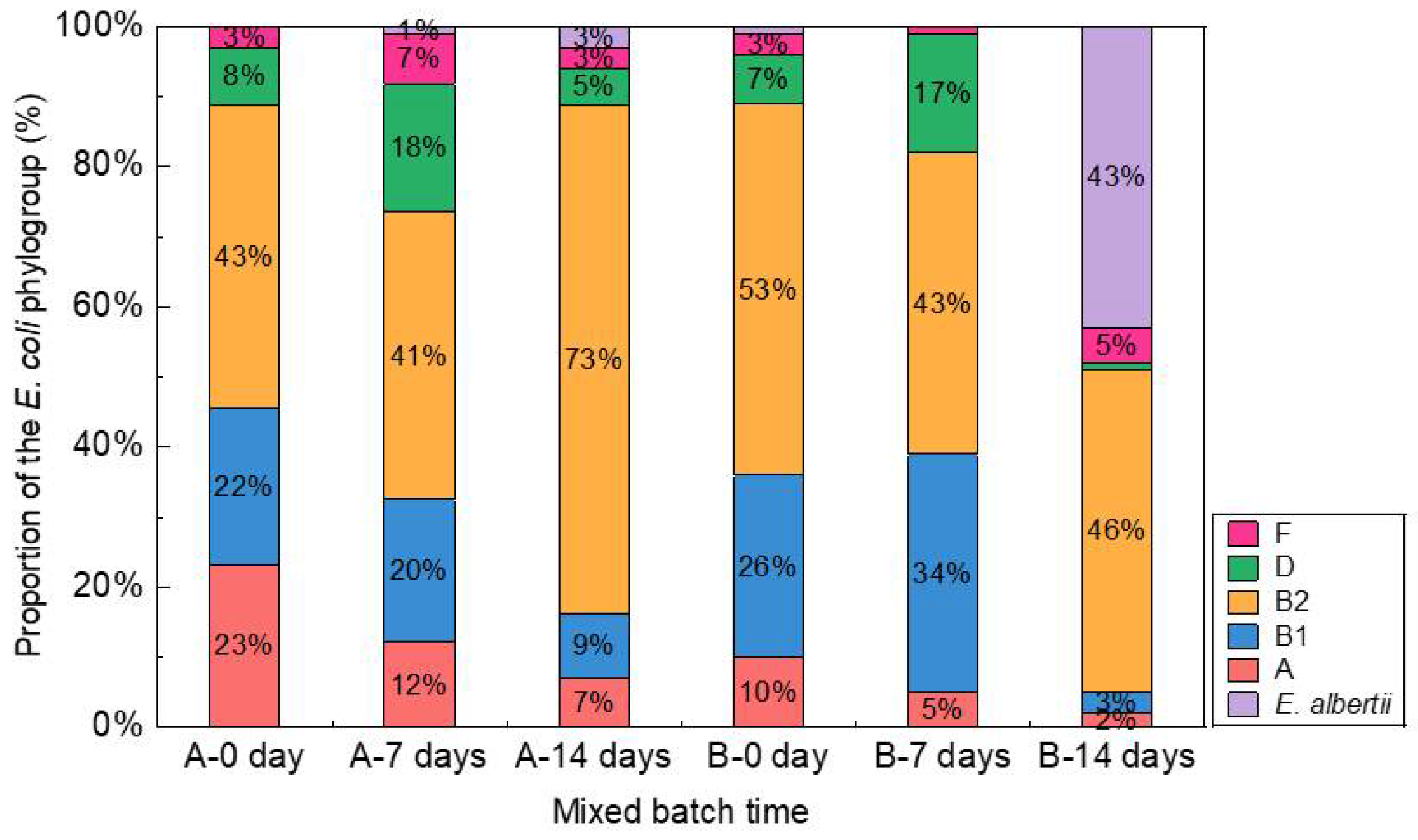
2.3. Phylogroups of E. coli
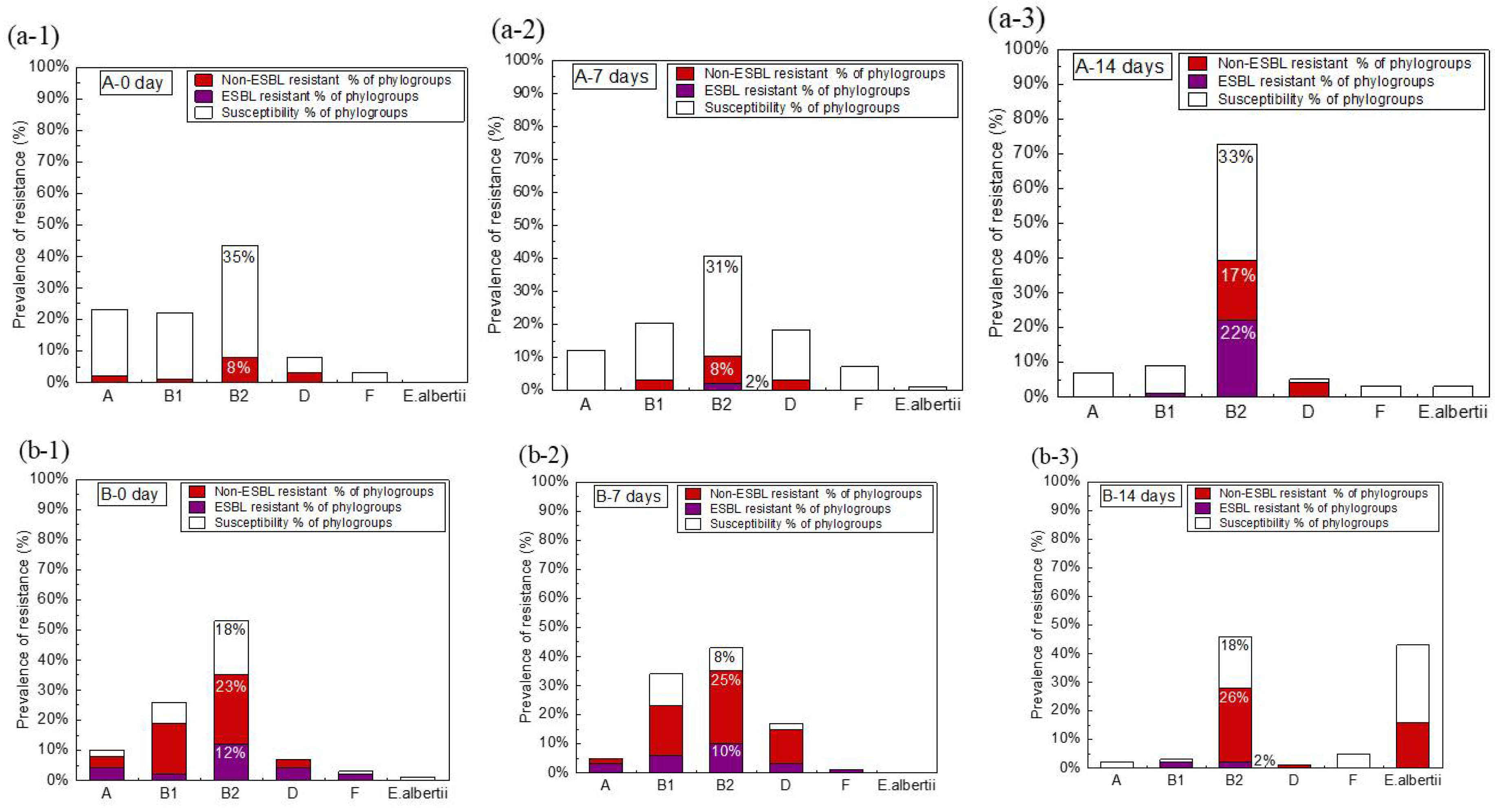
2.4. Antibiotic Susceptibility of E. coli
2.5. ESBL-Producing E. coli Isolates and Genotypes of E. coli
3. Discussion
3.1. Changes to the Composition of E. coli Phylogroups
3.2. Relationship between Phylogroups and Antibiotic Resistance of E. coli
4. Materials and Methods
4.1. Sampling
4.2. Batch Mixing Experiment and Isolation of E. coli
4.3. Identification of E. coli by Matrix-Assisted Laser Desorption/Ionization Coupled to Time-of-Flight Mass Spectrometry (MALDI-TOF MS)
4.4. Classification of Phylogroups of E. coli by Multiplex PCR
4.5. Antimicrobial Susceptibility Testing
4.6. ESBL Genotypes of E. coli by Multiplex PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- O’Neill, J. Review on antimicrobial resistance. In Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations; The UK Department of Health: London, UK, 2014; pp. 1–20. [Google Scholar]
- Tsuzuki, S.; Matsunaga, N.; Yahara, K.; Gu, Y.; Hayakawa, K.; Hirabayashi, A.; Kajihara, T.; Sugai, M.; Shibayama, K.; Ohmagari, N. National trend of blood-stream infection attributable deaths caused by Staphylococcus aureus and Escherichia coli in Japan. J. Infect. Chemother. 2020, 26, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.D.; Garrett, V.D.; Sobel, J.; Monroe, S.S.; Fankhauser, R.L.; Schwab, K.J.; Bresee, J.S.; Mead, P.S.; Higgins, C.; Campana, J.; et al. Multistate outbreak of Norwalk-like virus gastroenteritis associated with a common caterer. Am. J. Epidemiol. 2001, 154, 1013–1019. [Google Scholar] [CrossRef]
- Walters, S.P.; Thebo, A.L.; Boehm, A.B. Impact of urbanization and agriculture on the occurrence of bacterial pathogens and stx genes in coastal waterbodies of central California. Water Res. 2011, 45, 1752–1762. [Google Scholar] [CrossRef]
- Daniel, D.S.; Lee, S.M.; Gan, H.M.; Dykes, G.A.; Rahman, S. Genetic diversity of Enterococcus faecalis isolated from environmental, animal and clinical sources in Malaysia. J. Infect. Public Health 2017, 10, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg-Goldstein, R.E.; Micallef, S.A.; Gibbs, S.G.; George, A.; Claye, E.; Sapkota, A.; Joseph, S.W.; Sapkota, A.R. Detection of vancomycin-resistant enterococci (VRE) at four U.S. wastewater treatment plants that provide effluent for reuse. Sci. Total Environ. 2014, 466–467, 404–411. [Google Scholar] [CrossRef]
- Diwan, V.; Hanna, N.; Purohit, M.; Chandran, S.; Riggi, E.; Parashar, V.; Tamhankar, A.J.; Stålsby Lundborg, C. Seasonal variations in water-quality, antibiotic residues, resistant bacteria and antibiotic resistance genes of Escherichia coli isolates from water and sediments of the Kshipra river in central India. Int. J. Environ. Res. Public Health 2018, 15, 1281. [Google Scholar] [CrossRef] [PubMed]
- Gelband, H.; Miller-Petrie, M.; Pant, S.; Gandra, S.; Levinson, J.; Barter, D.; White, A.; Laxminarayan, R. Center for Disease Dynamics, Economics & Policy (CDDEP). State of the World’s Antibiotics; CDDEP: Washington, DC, USA, 2015. [Google Scholar]
- ECDC/EMEA. ECDC/EMEA Joint Technical Report: The Bacterial Challenge: Time to React. In A Call to Narrow the Gap between Multidrug-Resistant Bacteria in the EU and the Development of New Antibacterial Agents; European Centre for Disease Prevention and Control (ECDPC): Stockholm, Sweden, 2009; pp. 1–54. [Google Scholar]
- Aristizábal-Hoyos, A.M.; Rodríguez, E.A.; Arias, L.; Jiménez, J.N. High clonal diversity of multidrug-resistant and extended spectrum beta-lactamase-producing Escherichia coli in a wastewater treatment plant. J. Environ. Manag. 2019, 245, 37–47. [Google Scholar] [CrossRef]
- Adefisoye, M.A.; Okoh, A.I. Identification and antimicrobial resistance prevalence of pathogenic Escherichia coli strains from treated wastewater effluents in Eastern Cape, South Africa. MicrobiologyOpen 2016, 5, 143–151. [Google Scholar] [CrossRef]
- Kümmerer, K. Antibiotics in the aquatic environment—a review—Part II. Chemosphere 2009, 75, 435–441. [Google Scholar] [CrossRef]
- Müller, A.; Stephan, R.; Nüesch-Inderbinen, M. Distribution of virulence factors in ESBL-producing Escherichia coli isolated from the environment, livestock, food and humans. Sci. Total Environ. 2016, 541, 667–672. [Google Scholar] [CrossRef]
- Abia, A.L.K.; Ubomba-Jaswa, E.; Momba, M.N.B. Occurrence of diarrhoeagenic Escherichia coli virulence genes in water and bed sediments of a river used by communities in Gauteng, South Africa. Environ. Sci. Pollution Res. Int. 2016, 23, 15665–15674. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Antibiotic Resistance Threats in the United States; Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019; pp. 1–148. [Google Scholar]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef]
- Woerther, P.L.; Burdet, C.; Chachaty, E.; Andremont, A. Trends in human fecal carriage of extended-spectrum β-lactamases in the community: Toward the globalization of CTX-M. Clin. Microbiol. Rev. 2013, 26, 744–758. [Google Scholar] [CrossRef] [PubMed]
- Manges, A.R.; Johnson, J.R.; Foxman, B.; O’Bryan, T.T.; Fullerton, K.E.; Riley, L.W. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N. Engl. J. Med. 2001, 345, 1007–1013. [Google Scholar] [CrossRef]
- Chaudhuri, R.R.; Henderson, I.R. The evolution of the Escherichia coli phylogeny. Infect. Genet. Evolut. 2012, 12, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Duriez, P.; Clermont, O.; Bonacorsi, S.; Bingen, E.; Chaventré, A.; Elion, J.; Picard, B.; Denamur, E. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology 2001, 147, 1671–1676. [Google Scholar] [CrossRef]
- Escobar-Páramo, P.; Le Menac’H, A.; Le Gall, T.; Amorin, C.; Gouriou, S.; Picard, B.; Skurnik, D.; Denamur, E. Identification of forces shaping the commensal Escherichia coli genetic structure by comparing animal and human isolates. Environ. Microbiol. 2006, 8, 1975–1984. [Google Scholar] [CrossRef]
- Orsi, R.H.; Stoppe, N.C.; Sato, M.I.Z.; Gomes, T.A.T.; Prado, P.I.; Manfio, G.P.; Ottoboni, L.M.M. Genetic variability and pathogenicity potential of Escherichia coli isolated from recreational water reservoirs. Res. Microbiol. 2007, 158, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.M.; Clermont, O.; Tolley, H.; Denamur, E. Assigning Escherichia coli strains to phylogenetic groups: Multi-locus sequence typing versus the PCR triplex method. Environ. Microbiol. 2008, 10, 2484–2496. [Google Scholar] [CrossRef]
- Higgins, J.; Hohn, C.; Hornor, S.; Frana, M.; Denver, M.; Joerger, R. Genotyping of Escherichia coli from environmental and animal samples. J. Microbiol. Methods 2007, 70, 227–235. [Google Scholar] [CrossRef]
- Anastasi, E.M.; Matthews, B.; Gundogdu, A.; Vollmerhausen, T.L.; Ramos, N.L.; Stratton, H.; Ahmed, W.; Katouli, M. Prevalence and persistence of Escherichia coli strains with uropathogenic virulence characteristics in sewage treatment plants. Appl. Environ. Microbiol. 2010, 76, 5882–5886. [Google Scholar] [CrossRef] [PubMed]
- Bojar, B.; Sheridan, J.; Beattie, R.; Cahak, C.; Liedhegner, E.; Munoz-Price, L.S.; Hristova, K.R.; Skwor, T. Antibiotic resistance patterns of Escherichia coli isolates from the clinic through the wastewater pathway. Int. J. Hygiene Environ. Health 2021, 238, 113863. [Google Scholar] [CrossRef] [PubMed]
- Paulshus, E.; Kühn, I.; Möllby, R.; Colque, P.; O’Sullivan, K.; Midtvedt, T.; Lingaas, E.; Holmstad, R.; Sørum, H. Diversity and antibiotic resistance among Escherichia coli populations in hospital and community wastewater compared to wastewater at the receiving urban treatment plant. Water Res. 2019, 161, 232–241. [Google Scholar] [CrossRef]
- The AMR One Health Surveillance Committee. Nippon AMR One Health Report (NAOR); Tokyo: Tuberculosis and Infectious Diseases Control Division, Health Service Bureau, Ministry of Health, Labour and Welfare: Tokyo, Japan, 2019; pp. 1–109.
- Briran, D.Z.; Ron, E. Extraintestinal pathogenic Escherichia coli. Curr. Top. Microbiol. Immunol. 2018, 416, 149–161. [Google Scholar]
- Sarowska, J.; Futoma-Koloch, B.; Jama-Kmiecik, A.; Frej-Madrzak, M.; Ksiazczyk, M.; Bugla-PLoSkonska, G.; Choroszy-Krol, I. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: Recent reports. Gut Pathog. 2019, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Kuskowski, M.A.; O’Bryan, T.T.; Maslow, J.N. Epidemiological correlates of virulence genotype and phylogenetic background among Escherichia coli blood isolates from adults with diverse-source bacteremia. J. Infect. Dis. 2002, 185, 1439–1447. [Google Scholar] [CrossRef][Green Version]
- Mokracka, J.; Koczura, R.; Jabłońska, L.; Kaznowski, A. Phylogenetic groups, virulence genes and quinolone resistance of integron-bearing Escherichia coli strains isolated from a wastewater treatment plant. Antonie Leeuwenhoek 2011, 99, 817–824. [Google Scholar] [CrossRef]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- Inglis, T.J.J.; Merritt, A.J.; Bzdyl, N.; Lansley, S.; Urosevic, M.N. First bacteraemic human infection with Escherichia albertii. N. Microb. New Infect. 2015, 8, 171–173. [Google Scholar] [CrossRef]
- Hyma, K.E.; Lacher, D.W.; Nelson, A.M.; Bumbaugh, A.C.; Janda, J.M.; Strockbine, N.A.; Young, V.B.; Whittam, T.S. Evolutionary genetics of a new pathogenic Escherichia species: Escherichia albertii and related Shigella boydii strains. J. Bacteriol. 2005, 187, 619–628. [Google Scholar] [CrossRef]
- Ooka, T.; Seto, K.; Kawano, K.; Kobayashi, H.; Etoh, Y.; Ichihara, S.; Kaneko, A.; Isobe, J.; Yamaguchi, K.; Horikawa, K.; et al. Clinical significance of Escherichia albertii. Emerg. Infect. Dis. 2012, 18, 488–492. [Google Scholar] [CrossRef]
- Walk, S.T.; Alm, E.W.; Gordon, D.M.; Ram, J.L.; Toranzos, G.A.; Tiedje, J.M.; Whittam, T.S. Cryptic lineages of the genus Escherichia. Appl. Environ. Microbiol. 2009, 75, 6534–6544. [Google Scholar] [CrossRef]
- Abbott, S.L.; O’Connor, J.; Robin, T.; Zimmer, B.L.; Janda, J.M. Biochemical properties of a newly described Escherichia species, Escherichia albertii. J. Clin. Microbiol. 2003, 41, 4852–4854. [Google Scholar] [CrossRef] [PubMed]
- Gomes, T.A.; Ooka, T.; Hernandes, R.T.; Yamamoto, D.; Hayashi, T. Escherichia albertii pathogenesis. EcoSalPlus 2020, 9, 1–18. [Google Scholar]
- Bingen, E.; Picard, B.; Brahimi, N.; Mathy, S.; Desjardins, P.; Elion, J.; Denamur, E. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J. Infect. Dis. 1998, 177, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Desjardins, P.; Picard, B.; Kaltenböck, B.; Elion, J.; Denamurl, E. Sex in Escherichia coli does not disrupt the clonal structure of the population: Evidence from random amplified polymorphic DNA and restriction-fragment-length polymorphism. J. Molec. Evolut. 1995, 41, 440–448. [Google Scholar] [CrossRef]
- Picard, B.; Garcia, J.S.; Gouriou, S.P.; Duriez, P.; Brahimi, N.M.; Bingen, E.; Elion, J.; Denamur, E. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 1999, 67, 546–553. [Google Scholar] [CrossRef]
- Le Gall, T.; Clermont, O.; Gouriou, S.; Picard, B.; Nassif, X.; Denamur, E.; Tenaillon, O. Extraintestinal virulence is a coincidental by-product of commensalism in B2 phylogenetic group Escherichia coli strains. Molec. Biol. Evolut. 2007, 24, 2373–2384. [Google Scholar] [CrossRef]
- Mugnaioli, C.; Luzzaro, F.; De Luca, F.; Brigante, G.; Perilli, M.; Amicosante, G.; Stefani, S.; Toniolo, A.; Rossolini, G.M. CTX-M-type extended-spectrum β-lactamases in Italy: Molecular epidemiology of an emerging countrywide problem. Antimicrob. Agents Chemother. 2006, 50, 2700–2706. [Google Scholar] [CrossRef]
- Nicolas-Chanoine, M.H.; Blanco, J.; Leflon-Guibout, V.; Demarty, R.; Alonso, M.P.; Caniça, M.M.; Park, Y.J.; Lavigne, J.P.; Pitout, J.; Johnson, J.R. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 2008, 61, 273–281. [Google Scholar] [CrossRef]
- Suzuki, Y.; Niina, K.; Matsuwaki, T.; Nukazawa, K.; Iguchi, A. Bacterial flora analysis of coliforms in sewage, river water, and ground water using MALDI-TOF mass spectrometry. J. Environ. Sci. Health Part A Toxic/Hazard. Subst. Environ. Eng. 2018, 53, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Sauget, M.; Nicolas-Chanoine, M.-H.; Cabrolier, N.; Bertrand, X.; Hocquet, D. Matrix-assisted laser desorption ionization-time of flight mass spectrometry assigns Escherichia coli to the phylogroups A, B1, B2 and D. Int. J. Med. Microbiol. 2014, 304, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017; pp. 1–282. ISBN 1-56238-804-5. [Google Scholar]
- Shibata, N.; Kurokawa, H.; Doi, Y.; Yagi, T.; Yamane, K.; Wachino, J.-I.; Suzuki, S.; Kimura, K.; Ishikawa, S.; Kato, H.; et al. PCR classification of CTX-M-Type β-lactamase genes identified in clinically isolated gram-negative bacilli in Japan. Antimicrob. Agents Chemother. 2006, 50, 791–795. [Google Scholar] [CrossRef] [PubMed]
- Yagi, T.; Kurokawa, H.; Shibata, N.; Shibayama, K.; Arakawa, Y. A preliminary survey of extended-spectrum β-lactamases (ESBLs) in clinical isolates of Klebsiella pneumoniae and Escherichia coli in Japan. FEMS Microbiol. Lett. 2000, 184, 53–56. [Google Scholar] [CrossRef]

| Isolates from Plant A and B | ESBL Types by the Following PCR Type | Phylogroup | ||||
|---|---|---|---|---|---|---|
| TEM | SHV | CTX-M-1 | CTX-M-2 | CTX-M-9 | ||
| A-7 days-91 | N | N | N | N | Y | B2 |
| A-7 days-100 | N | N | N | Y | Y | B2 |
| A-14 days-5 | N | N | Y | N | N | B2 |
| A-14 days-7 | N | N | N | Y | N | B2 |
| A-14 days-8 | N | N | N | N | Y | B2 |
| A-14 days-9 | N | N | N | N | Y | B2 |
| A-14 days-21 | N | N | N | Y | Y | B2 |
| A-14 days-23 | N | N | N | N | Y | B2 |
| A-14 days-42 | N | N | N | N | Y | B2 |
| A-14 days-54 | N | N | N | N | Y | B2 |
| A-14 days-59 | N | N | N | N | Y | B2 |
| A-14 days-62 | N | N | N | N | Y | B2 |
| A-14 days-71 | N | N | N | N | Y | B2 |
| A-14 days-73 | N | N | N | N | Y | B2 |
| A-14 days-80 | N | Y | N | N | N | B2 |
| A-14 days-82 | N | Y | N | N | N | B2 |
| A-14 days-83 | N | N | Y | Y | N | B2 |
| A-14 days-84 | N | N | Y | Y | N | B2 |
| A-14 days-85 | N | N | Y | N | N | B2 |
| A-14 days-86 | N | N | N | N | Y | B2 |
| A-14 days-87 | N | N | N | N | Y | B2 |
| A-14 days-88 | N | N | N | N | Y | B2 |
| A-14 days-92 | N | N | N | N | Y | B2 |
| A-14 days-94 | N | N | N | N | Y | B2 |
| A-14 days-100 | N | Y | N | N | N | B1 |
| B-0 day-1 | N | N | N | N | Y | B2 |
| B-0 day-2 | N | N | N | N | Y | D |
| B-0 day-3 | N | N | N | Y | N | B2 |
| B-0 day-4 | N | Y | N | N | N | B2 |
| B-0 day-12 | N | N | N | N | Y | D |
| B-0 day-13 | N | Y | N | N | N | D |
| B-0 day-14 | N | N | N | N | Y | F |
| B-0 day-23 | N | N | N | N | Y | B2 |
| B-0 day-24 | N | N | Y | N | N | B2 |
| B-0 day-25 | N | N | N | N | Y | B2 |
| B-0 day-26 | N | Y | N | N | N | A |
| B-0 day-38 | N | N | N | Y | Y | B2 |
| B-0 day-39 | N | Y | N | N | N | B1 |
| B-0 day-43 | N | Y | N | N | N | A |
| B-0 day-60 | N | N | N | Y | Y | B2 |
| B-0 day-62 | N | Y | N | N | N | B2 |
| B-0 day-64 | N | Y | N | N | N | B2 |
| B-0 day-65 | N | Y | N | N | N | A |
| B-0 day-66 | N | Y | N | N | N | A |
| B-0 day-85 | N | N | N | N | Y | D |
| B-0 day-86 | N | Y | N | N | N | B1 |
| B-0 day-87 | N | N | N | N | Y | B2 |
| B-0 day-88 | N | Y | N | N | N | F |
| B-0 day-89 | N | Y | N | N | N | B2 |
| B-7 days-17 | N | N | N | Y | N | B2 |
| B-7 days-18 | N | Y | N | N | N | B1 |
| B-7 days-19 | N | N | Y | N | Y | B1 |
| B-7 days-20 | N | Y | N | N | Y | F |
| B-7 days-26 | N | Y | N | N | Y | D |
| B-7 days-40 | N | Y | Y | N | N | B2 |
| B-7 days-45 | N | Y | N | N | Y | B2 |
| B-7 days-46 | N | Y | N | N | Y | B2 |
| B-7 days-47 | Y | Y | N | N | N | A |
| B-7 days-48 | N | Y | N | N | N | D |
| B-7 days-49 | N | N | N | Y | N | B2 |
| B-7 days-50 | N | N | N | N | Y | A |
| B-7 days-51 | N | Y | Y | N | N | A |
| B-7 days-53 | N | Y | N | N | N | B1 |
| B-7 days-58 | N | N | N | N | Y | D |
| B-7 days-67 | N | N | N | N | Y | B2 |
| B-7 days-68 | N | N | N | N | Y | B2 |
| B-7 days-69 | N | N | N | Y | Y | B1 |
| B-7 days-70 | N | Y | Y | N | N | B1 |
| B-7 days-71 | N | N | N | N | Y | B1 |
| B-7 days-84 | N | Y | N | N | N | B2 |
| B-7 days-88 | N | Y | N | N | N | B2 |
| B-7 days-91 | Y | Y | N | N | Y | B2 |
| B-14 days-27 | N | Y | N | N | N | B2 |
| B-14 days-28 | N | N | N | Y | N | B1 |
| B-14 days-30 | N | Y | N | N | N | B1 |
| B-14 days-37 | N | N | N | Y | Y | B2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, H.; Ogura, Y.; Suzuki, Y. Persistence of Antibiotic-Resistant Escherichia coli Strains Belonging to the B2 Phylogroup in Municipal Wastewater under Aerobic Conditions. Antibiotics 2022, 11, 202. https://doi.org/10.3390/antibiotics11020202
Xie H, Ogura Y, Suzuki Y. Persistence of Antibiotic-Resistant Escherichia coli Strains Belonging to the B2 Phylogroup in Municipal Wastewater under Aerobic Conditions. Antibiotics. 2022; 11(2):202. https://doi.org/10.3390/antibiotics11020202
Chicago/Turabian StyleXie, Hui, Yoshitoshi Ogura, and Yoshihiro Suzuki. 2022. "Persistence of Antibiotic-Resistant Escherichia coli Strains Belonging to the B2 Phylogroup in Municipal Wastewater under Aerobic Conditions" Antibiotics 11, no. 2: 202. https://doi.org/10.3390/antibiotics11020202
APA StyleXie, H., Ogura, Y., & Suzuki, Y. (2022). Persistence of Antibiotic-Resistant Escherichia coli Strains Belonging to the B2 Phylogroup in Municipal Wastewater under Aerobic Conditions. Antibiotics, 11(2), 202. https://doi.org/10.3390/antibiotics11020202







