Abstract
In the recent decades, antibiotic resistance has emerged and spread rapidly among clinically relevant pathogens. The natural ability of bacteria to transmit resistance determinants through horizontal gene transfer poses constant challenges to drug development. Natural molecules produced by soil microorganisms continue to be a key source of new antimicrobial agents. In this context, bacteria from the Geobacillus and Parageobacillus genera deserve special attention. Although there is commercial and industrial interest in these microorganisms, the full range of antibacterial compounds biosynthesized by the Geobacillus and Parageobacillus species remains largely unexplored. The aim of this review is to present the strong antimicrobial potential of these bacteria and endolysins produced by their bacteriophages.
1. Introduction
Geobacillus and Parageobacillus genera belong to the Bacillaceae family, which is a large and heterogeneous group that includes many mesophilic, facultative thermophilic and thermophilic species, widely distributed in different habitats [1]. The current taxonomic classification of Geobacillus and Parageobacillus is presented in Figure 1. Their closest evolutionary relatives are bacteria of the genus Bacillus, exemplified by Bacillus subtilis (B. subtilis) [1]. B. subtilis strains and other species from the genus Bacillus are well known for their probiotic properties and production of various antimicrobial compounds [2,3,4].
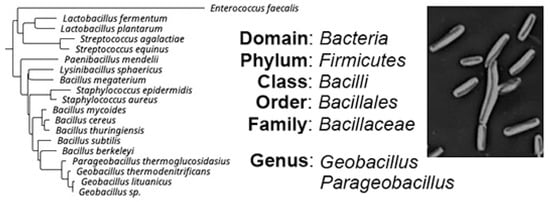
Figure 1.
Current taxonomic classification of the genera Geobacillus and Parageobacillus.
Geobacillus and Parageobacillus have been found in various locations, differing geographically and environmentally. They have been detected on all continents and also in the seas, oceans and even in the atmosphere [5,6]. Representatives of these genera can be isolated from extreme places such as hot springs, hydrothermal vents, high-temperature oil fields, composts, and greenhouse soil [1,6]. Interestingly, they have also been found (in large numbers) in cold places, much below their minimal growth temperatures, such as ocean sediments or soil samples [6].
The genera Geobacillus and Parageobacillus comprise thermophile, endospore-forming, chemo-organotrophic rods. The structure of their cell walls is Gram-positive, but the result of the gram-stain reaction may be varied. Cells are motile or non-motile and can occur individually or in chains [1,5,7]. Cellular fatty acids are characterized by iso-15:0, iso-16:0 and iso-17:0. The G + C content of DNA ranges between 48.2 and 58 mol% [7]. Depending on the strain, these bacteria are aerobic or facultatively anaerobic. Their growth is observed in a pH range between 6.0 and 8.5 and temperature range of 35–80 °C (optimum 55–65 °C) [1,6,7].
Until 2001, Geobacillus and Parageobacillus were classified on the basis of 16s rRNA gene sequence analysis as thermophilic variants of Bacillus spp. [8]. The genus Bacillus initially comprised five phylogenetically distinct groups, with the future Geo- and Parageobacillus included in group 5 [8]. Subsequently, according to physiological characteristics, 16S rRNA gene sequences analyses, fatty acid composition analyses, G-C contents and DNA–DNA homology studies, Geobacillus and Parageobacillus were reclassified together as Geobacillus gen. nov. [7]. In 2016, Aliyu and colleagues separated Parageobacillus from Geobacillus [9]. In their research, they used multiple phylogenomic strategies to estimate relatedness between sixty-three Geobacillus strains, whose genome sequences were available at the time. Their analysis allowed them to distinguish two clades on the basis of differences in nucleotide base composition. Clade I with G + C content 48.8–53.1% (Geobacillus species) and clade II with 42.1–44.4% (new, Parageobacillus species). Species belonging at the baseline of Geobacillus, which have been moved to Parageobacillus, include: P. caldoxylosilyticus, P. thermoglucosidasius, P. thermantarcticus, P. toebii and P. genomospecies 1 (NUB3621) [9]. Subsequently, in 2020, Najar and colleagues presented the analysis that supports their proposal of including two other species: G. galactosidasius and G. yumthangensis in the genus Parageobacillus [10].
As genera derived from extreme environments, Geobacillus and Parageobacillus are sources of proteins that are stable at high temperatures and are functional under other extreme conditions, especially compared to mesophilic homologues. Over the past few years, an extensive exploration of the Geobacillus and Parageobacillus transcriptomes and secretomes has revealed many proteins with either proven or potential industrial and medicinal applications [1,11,12]. The strong metabolism and cellular propagation of these organisms make them appropriate hosts for different bioprocesses (whole-cell applications) [5]. Thus, these bacteria are exploited in various biotechnological and industrial applications, such as food production, the textile industry, the paper industry, bio-detergent technology, cosmetics, drugs and the pharmaceutical industry, biofuel or chemical production, bioremediation and many others [6,13,14]. Recently, Geobacillus has been investigated as a source of thermostable L-asparaginase with potential therapeutic properties [15]. There are also promising reports about the antimicrobial applications of Geobacillus. Alkhalili and colleagues describe Geobacillus sp. strain ZGt-1, isolated in Jordan, which demonstrated an antimicrobial activity against G. stearothermophilus, B. subtilis and Salmonella typhimurium (S. typhimurium) [16]. Additionally, Pokusaeva and colleagues reported that they had identified and partially purified bacteriocins (ribosomally produced proteins that inhibit other strains or species) synthesized by G. stearothermophilus strains isolated from oil-wells in Lithuania [17]. Such research may indicate many new valuable applications for the genera Geobacillus and Parageobacillus.
2. Antimicrobial Potential of Geobacillus and Parageobacillus
2.1. Geobacillus and Parageobacillus as a Source of Novel Antimicrobial Compounds
The potential of Geobacillus and Parageobacillus to produce a wide range of bioactive metabolites (that mediate antibiosis) is not fully explored. In this section we list, and briefly characterize, the antimicrobial molecules produced by Geobacillus/Parageobacillus that have been described in the literature so far. Types of the Geobacillus and Parageobacillus antimicrobial compounds are exemplified by several bacteriocins, bacteriocin-like inhibitory substances (BLISes), volatile organic substances (VOCs), and antibiotic pigments (Figure 2).
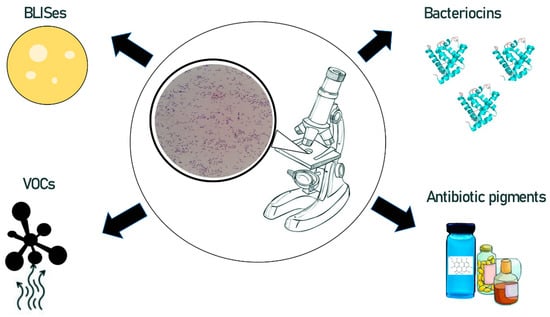
Figure 2.
Types of Geobacillus and Parageobacillus derived antimicrobial compounds.
2.1.1. Geobacillus-Derived Volatile Organic Substances and Antibiotic Pigments
Geobacillus sp. (M-7) appears to have a unique approach to combatting competitive bacteria by producing a range of volatile organic substances (benzaldehyde, acetic acid, amongst others). The Geobacillus strain, as well as a mixture of the detected chemicals, was able to inhibit the growth after 48 h and cause death after 72 h of Aspergillus fumigatus, Botrytis cinerea, Verticillium dahliae and Geotrichum candidum [18].
Geobacillus sp. Iso5 produces a unique cyanoxanthomycin-type antibiotic pigment. This fluorescent pigment shows potent antimicrobial activity against selected Gram-positive and Gram-negative bacterial species: B. subtilis (MTCC 3053), E. coli (MTCC 1698), Pseudomonas aeruginosa (P. aeruginoasa) (MTCC 6458), Staphylococus aureus (S. aureus) (MTCC 6908) and Streptococcus sp. (MTCC 9724) [19].
Geobacillus sp. LEMMJ02 is a thermophilic bacterial species isolated from the sediments of an Antarctic volcano on Deception Island. Annotation of the Geobacillus sp. LEMMJ02 genome revealed the presence of genes associated with the production of secondary metabolites with antimicrobial properties. The strain is likely to produce phengycin (antifungal lipopeptide), bacteriocin, and terpene [20].
2.1.2. Geobacillus and Parageobacillus Derived Bacteriocins
Bacteriocins are peptides with antimicrobial activity. They are synthesized by many bacteria and archaea strains. Various types of these molecules have been identified and purified since their discovery in 1925 by André Gratia [21].
The synthesis of bacteriocins is carefully regulated. Bacteriocins are synthesized by the ribosome in an inactive form. Posttranslational modifications and cleavage of the leader sequence help the cell to avoid self-inflicted damage. Specific defense proteins and efflux pumps are also employed. Bacteriocins are secreted during the logarithmic growth phase of bacteria. Their secretion is stimulated by various environmental factors, including bacterial cell density, nutrient availability, the presence of acetic acid, and signal peptides such as competence-enhancing peptides [22,23]. For example, Firmicutes commonly regulate the expression of the bacteriocin encoded genes through quorum sensing [24].
Bacteriocins are a very broad group and are classified into several different families, hence their diverse mode of action and a wide variety of affected strains [23,25]. The growing interest in exploiting the potential of these molecules led to the development of databases and search engines capable of identifying plausible bacteriocin genes and providing information about already characterized species [26,27,28,29,30].
Currently, bacteriocins are used mainly in the food processing industry as food preservatives. Nisin is one of many bacteriocins derived from lactic acid bacteria (LAB)-Lactococcus lacti. It is a lantibiotic—a polycyclic peptide with incorporated lanthionine and methyllanthionine and antimicrobial activity [31]. It is legally approved as a food preservative and was also investigated for its biomedical applications. Some bacteriocins are capable of killing bacterial spores [32]. More recently, the effects of bacteriocins on fungi and pathogenic bacteria, including antibiotic-resistant strains, were studied [33,34,35,36,37,38]. Some bacteriocins were found to affect specific bacterial strains; therefore, it is possible to use them for treatments on human microbiomes including the gut microbiome [39,40].
Thermophilic bacteria from the genus Geobacillus and Parageobacillus are a source of vital enzymes, and widely employed in biotechnology and industry. As most bacteriocins are derived from Gram-positive bacteria, Geobacillus and Parageobacillus can be also considered as a rich source of such antimicrobial peptides.
There are already several characterized and semi-characterized bacteriocins derived from Geobacillus and Parageobacillus strains (Table 1). However, their modes of action and mechanisms of secretion have not been thoroughly investigated. The two most investigated—geobacillin I and geobacillin II—are nisin analogs. Their N-terminal structure is very similar to that of nisin rings but the C-terminal sites show no homology. The mode of action of geobacillin I, similarly to nisin, includes the formation of the pores in the cell membrane [41]. Geobacillin I and geobacillin II were identified in the genome of an oil-well-derived strain G. thermodenitrificans NG-80-2 [42]. Several other strains were confirmed to produce geobacillin I. Geobacillin I showed a comparable bioactivity range to nisin but higher stability in the pH 7 and 8 range at 37 °C and 60 °C. Recombinant geobacillin II showed antimicrobial activity only towards Bacillus species [42,43].

Table 1.
Characterized and semi-characterized bacteriocins from the Geobacillus and Parageobacillus species.
Several Geobacillus spp. were tested for bacteriocin activity only against closely related strains, for example: G. stearothermophilus 15 [51], G. stearothermophilus 31 [17], G. stearothermophilus NU-1, NU-2, NU-4, NU-7, NU-23W, NU-34, NU-37, NU-41, NU-44, NCA-1373, NCA-1492 [52], G. stearothermophilus DSM 458 [53]. Further testing of these compounds might reveal their utility in the fight against pathogenic and antibiotic-resistant strains. Such an investigation of thermocin from strain G. stearothermophilus NU-10, previously reported to be active against other Geobacillus strains [52], led to the identification of thermocin 10 activity against B. circulans 4516 [48]. Strong proof of this concept was observed in the study of Pranckutė et al. Screening of 101 strains of Geobacillus revealed that each of them produces bacteriocins active against other Geobacillus spp. bacteria. Moreover, all the investigated strains were able to inhibit the growth of at least one pathogenic strain (out of the 19 tested) [50].
An in silico search for novel bacteriocins, employing genome mining tools, resulted in the identification of several putative Geobacillus bacteriocins and their genetic structures [53,54,55,56,57]. These findings highlight the potential, which is yet to be revealed for Geobacillus and Parageobacillus, as a source of antimicrobials.
2.1.3. Geobacillus and Parageobacillus Derived Bacteriocin-like Inhibitory Substances
BLISes are bacteriocin-like inhibitory substances, though the line between the bacteriocins and BLISes is blurred. Mostly, BLISes are classified as inhibitory bacterial substances, active against a wider spectrum of microbes than bacteriocins, while also being proteinaceous [58,59]. Though there are precious few articles on the matter, Geobacillus- and Parageobacillus-derived BLISes are still worth discussing.
Turkish thermal springs/soils delivered the G. toebii HBB-247 strain. A 38 kDa BLIS from P. toebii was found to be stable up to 60 °C but also susceptible to proteolysis. It has shown bioactivity towards the growth inhibition of several strains including G. stearothermophilus (DSMZ 22), Listeria sp. (food isolate), Enterococcus faecalis (E. faecalis) (ATCC 51299), Enterococcus avium AS-3, Anoxybacillus sp. HBB-134, Geobacillus sp. HBB-269, Geobacillus sp. HBB-270, Anoxybacillus sp. HBB-229, Clostridium pasteurianum (DSM 525) and Cellulomonas fimi (DSM 20114). Its activity against C. pasteurianum (DSM 525) was as high as against G. stearothermophilus (DSMZ 22), which adds a point for classifying this antimicrobial agent as a BLIS rather than bacteriocin [60].
G. stearothermophilus 32A, 17, 30 and 31 strains, obtained from oil wells in Lithuania, were evaluated for their antibacterial activity and while their prevalent inhibition was centered around various Geobacillus strains, their activity was also recorded for three pathogenic microorganisms: B. cereus DSM 12001, Staphylococcus haemolyticus (S. haemolyticus) P903, as well as P. aeruginosa ATCC 27853 where there was observed a slight inhibition, by no more than two of the studied strains, in varied combinations. This brings back the question of the already mentioned artificial classification, which distinguishes between bacteriocins and BLISes. The range of organisms affected by the 6–7.5 kDa proteins produced by the considered strains is unquestionably beyond the scope of the bacteriocin definition and, yet the efficacy is in comparison much weaker than against the related Geobacillus strains [17].
The G. pallidus strain SAT4, found in a Pakistani desert, was reported to produce a polypeptide secondary metabolite of most significant antagonistic activity against Micrococcus luteus ATCC 10240, Staphylococcus aureus (S. aureus) subsp. aureus Rosenbach ATCC 6538, B. subtilis NCTC 10400 and P. aeruginosa ATCC 49189 at 55 °C, with a noted activity at as low as 45 °C and as high as 60 °C, as expected for a thermophilic bacterial strain [61].
A study of the Jordan Zara hot spring Geobacillus sp. ZGt-1 revealed a thermostable 15–20 kDa peptide of growth inhibition properties against not only G. stearothermophilus strain 10, but also against mesophilic B. subtilis and S. typhimurium CCUG 31969 grown at 37 °C, while no inhibition effect was found for E. coli 1005, S. aureus NCTC 83254, S. epidermidis or Proteus vulgaris [16].
2.1.4. Challenges in the Use of Geobacillus/Parageobacillus Bacteriocins and BLISes
One of the main challenges in the use of bacteriocin/BLISes seems to be the appearance and transfer of potential bacteriocin resistance. Bacteria can develop antibiotic or bacteriocin resistance through spontaneous mutations in their DNA or horizontal gene transfer. It is believed that bacterial strains can even reach a state of dual resistance for both types of antimicrobial compounds. Additional limitations to the wide use of bacteriocins/BLISes may be their non-specificity or narrow spectrum of activity. However, they can be highly effective when used together with antibiotics in synergistic therapies to improve efficacy and to minimize antibiotic concentrations. A route of bacteriocin administration, especially oral or intravenous, can also pose a problem. Most bacteriocins and BLISes are smaller than 10 kDa and easily degraded by proteases. This leads to their poor bioavailability [23,62]. The short plasma half-life of bacteriocins/BLISes causes the necessity of bioengineering their properties. Moreover, the synthesis of these compounds is inefficient and depends on many conditions. For that reason, research should aim at increasing their synthesis efficiency and improving their stability. Other problematic issues concerning bacteriocins are: (i) high production cost, (ii) complicated purification process, (iii) route of administration, (iv) low solubility, (v) fast biotransformation, and (vi) low half-life. Many aspects of bacteriocin effectiveness are not yet well understood [23,62]. Geobacillus/Parageobacillus bacteriocins/BLISes withstand a wide range of pH and temperatures. Thus, they are naturally more stable when compared to bacteriocins derived from mesophilic strains. It is also worth mentioning that their spectrum of activity is quite narrow, which can be either a challenge or an advantage, depending on the potential use. Hopefully, future investigation and usage of the Geobacillus/Parageobacillus derived bacteriocins and BLISes will help overcome some of the challenges described above.
2.2. The Potential of Geobacillus and Parageobacillus as Probiotics
The probiotic definition has evolved through the years [2,63]. The most commonly used definition is based on the International Life Sciences Institute–Europe and World Health Organization (WHO) work and states that probiotics are “live microorganisms which when administered in adequate amounts confer a health benefit on the host” [2,64,65]. In some countries, including Japan, a different definition is used. The term probiotic includes not only viable microorganisms but also cells of non-viable microorganisms that confer health benefits [63,65]. It is now understood that microbial viability is not always directly related to their culturability.
The probiotics are often antagonistic against other microorganisms. They are believed to exert their effects through the secretion of vitamins, exoenzymes or bacteriocins, among others. Scientific research into probiotics has recently made significant advances and the scale of the probiotic market is constantly expanding. The use of probiotics has significantly increased, especially in food animal production [66,67,68]. Many studies have shown that probiotics improve growth outcomes, survivability, and resilience to diseases [66,68,69]. Many members of the phylum Firmicutes have been demonstrated as beneficial microbes used as bimodal probiotic microbiota for humans and animals [2,70].
In a post-antibiotic era, and during a time of growing concerns about antimicrobial resistance, there is a constant need for new candidate probiotics, exhibiting greater inhibitory activity against pathogenic bacteria. Since Geobacillus and Parageobacillus also belong to the phylum Firmicutes (Figure 1), they may provide a rich source of novel candidate probiotic strains with unique properties [71]. These bacteria are known to biosynthesize several antibacterial compounds, such as antibiotics, bacteriocins, and BLISes (see Section 2.1). For example, it was demonstrated that G. thermoleovorans could inhibit the growth of some reference pathogenic strains, including S. typhimurium (ATCC1408), Vibrio parahaemolytiticus (ATCC 17802), Vibrio alginolyticus (ATCC 17749), S. aureus (ATCC 25923) and a β-lactamase-producing E. coli strain (ATCC 35218) [45,71]. Additionally, some Geobacillus/Parageobacillus species are able to interfere with quorum sensing in Gram-negative bacteria [70,72]. Last but not least, their adherence ability to abiotic surfaces may help these bacteria to eliminate potential pathogens and create a bio-secure environment, preventing the introduction and/or spread of harmful microorganisms [71].
Due to the above-mentioned properties, Geobacillus and Parageobacillus should certainly be considered as beneficial candidate environmental probiotics. Their advantage over other probiotic strains could be their high survival potential. They are capable of surviving in harsh conditions, such as: draughts, UV radiation, extreme pH and temperatures, organic and inorganic chemicals, salts, detergents and the presence of heavy metal ions [73]. In fact, their spores, derived from terrestrial soils, can persist through cold, extreme aridity and UV radiation at high altitudes. Thus, they can be carried in the atmosphere and deposited far from their origins [6,74]. According to Nicholson’s estimation, the longevity of Geobacillus spores at 40 °C could be 1.9 billion years [6,75]. Moreover, these non-pathogenic bacteria should pose no risk to humans or animals, as they constitute an essential part of the microbial soil community. For many years, both people and animals have had constant exposure to the soil and the aerosoled dust, and a large variety of microorganisms (or their spores), which originally inhabit soil [74]. They even ate soil. According to archeological evidence, Homo erectus and other hominids were already practicing geophagy [74]. It is also worth noting that most primate species are geophageous [74,76]. Nowadays, soil and its microbial components still can enter humans and animals through direct or indirect pathways. Accidental geophagy occurs when improperly washed vegetables are eaten. Contemporary geophagy is defined as willful ingestion of specific types of soil for their nutritive or medicinal value. In several regions of Africa, Asia, South and Latin America, and the pacific Islands, geophagy still remains common practice, among pregnant and nonpregnant women and even men [74,77,78,79]. Interestingly, consumption of soil is seen by geophagic people as a natural stimulant, having a positive and euphoric effect on humans, easing their upsetting thoughts, helping them to relax and struggle with stress [79]. Golokhvast et al. demonstrated that experimental geophagy resulted in significant improvement in the behavior of laboratory rats, subjected to instrumental stress [80]. At this point, it is also worth recalling an intriguing environmental microbiome hypothesis, proposed by Blum et al. [81]. According to this hypothesis, there is a close link between the human intestinal and soil microbiomes. This linkage was established during evolution and is still developing. Both microbiomes can be regarded as “superorganisms” that complement each other with inoculants, genes and growth-sustaining molecules [81]. Extensive research into the geographical and functional links between soils, plant and gut microbiota could benefit human health, sustainable agriculture as well as food industry [81,82].
However, there is another side to the coin: improper selection and inadequate colonization of probiotics may lead to a change in the particular microbiome, not its protection and support [2,83]. Additionally, environmental probiotics with a high survival potential may be difficult to remove if necessary. Thus, the probiotics should be always used with caution. To overcome the technological, economic, and clinical problems, which can be associated with probiotics, scientists adopted postbiotics as a no-viable form. Since postbiotics do not include live microorganisms, the risks associated with their use are minimized [84].
2.3. A Fusion and Display System Based on Proteins Derived from Geobacillus
Both Geobacillus and Parageobacillus genera are known to be a rich source of thermostable, biotechnologically useful proteins. An interesting example of such molecules is a multimeric enzyme—a pyruvate dehydrogenase (PDH)—derived from G. stearothermophilus. This multienzyme complex catalyzes the oxidative decarboxylation of pyruvate, which results in acetyl coenzyme A biosynthesis [85,86]. The PDH is composed of three different subunits: E1, E2 and E3. One of the PDH subunits—dihydrolipoamide acetyltransferase (E2 subunit)—has a unique ability to self-assembly into trimers, which integrate into a 60-meric spherical protein cage (molecular mass 150 MDa). Both the E1 (150 kDa) and E3 (90 kDa) subunits are noncovalently bound into the scaffold formed by E2 and are displayed peripherally in the number of 60 copies [87,88,89,90]. The denatured E2 protein can be renatured in vitro, without the participation of chaperonins, to form a catalytically active 60-mer structure with icosahedral symmetry [91,92,93].
Rationally designed, recombinant proteins, peptides or antigens can be attached to the N-terminal end of the E2 polypeptide or can be displayed on its surface in place of the lipoyl domains. The obtained, recombinant E2 fusion protein variants still have the ability to self-assemble into the spherical protein cage structure [87,88,94]. Moreover, the sizes of the recombinant fusion partners are not limited to small proteins or short peptides.
These unique properties of the dihydrolipoamide acetyltransferase were used to obtain a modified E2 variant with an increased number of phenylalanines per subunit. The modified scaffold served as a nanocarrier for Doxorubicin delivery to human breast cancer cells [95].
Additionally, the recombinant Env V3 or Gag(p17) proteins, derived from HIV-1, were fused to the E2 domain and used for vaccine development. The obtained multimeric structures induced T cell response in mice [87,96,97,98]. The modified E2 cage protein was also used for the construction of biosensors, which were able to detect HeLa cells or to measure the concentration of thrombin. The biosensors were obtained by the modular conjugation of the E2 nanoparticles with various functional moieties, enabling antibody, enzyme, DNA aptamer, or fluorescent dye immobilization [99].
The E2 fusion and display system creates opportunities to develop novel applications in biotechnology and medicine, such as new vaccines [100]. The modified E2 scaffolds also have a potential to be used as nanocarriers in antimicrobial therapies, by fusion or encapsulation with recombinant molecules exhibiting antimicrobial properties.
3. Antimicrobial Potential of Endolysins Derived from Bacteriophages Infecting Geobacillus and Parageobacillus
3.1. Historical Perspectives
The history of phages starts in 1896 when Ernest Hankin observed the antibacterial activity of water from the Ganges against Vibrio cholerae [101]. Nineteen years later, Frederik Twort described the activity of phages in detail but at that time he was not sure whether he observed the activity of an “ultra-microscopic virus”, “a living protoplasm” or “an enzyme with power to growth” [102]. It was not until 1917 when Félix d’Hérelle realized that what he was observing were bacterial viruses and gave them a name—bacteriophages [102]. D’Hérelle was also the first researcher to introduce phage therapies. First, he proved phage therapy to be effective in extinguishing avian typhoid epidemics in poultry and then he shifted his focus to the application of phage therapy in humans. The first successful phage therapy in humans took place in 1919 in France, where d’Hérelle treated several children suffering from dysentery. [103]. More or less successful attempts to use phage therapies continued until after World War II, but most of the Western world abandoned research in this topic as antibiotics were discovered and seemed to be a much more promising option for antibacterial treatment [104].
Today, as we enter the so-called post-antibiotic era, with antibiotic-resistance growing faster than new antibiotics can be discovered, there is an urgent need for alternative antimicrobial treatments. Obviously, phage therapy is one such option, but as far as the phages of the Geobacillus or Parageobacillus genera are considered, they are not expected to be able to target bacteria typically responsible for infections in humans as phages tend to have a rather narrow host range. However, phages of Geobacillus or Parageobacillus genera could offer biotechnology many other opportunities to support advances in antimicrobial treatments. They could be: (1) a source of lytic enzymes—some endolysins such as SAL200 have already been tested via intravenous administration in humans [105] and some formulations like Staphefekt ™ are already products ready to be used on human skin, or (2) a platform for the presentation of antigens, i.e., vaccines—so far at least 16 bacteriophage-based vaccines have been tested on animals [106], but none of them were based on a phage of the Geobacillus or Parageobacillus genera.
3.2. Endolysins as Novel Antimicrobials
Most of the bacteriophages, apart from the filamentous ones, encode proteins that allow the phages to escape the host cell via disruption of the cell wall. The proteins are supposed either to stop the synthesis of peptidoglycan or to digest it enzymatically. The latter is performed by a group of enzymes called endolysins, which can be further divided into five groups, depending on the exact type of bond being cleaved [107,108,109]. Figure 3 shows a schematic representation of the mechanisms of action of the endolysins on Gram-positive and Gram-negative bacteria:
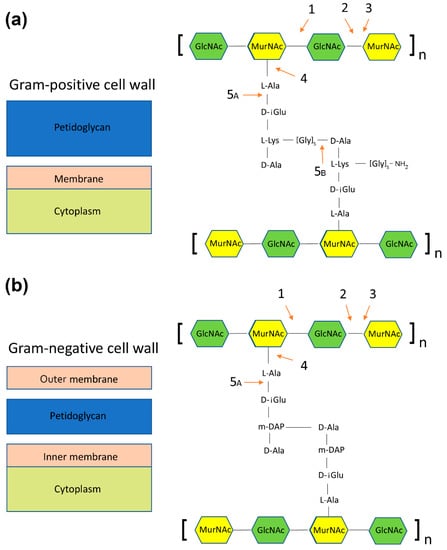
Figure 3.
Schematic representation of the mechanism of action of endolysins on Gram-positive (panel a) and Gram-negative bacteria (panel b). (1) N-acetyl-beta-d-glucosaminidase; (2) N-acetyl-b-d-muramidase; (3) lytic transglycosylase; (4) N-acetylmuramoyl-L-alanine amidase, and (5) endopeptidases: 5A, L-alanoyl-d-glutamate endopeptidase; 5B, d-alanyl-glycyl endopeptidase.
- N-acetylmuramidases (bond between N-acetylglucosamine and N-acetylmuramic acid) (Figure 3, panels a and b),
- N-acetylglucosaminidases (bond between N-acetylmuramic acid and N-acetylglucosamine) (Figure 3, panels a and b),
- Transglycosylases (bond between N-acetylmuramic acid and N-acetylglucosamine, a different mechanism than in N-acetylglucosaminidases) (Figure 3, panels a and b),
- Amidases (bond between N-acetylomuramic acid and L-alanine) (Figure 3, panels a and b)
- Endopeptidases:
As endolysins digest peptidoglycan, they first of all need to access it, i.e., crossing the barrier formed by the inner membrane of a bacterium. Usually this is achieved through cooperation with another protein—a holin, which causes permeabilization of the inner membrane and allows endolysins into the periplasm. Some of the endolysins possess a signal–arrest–release sequence (SAR), which enables their anchoring to the inner membrane without the help of pinholins. However, this mode of action requires depolarization of the membrane to release and activate an endolysin. Such depolarization can be either spontaneous or caused by holins [110].
As far as the structure of endolysins is concerned, endolysins of Gram-negative specific phages are simple, globular proteins consisting of one catalytic domain. On the other hand, a modular structure is common for endolysins of Gram-positive specific phages: usually there is one binding domain responsible for the recognition of the target peptidoglycan and at least one catalytic domain. This difference in the endolysin structure (between phages specific to Gram-positive and Gram-negative bacteria) is probably caused by the lack of an outer membrane in Gram-positive bacteria and the need to prevent ill-timed digestion of other bacteria from the outside [111].
3.3. Thermophilic Endolysins from Bacteriophage Infected Geobacillus Species
Tail bacteriophages, with double-stranded DNA genomes, are often equipped with enzymes called endolysins, which cause lysis of the host by digestion of the peptidoglycan wall. Such bacteriophages usually dominate the environment. Endolysins lacking special signal sequences cannot cross the cytoplasmic membrane unaided. These enzymes access the peptidoglycan by large pores (called “holes”), created in the membrane by holins. Some endolysins do not require holins due to the N-terminus SAR sequence, which allows for their transport by the bacterial Sec system [107].
Endolysins have acquired high substrate specificity. They interact selectively with the potential substrate, which is the peptidoglycan layer of the cell wall of bacteria species. As previously mentioned, such a high affinity of the enzyme for the substrate is usually conditioned by the presence of the cell wall binding domain (CBD), typically located at the C-terminus of the protein. CBD recognizes and binds specific ligands (carbohydrates or teichoic acids) [112]. In addition to the CBD domain, phage endolysins also have the EAD domain, located at the N-terminus [112]. Thus far, 24 variants of the EAD domain and 13 variants of the CBD domain have been characterized [113]. The EAD and CBD domain structures relate to endolysins derived from mycobacteria and the bacteriophages infecting Gram-positive microorganisms. Gram-negative phage endolysins often lack the CBD domain [107,114].
Resistance to antibiotics of many pathogenic bacteria has become a serious clinical problem; therefore, a continuous search for new therapeutic strategies is necessary [115]. The World Health Organization (WHO) warns that, up to the year 2050, antibiotic-resistant bacteria could kill more than 10 million people. The most promising solutions are bacteriophages, phage endolysins and antimicrobial peptides.
Endolysins encoded by bacteriophages are of a great interest because of their potential as antimicrobial agents, useful for controlling bacterial infections and preventing biofilm formation. They can also be used in the case of unwanted contamination of food with opportunistic or pathogenic bacteria [115,116]. According to the literature, bacteriophages, endolysins, and antimicrobial peptides can be used in combination therapy. Such an approach negates many of the limitations of their specificity as single antimicrobial agents [116,117].
Only a few functionally known, thermostable endolysins have been isolated from Geobacillus spp. infecting bacteriophages so far. Here, we focus on their characteristics.
3.3.1. Thermostable GVE2 Endolysin
The bacteriophage from the Siphovirus GVE2 family is one of a few isolated from Geobacillus. GVE2 is a virulent bacteriophage that infects thermophilic deep-sea Geobacillus sp. E263 (CGMCC1.7046), capable of growing at 45–80 °C, with an optimal temperature between 60 and 65 °C. The bacteriophage genome is 40,863 bp, 44.8% G + C, linear dsDNA with 62 ORFs. Proteomic analysis characterized six GVE2 proteins. A gene encoding endolysin was cloned, expressed in E. coli, purified and characterized. In addition, the holin–endolysin system was tested. Confocal microscopy data showed that GFP–endolysin aggregates in Geobacillus sp. E263 were infected with GVE2. The results revealed that GVE2 endolysin interacts directly with the phage holin. It was found that endolysin can interact with the host protein ABC transporter, suggesting that host proteins may be involved in the regulation of the lysis process [70,118].
Clostridium perfringens (C. perfringens) is a bacterial pathogen that causes necrotizing enteritis in poultry and livestock and is a source of food poisoning and gas gangrene in humans. With the decreasing use of antibiotics in feed, alternatives to antibiotics are needed. Bacteriophage endolysins are efficient at eliminating the pathogenic bacterial host. This type of enzyme is a potential replacement for the antibiotics that control C. perfringens. Animal feed is heat treated during pellet production. This treatment consists of the mixture of feed ingredients and conditioning with steam at temperatures up to 95 °C, with an exposure time from 20 s to 4 min. Thus, endolysins used in the production of animal feed pellets should be thermostable or thermotolerant. Additionally, their EAD domains can often be modified to target different bacterial species [119]. That makes the thermostable lytic enzymes highly desirable.
Swift et al. generated thermostable endolysins directed against C. perfringens. Thermostable, catalytic endolysin domains were fused to CBD domains derived from various C. perfringens prophage endolysins. Three thermostable catalytic domains were used. Two of them were prophage endolysins, identified in Geobacillus genomes. The third endolysin was isolated from a deep-sea thermophilic, Geobacillus bacteriophage E2 (GVE2). These catalytic domains carried the activities of L-alanine-amidase, glucosaminidase, and L-alanine-amidase, respectively. All were able to degrade bacterial cell wall peptidoglycan [119].
3.3.2. Thermostable G05 Endolysin
Geobacillus bacteriophages were isolated by Micreos BV (Hague, Netherlands) and designated G01-G09. Their genomes were sequenced to determine the sequence identity among bacteriophages and to identify endolysin encoding genes. Six of the nine analyzed genomes contained easily identifiable genes coding for endolysins. Out of the six identified endolysins, three: G05, G08, G09 exhibited 99% sequence similarity. The endolysin genes were cloned into an E. coli expression vector and the recombinant proteins were then isolated. The lytic activity of the recombinant endolysins was determined at 50 °C, by measuring the OD at 600 nm. The recombinant enzymes showed high lytic activity against Geobacillus sp. in the concentration range of 2–100 µg/mL. Geobacillus is a bacterial organism that produces a biofilm that is strictly thermophilic and facultatively anaerobic. The authors of the patent sought a solution to remove the Geobacillus bacteria biofilms that interfere with the growth and processing of tobacco [120].
Geobacillus biofilms were found in various environments (e.g., hospitals, kitchens, bathrooms, fluid pipes, water, milk, oil, fuel, sewage, boat hulls, plants or trees, animal mouths and in paper or pulp mills). Thus, endolysins specific to the Geobacillus containing biofilm can be helpful in the reduction or elimination of these bacteria.
3.3.3. Thermostable GVE3 Endolysin
The GVE3 bacteriophage, infecting G. thermoglucosidasius, is a member of the Siphoviridae family. Genome bioinformatics analysis revealed the presence of genes encoding lytic enzymes: endolysin and holin, but their lysing properties for host bacteria and other strains were not determined. The thermostability of both the endolysin and holin were also not investigated. However, the proteins were considered stable at high temperatures. Such endolysins could be used for example, for milk treatment, to eliminate Geobacillus sp. Thus, they can be of commercial value. [121].
3.3.4. Thermostable TP-84_28 Endolysin
Endolysin TP-84_28 was isolated from the G. stearothermophilus strain 10 infected with the TP-84 bacteriophage [122]. The enzyme is thermostable. Its optimal reaction temperature is 55 °C. The protein melting point is at 77.6 °C, which was established by physicochemical analyses: differential scanning calorimetry (DSC) and circular dichroism spectroscopy (CD). Interestingly, above this temperature the enzyme still exhibits lytic activity, albeit at a lower level. Incubation of the protein at 100 °C for 30 min causes loss of activity, but not completely [Zebrowska J (unpublished)].
Although the endolysin is derived from the TP-84 bacteriophage, which infects specific Geobacillus strains, it also shows activity against other bacterial species. The antimicrobial effect of the endolysin was tested against two groups of bacteria: Gram-positive and Gram-negative. The enzyme was added directly to the culture of the investigated bacterial strains. Since the optimal temperature for the TP-84_28 endolysin activity is 55 °C, thermophilic bacteria were chosen for testing. The endolysin was highly active against Gram-positive bacteria such as G. stearothermophilus, G. thermoleovorans, and Geobacillus sp. ICI. Surprisingly, the endolysin was found to be active against mesophilic Gram-negative strains, but to a lesser extent [Zebrowska J (unpublished)].
The optimal growth temperature of mesophilic strains did not allow the testing of the endolysin activity directly in a growing microbial culture. However, simple incubation of the selected mesophilic bacteria with the endolysin at 55 °C showed its lytic activity against both Gram-positive and Gram-negative mesophilic strains, with a predominance of Gram-positive strains such as B. cereus and B. subtilis [Zebrowska J (unpublished)].
A thermostable endolysin TP-84_28 may be used for the disinfection of surfaces exposed to high temperatures or as a component of an antimicrobial wound healing preparation. Such preparations could be particularly desirable against hospital-acquired pathogenic strains.
The thermostable endolysin, isolated from the Thermus scotoductus MAT2119 strain infected with Ph2119 bacteriophage, is worth mentioning. The enzyme is the first type 2 N-acetylmuramyl-L-alanine amidase isolated from a thermophilic phage. Plotka et al. demonstrated for the first time that the thermophilic Ph2119 endolysin exhibited lytic activity against the peptidoglycan of mesophilic Gram-negative bacteria, such as: E. coli, Serratia marcescens, Pseudomonas fluorescens and Salmonella enterica serovar Panama. The Ph2119 endolysin exhibits 22% identity with the bacteriophage T7 lysozyme and 23% identity with the T3 lysozyme. The enzyme also showed similarity to the eukaryotic peptidoglycan recognition proteins involved in the innate immune defense found in both insects and mammals. This observation brings interesting conclusions leading to the possibility of using such a protein type for the recognition and digestion of peptidoglycan of eukaryotic origin [123].
Endolysins are noteworthy proteins due to their strong antimicrobial potency. A very small amount of an endolysin is able to eliminate the bacteria from a bacterial suspension. To date, there are no other biological compounds capable of killing microorganisms as quickly and effectively as endolysins. The most promising aspect of these enzymes is their ability to combat antibiotic-resistant bacteria. The efficacy of endolysins has been demonstrated against multi-drug, penicillin- and vancomycin-resistant bacterial strains. A cocktail of different endolysins, with or without antibiotics, could further enhance their antimicrobial activity. High temperatures inactivate most of the available antibiotics. Therefore, such cocktails of endolysins can be particularly useful against thermophilic bacteria [108,117].
3.4. Challenges in the Application of the Geobacillus Bacteriophage Derived Endolysins
Endolysins are a novel class of drugs and so their journey from the laboratory to the market poses a challenge. Thus far, only one endolysin-based product group is available on the European market: Gladskin are products for topical administration based on the active enzyme Staphefekt ™(SA.100). However, more products might be underway as research on the clinical application of various endolysins continues on endolysins such as XZ.700, Artilysin Art-175, SAL200, Exebacase, Ectolysin P128 [124].
What seems to be the biggest obstacle in the practical use of endolysins is their proteinaceous nature. Two aspects seem to be crucial: the administration route and reactions of the immune system.
Not surprisingly, the first commercially available endolysin-based products are meant for topical application—remaining on the surface of the skin, the applied protein is subject to enzymatic digestion or reaction of the immune system to a much lesser extent than when applied orally or intravenously. As far as the oral delivery route is considered, it could most probably only be used for targets within the digestive tract, because proteins are believed not to be absorbable in relevant amounts by enterocytes if not digested to peptides [125]. If an endolysin was expected to act within the digestive tract, highly variable conditions inside it have to be born in mind. The pH varies from highly acidic in the stomach to 7, 4 in the terminal ileum [126] and an inappropriate pH could readily deactivate an enzyme. Even if the influence of gastric acid is omitted via encapsulation in gastro-resistant capsules, an endolysin could be digested by enzymes such as trypsin, chymotrypsin or carboxypeptidase within the intestine. Additionally, interaction with products of the gut microbiota metabolism or ingested food could affect the activity of such enzymatic drugs as endolysins. Another possible mode of delivery of a proteinaceous drug is intravenous administration. It offers a much better bioavailability than oral intake; however, it severely limits accessibility of such a treatment to patients. Inserting a peripheral venous catheter is a simple medical procedure but requires a patient to see medical professionals for each dose of a drug to be administered. Proposed delivery routes such as nasal route and inhalation route have their own limitations, too: probably a high proteolytic activity of the enzymes of macrophage origin in the lungs and size limit absorption of a protein via mucosa in the nose [124].
As all proteins, endolysins could evoke the reaction of the human immune system. Three theoretically expected challenging phenomena could be: allergic reactions, neutralization of drug particles by raised antibodies, and the formation of auto-antibodies. Until now, there are little data on clinical trials in humans, but one worth mentioning is the human safety evaluation of endolysin SAL200 by Jun et al. [127]. Although there were no serious adverse effects including severe allergic reactions in this study, the emergence of antibodies against SAL200 was confirmed in 37% of the study group. The level of serum antibodies seemed to be proportional to the dose. An interesting observation has been made that the blood antibacterial activity was much lower than expected based on the concentration of the drug in blood and antibacterial activity of the blood samples varied between samples with similar serum concentrations of the drug. The authors indicate a possible reason for this phenomenon related to technical issues, but the presence of antibodies should also be taken into consideration as a factor for lowering the enzyme activity. Thinking about immune reactions, one should also bear in mind the possibility of the emergence of auto-antibodies and triggering of auto-immune diseases if an administered endolysin is similar to a protein present in humans. The tendency to evoke antibodies formation will vary among endolysins greatly, depending on their structure; therefore, the abovementioned issue will have to be considered for each drug separately.
Challenges that might arise due to the use of endolysins derived specifically from Geobacillus phages include their temperature optimum and substrate specificity. As Geobacilli are thermophilic, endolysins derived from phages of these bacteria can be expected to have their temperature optimum above the temperature of a human body. Though unfavorable for use in humans, higher temperature optima do not exclude industrial applications. Another factor limiting the use of endolysins derived from Geobacillus phages is their affinity for bonds found in the peptidoglycan of these specific bacteria. If similar structures of peptidoglycan are present in another bacterial strain or genus, the activity of an endolysin will be present. Therefore, the experimental testing of endolysins against various bacterial strains might be required to confirm their activity.
4. Outlook for Using Antimicrobials Derived from Geobacillus, Parageobacillus, and Their Bacteriophages
The age of antibiotics in the fight against pathogenic microorganisms is almost over, mainly due to the increasing antibiotic resistance of contemporary pathogens. This situation causes a rapidly growing interest in antimicrobial compounds that could be alternatives to antibiotics. The Geobacillus and Parageobacillus genera, as well as their bacteriophages, could offer many antagonistic compounds (such as: bacteriocins, probiotics, postbiotics, or endolysins) with a wide range of biological functions. Currently, the bacteriocins seem to be the best-studied group of the antimicrobial substances. However, their routine use requires further research, especially to improve their stability, bioavailability, solubility and performance in vivo. The Geobacillus and Parageobacillus derived bacteriocins may help to solve several problematic issues. Such thermostable antimicrobial compounds could be particularly useful in the food industry, in veterinary treatment, or to disinfect surfaces exposed to high temperatures. Additionally, screening for novel, beneficial environmental strains with probiotic qualities within the Geobacillus and Parageobacillus genera could be a promising future trend in new probiotic preparations. The industrial and environmental interest in these genera is worth greater investigation, especially given their bio-safety, resistance properties, and their wide range of action against mesophilic pathogenic bacteria. Both genera are largely unexplored and could reveal new functionalities and products of medicinal or industrial value. However, it should be noted that in some cases there is still a lack of specific and concrete data on Geobacillus/Parageobacillus derived antimicrobial compounds, making it difficult to assess the true antimicrobial potential of these bacteria.
Author Contributions
Conceptualization, A.Z.-S., J.Z.; writing—original draft preparation, J.Z., M.W., P.E.L., M.P., A.S., E.R., P.M.S., A.Z.-S.; writing—review and editing, A.Z.-S.; funding acquisition, P.M.S., J.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the National Center for Research and Development (Poland) grant TECHMATSTRATEG2/410747/11/NCBR/2019 and National Science Centre (Poland) Miniatura 3 grant 2019/03/X/NZ1/01903.
Acknowledgments
Patrick Groves is appreciated for critical reading of the manuscript and English corrections.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Najar, I.N.; Thakur, N. A systematic review of the genera Geobacillus and Parageobacillus: Their evolution, current taxonomic status and major applications. Microbiology 2020, 166, 800–816. [Google Scholar] [CrossRef] [PubMed]
- Jeżewska-Frąckowiak, J.; Seroczyńska, K.; Banaszczyk, J.; Jedrzejczak, G.; Żylicz-Stachula, A.; Skowron, P.M. The promises and risks of probiotic Bacillus species. Acta Biochim. Pol. 2018, 65, 509–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stein, T. Bacillus subtilis antibiotics: Structures, syntheses and specific functions. Mol. Microbiol. 2005, 56, 845–857. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yi, H. Potential antitumor and anti-inflammatory activities of an extracellular polymeric substance (EPS) from Bacillus subtilis isolated from a housefly. Sci. Rep. 2022, 12, 1383. [Google Scholar] [CrossRef] [PubMed]
- Wada, K.; Suzuki, H. Chapter 15—Biotechnological platforms of the moderate thermophiles, Geobacillus species: Notable properties and genetic tools. In Physiological and Biotechnological Aspects of Extremophiles; Salwan, R., Sharma, V., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 195–218. [Google Scholar] [CrossRef]
- Zeigler, D.R. The Geobacillus paradox: Why is a thermophilic bacterial genus so prevalent on a mesophilic planet? Microbiology 2014, 160 Pt 1, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nazina, T.N.; Tourova, T.P.; Poltaraus, A.B.; Novikova, E.V.; Grigoryan, A.A.; Ivanova, A.E.; Lysenko, A.M.; Petrunyaka, V.V.; Osipov, G.A.; Belyaev, S.S.; et al. Taxonomic study of aerobic thermophilic bacilli: Descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. th. Int. J. Syst. Evol. Microbiol. 2001, 51 Pt 2, 433–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ash, C.; Farrow, J.A.E.; Wallbanks, S.; Collins, M.D. Phylogenetic heterogeneity of the genus Bacillus revealed by comparative analysis of small-subunit-ribosomal RNA sequences. Lett. Appl. Microbiol. 1991, 13, 202–206. [Google Scholar] [CrossRef]
- Aliyu, H.; Lebre, P.; Blom, J.; Cowan, D.; De Maayer, P. Phylogenomic re-assessment of the thermophilic genus Geobacillus. Syst. Appl. Microbiol. 2016, 39, 527–533, Erratum in Syst. Appl. Microbiol. 2018, 41, 529–530. [Google Scholar] [CrossRef] [Green Version]
- Najar, I.N.; Das, S.; Thakur, N. Reclassification of Geobacillus galactosidasius and Geobacillus yumthangensis as Parageobacillus galactosidasius comb. nov. and Parageobacillus yumthangensis comb. nov., respectively. Int. J. Syst. Evol. Microbiol. 2020, 70, 6518–6523. [Google Scholar] [CrossRef]
- Lebre, P.H.; Aliyu, H.; De Maayer, P.; Cowan, D.A. In silico characterization of the global Geobacillus and Parageobacillus secretome. Microb. Cell Fact. 2018, 17, 156. [Google Scholar] [CrossRef]
- Aliyu, H.; Mohr, T.; Cowan, D.; de Maayer, P.; Neumann, A. Time-Course transcriptome of Parageobacillus thermoglucosidasius DSM 6285 grown in the presence of carbon monoxide and air. Int. J. Mol. Sci. 2020, 21, 3870. [Google Scholar] [CrossRef] [PubMed]
- Studholme, D.J. Some (bacilli) like it hot: Genomics of Geobacillus species. Microb. Biotechnol. 2015, 8, 40–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussein, A.H.; Lisowska, B.K.; Leak, D.J. The genus Geobacillus and their biotechnological potential. Adv. Appl. Microbiol. 2015, 92, 1–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadeem, M.S.; Al-Ghamdi, M.A.; Khan, J.A. Studies on the recombinant production in E. coli and characterization of pharmaceutically important thermostable L-asparaginase from Geobacillus thermodenitrificans. Pak. J. Zool. 2019, 51, 1235–1241. [Google Scholar] [CrossRef]
- Alkhalili, R.N.; Bernfur, K.; Dishisha, T.; Mamo, G.; Schelin, J.; Canbäck, B.; Emanuelsson, C.; Hatti-Kaul, R. Antimicrobial protein candidates from the thermophilic Geobacillus sp. strain ZGt-1: Production, proteomics, and bioinformatics analysis. Int. J. Mol. Sci. 2016, 17, 1363. [Google Scholar] [CrossRef] [Green Version]
- Pokusaeva, K.; Kuisiene, N.; Jasinskyte, D.; Rutiene, K.; Saleikiene, J.; Chitavichius, D. Novel bacteriocins produced by Geobacillus stearothermophilus. Open Life Sci. 2009, 4, 196–203. [Google Scholar] [CrossRef]
- Ren, Y.; Strobel, G.; Sears, J.; Park, M. Geobacillus sp., a thermophilic soil bacterium producing volatile antibiotics. Microb. Ecol. 2010, 60, 130–136. [Google Scholar] [CrossRef] [Green Version]
- Gurumurthy, D.M.; Charanraj, T.P.; Faniband, B.; Tallur, P.N.; Bagewadi, Z.K.; Neelagund, S.E.; Mulla, S.I. Cyanoxanthomycin, a bacterial antimicrobial compound extracted from thermophilic Geobacillus sp. Iso5. Jordan J. Biol. Sci. 2020, 13, 725–729. [Google Scholar]
- Schultz, J.; Kallies, R.; Nunes da Rocha, U.; Rosado, A.S. Draft genome sequence of Geobacillus sp. strain LEMMJ02, a thermophile isolated from Deception Island, an active volcano in Antarctica. Microbiol. Resour. Announc. 2019, 8, e00920-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chikindas, M.L.; Weeks, R.; Drider, D.; Chistyakov, V.A.; Dicks, L.M. Functions and emerging applications of bacteriocins. Curr. Opin. Biotechnol. 2018, 49, 23–28. [Google Scholar] [CrossRef]
- Ge, J.; Kang, J.; Ping, W. Effect of acetic acid on bacteriocin production by gram-positive bacteria. J. Microbiol. Biotechnol. 2019, 29, 1341–1348. [Google Scholar] [CrossRef]
- Meade, E.; Slattery, M.A.; Garvery, M. Bacteriocins, potent antimicrobial peptides and the fight against multi drug resistant species: Resistance is futile? Antibiotics 2020, 9, 32. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, D.; Mavridou, D.A.I. Making the best of aggression: The many dimensions of bacterial toxin regulation. Trends Microbiol. 2019, 27, 897–905. [Google Scholar] [CrossRef] [Green Version]
- Zimina, M.; Babich, O.; Prosekov, A.; Sukhikh, S.; Ivanova, S.; Shevchenko, M.; Noskova, S. Overview of global trends in classification, methods of preparation and application of bacteriocins. Antibiotics 2020, 9, 553. [Google Scholar] [CrossRef]
- Hammami, R.; Zouhir, A.; Naghmouchi, K.; Ben Hamida, J.; Fliss, I. SciDBMaker: New software for computer-aided design of specialized biological databases. BMC Bioinform. 2008, 9, 121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morton, J.T.; Freed, S.D.; Lee, S.W.; Friedberg, I. A large scale prediction of bacteriocin gene blocks suggests a wide functional spectrum for bacteriocins. BMC Bioinform. 2015, 16, 381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [Green Version]
- Yount, N.Y.; Weaver, D.C.; de Anda, J.; Lee, E.Y.; Lee, M.W.; Wong, G.C.L.; Yeaman, M.R. Discovery of novel Type II bacteriocins using a new high-dimensional bioinformatic algorithm. Front. Immunol. 2020, 11, 1873. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Luo, L.; Wang, X.; Lu, Y.; Yi, Y.; Shan, Y.; Liu, B.; Zhou, Y.; Lü, X. Mining, Heterologous Expression, Purification, Antibactericidal Mechanism, and Application of Bacteriocins: A Review. Compr. Rev. Food Sci. Food Saf. 2021, 20, 863–899. [Google Scholar] [CrossRef]
- Cheigh, C.-I.; Pyun, Y.-R. Nisin biosynthesis and its properties. Biotechnol. Lett. 2005, 27, 1641–1648. [Google Scholar] [CrossRef]
- Egan, K.; Field, D.; Rea, M.C.; Ross, R.P.; Hill, C.; Cotter, P.D. Bacteriocins: Novel solutions to age old spore-related problems? Front. Microbiol. 2016, 7, 461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smaoui, S.; Elleuch, L.; Bejar, W.; Karray-Rebai, I.; Ayadi, I.; Jaouadi, B.; Mathieu, F.; Chouayekh, H.; Bejar, S.; Mellouli, L. Inhibition of fungi and gram-negative bacteria by bacteriocin BacTN635 produced by Lactobacillus plantarum sp. TN635. Appl. Biochem. Biotechnol. 2010, 162, 1132–1146. [Google Scholar] [CrossRef] [Green Version]
- Sang, Y.; Blecha, F. Antimicrobial peptides and bacteriocins: Alternatives to traditional antibiotics. Anim. Health Res. Rev. 2008, 9, 227–235. [Google Scholar] [CrossRef] [Green Version]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A viable alternative to antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef]
- Newstead, L.L.; Varjonen, K.; Nuttall, T.; Paterson, G.K. Staphylococcal-produced bacteriocins and antimicrobial peptides: Their potential as alternative treatments for Staphylococcus aureus infections. Antibiotics 2020, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Ovchinnikov, K.V.; Kranjec, C.; Thorstensen, T.; Carlsen, H.; Diep, D.B. Successful development of bacteriocins into therapeutic formulation for treatment of MRSA skin infection in a murine model. Antimicrob. Agents Chemother. 2020, 64, e00829-20. [Google Scholar] [CrossRef]
- Simons, A.; Alhanout, K.; Duval, R.E. Bacteriocins, antimicrobial peptides from bacterial origin: Overview of their biology and their impact against multidrug-resistant bacteria. Microorganisms 2020, 8, 639. [Google Scholar] [CrossRef] [PubMed]
- Lopetuso, L.R.; Giorgio, M.E.; Saviano, A.; Scaldaferri, F.; Gasbarrini, A.; Cammarota, G. Bacteriocins and bacteriophages: Therapeutic weapons for gastrointestinal diseases? Int. J. Mol. Sci. 2019, 20, 183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heilbronner, S.; Krismer, B.; Brötz-Oesterhelt, H.; Peschel, A. The microbiome-shaping roles of bacteriocins. Nat. Rev. Microbiol. 2021, 19, 726–739. [Google Scholar] [CrossRef]
- Garg, N.; Oman, T.J.; Andrew Wang, T.-S.; De Gonzalo, C.V.G.; Walker, S.; van der Donk, W.A. Mode of action and structure–activity relationship studies of geobacillin I. J. Antibiot. 2014, 67, 133–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garg, N.; Tang, W.; Goto, Y.; Nair, S.K.; van der Donk, W.A. Lantibiotics from Geobacillus thermodenitrificans. Proc. Natl. Acad. Sci. USA 2012, 109, 5241–5246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garg, N.; Goto, Y.; Chen, T.; van der Donk, W.A. Characterization of the stereochemical configuration of lanthionines formed by the lanthipeptide synthetase GeoM. Pept. Sci. 2016, 106, 834–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ceylan, A.; Başbülbül, G. The use of bacteriocin produced by Geobacillus toebii HBB 218 to prevent the growth of Bacillus coagulans and Geobacillus thermophilus in canned food. Türk Mikrobiyol. Cemiy. Derg. 2019, 49, 140–146. [Google Scholar] [CrossRef] [Green Version]
- Novotny, J.F.; Perry, J.J. Characterization of bacteriocins from two strains of Bacillus thermoleovorans, a thermophilic hydrocarbon-utilizing species. Appl. Environ. Microbiol. 1992, 58, 2393–2396. [Google Scholar] [CrossRef] [Green Version]
- Vaičikauskaitė, M.; Ger, M.; Valius, M.; Maneikis, A.; Lastauskienė, E.; Kalėdienė, L.; Kaunietis, A. Geobacillin 26—High molecular weight bacteriocin from a thermophilic bacterium. Int. J. Biol. Macromol. 2019, 141, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Sharp, R.J.; Bingham, A.H.A.; Comer, M.J.; Atkinson, A.Y. 1979 Partial characterization of a bacteriocin (thermocin) from Bacillus stearothermophilus RS93. Microbiology 1979, 111, 449–451. [Google Scholar] [CrossRef] [Green Version]
- Yule, R.; Barridge, B.D. Isolation and characterization of a bacteriocin produced by Bacillus stearothermophilus strain NU-10. Can. J. Microbiol. 1976, 22, 1743–1750. [Google Scholar] [CrossRef] [PubMed]
- Fikes, J.D.; Crabtree, B.L.; Barridge, B.D. Studies on the mode of action of a bacteriocin produced by Bacillus stearothermophilus. Can. J. Microbiol. 1983, 29, 1576–1582. [Google Scholar] [CrossRef] [PubMed]
- Pranckute, R.; Kaunietis, A.; Kananavičiute, R.; Lebedeva, J.; Kuisiene, N.; Šaleikiene, J.; Čitavičius, D. Differences of antibacterial activity spectra and properties of bacteriocins, produced by Geobacillus sp. bacteria isolated from different environments. J. Microbiol. Biotechnol. Food Sci. 2015, 5, 155–161. [Google Scholar] [CrossRef] [Green Version]
- Kaunietis, A.; Pranckute, R.; Lastauskienė, E.; Čitavičius, D. Medium optimization for bacteriocin production and bacterial cell growth of Geobacillus sp. 15 strain. J. Antimicrob. Agents 2017, 3, 1. [Google Scholar] [CrossRef] [Green Version]
- Shafia, F. Thermocins of Bacillus stearothermophilus. J. Bacteriol. 1966, 92, 524–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egan, K.; Kelleher, P.; Field, D.; Rea, M.C.; Ross, R.P.; Cotter, P.D.; Hill, C. Genome sequence of Geobacillus stearothermophilus DSM 458, an antimicrobial-producing thermophilic bacterium, isolated from a sugar beet factory. Genome Announc. 2017, 5, e01172-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Kuipers, O.P. Identification and classification of known and putative antimicrobial compounds produced by a wide variety of Bacillales species. BMC Genom. 2016, 17, 882. [Google Scholar] [CrossRef] [Green Version]
- Egan, K.; Field, D.; Ross, R.P.; Cotter, P.D.; Hill, C. In silico prediction and exploration of potential bacteriocin gene clusters within the bacterial genus Geobacillus. Front. Microbiol. 2018, 9, 2116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, M.C.; Eslami, S.M.; Hetrick, K.J.; Ackenhusen, S.E.; Mitchell, D.A.; van der Donk, W.A. Precursor peptide-targeted mining of more than one hundred thousand genomes expands the lanthipeptide natural product family. BMC Genom. 2020, 21, 387. [Google Scholar] [CrossRef]
- Alkhalili, R.N.; Canbäck, B. Identification of putative novel class-I lanthipeptides in Firmicutes: A combinatorial in silico analysis approach performed on genome sequenced bacteria and a close inspection of Z-geobacillin lanthipeptide biosynthesis gene cluster of the thermophilic Geobacillus sp. strain ZGt-1. Int. J. Mol. Sci. 2018, 19, 2650. [Google Scholar] [CrossRef] [Green Version]
- Zacharof, M.P.; Lovitt, R.W. Bacteriocins produced by lactic acid bacteria—A review article. APCBEE Procedia 2012, 2, 50–56. [Google Scholar] [CrossRef] [Green Version]
- Gurban oglu Gulahmadov, S.; Batdorj, B.; Dalgalarrondo, M.; Chobert, J.M.; Alekper oglu Kuliev, A.; Haertlé, T. Characterization of bacteriocin-like inhibitory substances (BLIS) from lactic acid bacteria isolated from traditional Azerbaijani cheeses. Eur. Food Res. Technol. 2006, 224, 229–235. [Google Scholar] [CrossRef]
- Başbülbül Özdemir, G.; Biyik, H.H. Isolation and characterization of a bacteriocin-like substance produced by Geobacillus toebii strain HBB-247. Indian J. Microbiol. 2011, 52, 104–108. [Google Scholar] [CrossRef] [Green Version]
- Muhammad, S.A.; Ahmed, S.; Ismail, T.; Hameed, A. Taguchi’s experimental design for optimizing the production of novel thermostable polypeptide antibiotic from Geobacillus pallidus SAT4. Pak. J. Pharm. Sci. 2014, 27, 11–23. [Google Scholar]
- Ołdak, A.; Zielińska, D. Bakteriocyny bakterii fermentacji mlekowej jako alternatywa antybiotyków. Postepy Hig. Med. Dosw. 2017, 71, 328–338. [Google Scholar]
- Van Loveren, H.; Sanz, Y.; Salminen, S. Health claims in Europe: Probiotics and prebiotics as case examples. Annu. Rev. Food Sci. Technol. 2012, 3, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Araya, M.; Gopal, P.; Lindgren, S.E.; Lodi, R.; Oliver, G.; Saxelin, M.L.; Servin, A.L.; Stanton, C.; Gilliland, S.E.; Morelli, L.; et al. Joint FAO/WHO expert consultation on evaluation of health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. In Probiotics in Food: Health and Nutritional Properties and Guidelines for Evaluation; FAO Food and Nutrition paper 85; FAO: Rome, Italy, 2001; Available online: http://www.fao.org/3/a-a0512e.pdf (accessed on 11 February 2022).
- Salminen, S.; Ouwehand, A.; Benno, Y.; Lee, Y.-K. Probiotics: How should they be defined? Trends Food Sci. Technol. 1999, 10, 107–110. [Google Scholar] [CrossRef]
- Siragusa, G. Modern Probiology—Direct Fed Microbials and the Avian Gut Microbiota. In Proceedings of the 23rd Annual Australian Poultry Science Symposium, Sydney, NSW, Australia, 19–22 February 2012. [Google Scholar]
- Vargas-Albores, F.; Porchas-Cornejo, M.A.; Martínez-Porchas, M.; Villalpando-Canchola, E.; Gollas-Galván, T.; Martínez-Córdova, L.R. Bacterial biota of shrimp intestine is significantly modified by the use of a probiotic mixture: A high throughput sequencing approach. Helgol. Mar. Res. 2017, 71, 5. [Google Scholar] [CrossRef] [Green Version]
- Jha, R.; Das, R.; Oak, S.; Mishra, P. Probiotics (Direct-Fed Microbials) in poultry nutrition and their effects on nutrient utilization, growth and laying performance, and gut health: A systematic review. Animals 2020, 10, 1863. [Google Scholar] [CrossRef]
- Susanti, D.; Volland, A.; Tawari, N.; Baxter, N.; Gangaiah, D.; Plata, G.; Nagireddy, A.; Hawkins, T.; Mane, S.P.; Kumar, A. Multi-Omics characterization of host-derived Bacillus spp. probiotics for improved growth performance in poultry. Front. Microbiol. 2021, 12, 747845. [Google Scholar] [CrossRef]
- Łubkowska, B.; Jeżewska-Frąckowiak, J.; Sobolewski, I.; Skowron, P.M. Bacteriophages of thermophilic ‘bacillus group’ bacteria—A review. Microorganisms 2021, 9, 1522. [Google Scholar] [CrossRef]
- Mahdhi, A.; Hmila, Z.; Behi, A.; Bakhrouf, A. Preliminary characterization of the probiotic properties of Candida famata and Geobacillus thermoleovorans. Iran. J. Microbiol. 2011, 3, 129–134. [Google Scholar] [PubMed]
- McMullan, G.; Christie, J.M.; Rahman, T.J.; Banat, I.M.; Ternan, N.G.; Marchant, R. Habitat, applications and genomics of the aerobic, thermophilic genus Geobacillus. Biochem. Soc. Trans. 2004, 32, 214–217. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.; Korza, G.; De Marco, A.M.; Kuipers, O.P.; Li, Y.Q.; Setlow, P. Properties of spores of Bacillus subtilis with or without a transposon that decreases spore germination and increases spore wet heat resistance. J. Appl. Microbiol. 2021, 131, 2918–2928. [Google Scholar] [CrossRef]
- Sing, D.; Sing, C.F. Impact of direct soil exposures from airborne dust and geophagy on human health. Int. J. Environ. Res. Public Health 2010, 7, 1205–1223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicholson, W.L. Using thermal inactivation kinetics to calculate the probability of extreme spore longevity: Implications for paleomicrobiology and lithopanspermia. Orig. Life Evol. Biosph. 2003, 33, 621–631. [Google Scholar] [CrossRef]
- Ketch, L.A.; Malloch, D.; Mahaney, W.C.; Huffman, M.A. Comparative microbial analysis and clay mineralogy of soils eaten by chimpanzees (Pan troglodytes schweinfurthii) in Tanzania. Soil Biol. Biochem. 2001, 33, 199–203. [Google Scholar] [CrossRef]
- Bisi-Johnson, M.A.; Obi, C.L.; Ekosse, G.E. Microbiological and health related perspectives of geophagia: An overview. Afr. J. Biotechnol. 2010, 9, 5784–5791. Available online: http://www.academicjournals.org/AJB (accessed on 11 February 2022).
- Nyanza, E.C.; Joseph, M.; Premji, S.S.; Thomas, D.S.; Mannion, C. Geophagy practices and the content of chemical elements in the soil eaten by pregnant women in artisanal and small scale gold mining communities in Tanzania. BMC Pregnancy Childbirth 2014, 14, 144. [Google Scholar] [CrossRef] [Green Version]
- Huebl, L.; Leick, S.; Guettl, L.; Akello, G.; Kutalek, R. Geophagy in Northern Uganda: Perspectives from consumers and clinicians. Am. J. Trop. Med. Hyg. 2016, 95, 1440–1449. [Google Scholar] [CrossRef]
- Golokhvast, K.; Sergievich, A.; Grigoriev, N. Geophagy (rock eating), experimental stress and cognitive idiosyncrasy. Asian Pac. J. Trop. Biomed. 2014, 4, 362–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blum, W.E.H.; Zechmeister-Boltenstern, S.; Keiblinger, K.M. Does Soil Contribute to the Human Gut Microbiome? Microorganisms 2019, 7, 287. [Google Scholar] [CrossRef] [Green Version]
- Hirt, H. Healthy soils for healthy plants for healthy humans: How beneficial microbes in the soil, food and gut are interconnected and how agriculture can contribute to human health. EMBO Rep. 2020, 21, e51069. [Google Scholar] [CrossRef]
- Abbasi, A.; Sheykhsaran, E.; Kafil, H.S. Obstacles and challenges in the use of probiotics. In Postbiotics: Science, Technology and Applications; Bentham Science Publishers: Al Sharjah, United Arab Emirates, 2021. [Google Scholar] [CrossRef]
- Żółkiewicz, J.; Marzec, A.; Ruszczyński, M.; Feleszko, W. Postbiotics-A step beyond pre- and probiotics. Nutrients 2020, 12, 2189. [Google Scholar] [CrossRef]
- Mande, S.S.; Sarfaty, S.; Allen, M.D.; Perham, R.N.; Hol, W.G.J. Protein-protein interactions in the pyruvate dehydrogenase multienzyme complex: Dihydrolipoamide dehydrogenase complexed with the binding domain of dihydrolipoamide acetyltransferase. Structure 1996, 4, 277–286. [Google Scholar] [CrossRef] [Green Version]
- Henderson, C.E.; Perham, R.N. Purification of the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus and resolution of its four component polypeptides. Biochem. J. 1980, 189, 161–172. [Google Scholar] [CrossRef] [Green Version]
- Domingo, G.J.; Caivano, A.; Sartorius, R.; Barba, P.; Bäckström, M.; Piatier-Tonneau, D.; Guardiola, J.; De Berardinis, P.; Perham, R.N. Induction of specific T-helper and cytolytic responses to epitopes displayed on a virus-like protein scaffold derived from the pyruvate dehydrogenase multienzyme complex. Vaccine 2003, 21, 1502–1509. [Google Scholar] [CrossRef]
- Domingo, G.J.; Orru’, S.; Perham, R.N. Multiple display of peptides and proteins on a macromolecular scaffold derived from a multienzyme complex. J. Mol. Biol. 2001, 305, 259–267. [Google Scholar] [CrossRef] [PubMed]
- D’Apice, L.; Sartorius, R.; Caivano, A.; Mascolo, D.; Del Pozzo, G.; Di Mase, D.S.; Ricca, E.; Li Pira, G.; Manca, F.; Malanga, D.; et al. Comparative analysis of new innovative vaccine formulations based on the use of procaryotic display systems. Vaccine 2007, 25, 1993–2000. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.D.; Perham, R.N. The catalytic domain of dihydrolipoyl acetyltransferase from the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus. Expression, purification and reversible denaturation. FEBS Lett. 1997, 413, 339–343. [Google Scholar] [CrossRef] [Green Version]
- Howard, M.J.; Chauhan, H.J.; Domingo, G.J.; Fuller, C.; Perham, R.N. Protein-protein interaction revealed by NMR T2 relaxation experiments: The lipoyl domain and E1 component of the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus. J. Mol. Biol. 2000, 295, 1023–1037. [Google Scholar] [CrossRef]
- Trovato, M. Novel antigen delivery systems. World J. Virol. 2015, 4, 156. [Google Scholar] [CrossRef]
- Trovato, M. Delivery strategies for novel vaccine formulations. World J. Virol. 2012, 1, 4. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Lee, H.; Lim, S. Isolating a trimer intermediate in the self-assembly of E2 protein cage. Biomacromolecules 2012, 13, 699–705. [Google Scholar] [CrossRef]
- Ren, D.; Dalmau, M.; Randall, A.; Shindel, M.M.; Baldi, P.; Wang, S.-W. Biomimetic design of protein nanomaterials for hydrophobic molecular transport. Adv. Funct. Mater. 2012, 15, 3170–3180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krebs, S.J.; McBurney, S.P.; Kovarik, D.N.; Waddell, C.D.; Jaworski, J.P.; Sutton, W.F.; Gomes, M.M.; Trovato, M.; Waagmeester, G.; Barnett, S.J.; et al. Multimeric scaffolds displaying the HIV-1 envelope MPER induce MPER-specific antibodies and cross-neutralizing antibodies when co-immunized with gp160 DNA. PLoS ONE 2014, 9, e113463, Erratum in PLoS ONE 2015, 10, e0120027. [Google Scholar] [CrossRef] [PubMed]
- Jaworski, J.P.; Krebs, S.J.; Trovato, M.; Kovarik, D.N.; Brower, Z.; Sutton, W.F.; Waagmeester, G.; Sartorius, R.; D’Apice, L.; Caivano, A.; et al. Co-immunization with multimeric scaffolds and DNA rapidly induces potent autologous HIV-1 neutralizing antibodies and CD8+ T cells. PLoS ONE 2012, 7, e31464. [Google Scholar] [CrossRef] [Green Version]
- Caivano, A.; Doria-Rose, N.A.; Buelow, B.; Sartorius, R.; Trovato, M.; D’Apice, L.; Domingo, G.J.; Sutton, W.F.; Haigwood, N.L.; De Berardinis, P. HIV-1 Gag p17 presented as virus-like particles on the E2 scaffold from Geobacillus stearothermophilus induces sustained humoral and cellular immune responses in the absence of IFNγ production by CD4+ T cells. Virology 2010, 407, 296–305. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Chen, Q.; Blackstock, D.; Chen, W. Post-translational modification of bionanoparticles as a modular platform for biosensor assembly. ACS Nano 2015, 9, 8554–8561. [Google Scholar] [CrossRef]
- Wichgers Schreur, P.J.; Tacken, M.; Gutjahr, B.; Keller, M.; van Keulen, L.; Kant, J.; van de Water, S.; Lin, Y.; Eiden, M.; Rissmann, M.; et al. Vaccine efficacy of self-assembled multimeric protein scaffold particles displaying the glycoprotein Gn Head Domain of Rift Valley Fever Virus. Vaccines 2021, 9, 301. [Google Scholar] [CrossRef] [PubMed]
- Hankin, E.H. L’action bactericide des eaux de la Jumna et du Gange sur le vibrion du cholera. Ann. Inst. Pasteur 1896, 10, 511. [Google Scholar]
- Dublanchet, A.; Bourne, S. The epic of phage therapy. Can. J. Infect. Dis. Med. Microbiol. 2007, 18, 15–18. [Google Scholar] [CrossRef] [Green Version]
- Abedon, S.T.; Kuhl, S.J.; Blasdel, B.G.; Kutter, E.M. Phage treatment of human infections. Bacteriophage 2011, 1, 66–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reuter, M.; Kruger, D.H. Approaches to optimize therapeutic bacteriophage and bacteriophage-derived products to combat bacterial infections. Virus Genes 2020, 56, 136–149. [Google Scholar] [CrossRef]
- Jun, S.Y.; Jang, I.J.; Yoon, S.; Jang, K.; Yu, K.S.; Cho, J.Y.; Seong, M.W.; Jung, G.M.; Yoon, S.J.; Kang, S.H. Pharmacokinetics and tolerance of the hage endolysin-based candidate drug SAL200 after a single intravenous administration among healthy volunteers. Antimicrob. Agents Chemother. 2017, 61, e02629-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Mora, A.; Hernández-Pérez, J.; Iqbal, H.M.N.; Rito-Palomares, M.; Benavides, J. Bacteriophage-based vaccines: A potent approach for antigen delivery. Vaccines 2020, 8, 504. [Google Scholar] [CrossRef] [PubMed]
- Woźnica, M.; Bigos, J.; Łobocka, M. Lysis of bacterial cells in the process of bacteriophage release—Canonical and newly discovered mechanisms. Postepy Hig. Med. Dosw. 2015, 69, 114–126. [Google Scholar]
- Oliveira, H.; Azeredo, J.; Lavigne, R.; Kluskens, L.D. Bacteriophage endolysins as a response to emerging foodborne pathogens. Trends Food Sci. Technol. 2012, 28, 103–115. [Google Scholar] [CrossRef] [Green Version]
- Borysowski, J.; Weber-Da.browska, B.; Górski, A. Bacteriophage endolysins as a novel class of antibacterial agents. Exp. Biol. Med. 2006, 231, 366–377. [Google Scholar] [CrossRef]
- Young, R. Phage lysis: Three steps, three choices, one outcome. J. Microbiol. 2014, 52, 243–258. [Google Scholar] [CrossRef]
- Maciejewska, B.; Olszak, T.; Drulis-Kawa, Z. Applications of bacteriophages versus phage enzymes to combat and cure bacterial infections: An ambitious and also a realistic application? Appl. Microbiol. Biotechnol. 2018, 102, 2563–2581. [Google Scholar] [CrossRef] [Green Version]
- Schmelcher, M.; Donovan, D.M.; Loessner, M.J. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012, 7, 1147–1171. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, H.; Melo, L.D.R.; Santos, S.B.; Nobrega, F.L.; Ferreira, E.C.; Cerca, N.; Azeredo, J.; Kluskens, L.D. Molecular aspects and comparative genomics of bacteriophage endolysins. J. Virol. 2013, 87, 4558–4570. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, H.; São-José, C.; Azeredo, J. Phage-derived peptidoglycan degrading enzymes: Challenges and future prospects for in vivo therapy. Viruses 2018, 10, 292. [Google Scholar] [CrossRef] [Green Version]
- Mirski, T.; Mizak, L.; Nakonieczna, A.; Gryko, R. Bacteriophages, phage endolysins and antimicrobial peptides—The possibilities for their common use to combat infections and in the design of new drugs. Ann. Agric. Environ. Med. 2019, 26, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.C.; Schmelcher, M.; Rodriguez-Rubio, L.; Klumpp, J.; Pritchard, D.G.; Dong, S.; Donovan, D.M. Endolysins as antimicrobials. Adv. Virus Res. 2012, 83, 299–365. [Google Scholar] [CrossRef] [Green Version]
- Rahman, M.; Wang, W.; Sun, Q.; Shah, J.A.; Li, C.; Sun, Y.; Li, Y.; Zhang, B.; Chen, W.; Wang, S. Endolysin, a promising solution against antimicrobial resistance. Antibiotics 2021, 10, 1277. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Ye, T.; Zhang, X. Roles of bacteriophage GVE2 endolysin in host lysis at high temperatures. Microbiology 2013, 159, 1597–1605. [Google Scholar] [CrossRef] [Green Version]
- Swift, S.M.; Reid, K.P.; Donovan, D.M.; Ramsay, T.G. Thermophile lytic enzyme fusion proteins that target Clostridium perfringens. Antibiotics 2019, 8, 214. [Google Scholar] [CrossRef] [Green Version]
- Miller, E.; Warek, U.; Xu, D. Endolysin from Bacteriophage against Geobacillus and Methods of Using. International Patent Application WIPO/PCT WO 2016/123425 Al, PCT/US2016/015516, 2016. [Google Scholar]
- Van Zyl, L.J.; Sunda, F.; Taylor, M.P.; Cowan, D.A.; Trindade, M.I. Identification and characterization of a novel Geobacillus thermoglucosidasius bacteriophage, GVE3. Arch. Virol. 2015, 160, 2269–2282. [Google Scholar] [CrossRef] [Green Version]
- Skowron, P.M.; Kropinski, A.M.; Zebrowska, J.; Janus, L.; Szemiako, K.; Czajkowska, E.; Maciejewska, N.; Skowron, M.; Łoś, J.; Łoś, M.; et al. Sequence, genome organization, annotation and proteomics of the thermophilic, 47.7-kb Geobacillus stearothermophilus bacteriophage TP-84 and its classification in the new Tp84virus genus. PLoS ONE 2018, 13, e0195449, Erratum in PLoS ONE 2018, 13, e0196798. [Google Scholar] [CrossRef]
- Plotka, M.; Kaczorowska, A.K.; Stefanska, A.; Morzywolek, A.; Fridjonsson, O.H.; Dunin-Horkawicz, S.; Kozlowski, L.; Hreggvidsson, G.O.; Kristjansson, J.K.; Dabrowski, S.; et al. Novel highly thermostable endolysin from Thermus scotoductus MAT2119 bacteriophage Ph2119 with amino acid sequence similarity to eukaryotic peptidoglycan recognition proteins. Appl. Environ. Microbiol. 2014, 80, 886–895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, E.; Draper, L.A.; Ross, R.P.; Hill, C. The advantages and challenges of using endolysins in a clinical setting. Viruses 2021, 13, 680. [Google Scholar] [CrossRef]
- Miner-Williams, W.M.; Stevens, B.R.; Moughan, P.J. Are intact peptides absorbed from the healthy gut in the adult human? Nutr. Res. Rev. 2014, 27, 308–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fallingborg, J. Intraluminal pH of the human gastrointestinal tract. Dan. Med. Bull. 1999, 46, 183–196. [Google Scholar] [PubMed]
- Jun, S.Y.; Jung, G.M.; Yoon, S.J.; Youm, S.Y.; Han, H.Y.; Lee, J.H.; Kang, S.H. Pharmacokinetics of the phage endolysin-based candidate drug SAL200 in monkeys and its appropriate intravenous dosing period. Clin. Exp. Pharmacol. Physiol. 2016, 43, 1013–1016. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).