Idiosyncratic Fitness Costs of Ampicillin-Resistant Mutants Derived from a Long-Term Experiment with Escherichia coli
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Fitness Assays
3. Results
3.1. Fitness Costs Significantly Vary among Ampicillin-Resistant Mutants
3.2. Level of Resistance Does Not Explain the Variation in Fitness Costs
3.3. Genetic Background Does Not Explain the Variation in Fitness Costs
3.4. Hitchhiking Does Not Explain the Variation in Fitness Costs
3.5. Genetic Basis for the Idiosyncratic Variation in Fitness Costs
3.6. Summary of Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wohlleben, W.; Mast, Y.; Stegmann, E.; Ziemert, N. Antibiotic drug discovery. Microb. Biotechnol. 2016, 9, 541–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez, G.V.; Fleming-Dutra, K.E.; Roberts, R.M.; Hicks, L.A. Core elements of outpatient antibiotic stewardship. Morb. Mortal. Wkly. Rep. Recomm. Rep. 2016, 65, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Baquero, F.; Martínez, J.L.; Lanza, V.F.; Rodríguez-Beltrán, J.; Galán, J.C.; San Millán, A.; Cantón, R.; Coque, T.M. Evolutionary pathways and trajectories in antibiotic resistance. Clin. Microbiol. Rev. 2021, 34, e00050-19. [Google Scholar] [CrossRef]
- Lenski, R.E.; Bouma, J.E. Effects of segregation and selection on instability of plasmid pACYC184 in Escherichia coli B. J. Bacteriol. 1987, 169, 5314–5316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, T.N.M.; Phan, Q.G.; Duong, L.P.; Bertrand, K.P.; Lenski, R.E. Effects of carriage and expression of the Tn10 tetracycline-resistance operon on the fitness of Escherichia coli K12. Mol. Biol. Evol. 1989, 6, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Hughes, D. Antibiotic resistance and its cost: Is it possible to reverse resistance? Nat. Rev. Microbiol. 2010, 8, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Vogwill, T.; MacLean, R.C. The genetic basis of the fitness costs of antimicrobial resistance: A meta-analysis approach. Evol. Appl. 2015, 8, 284–295. [Google Scholar] [CrossRef]
- Hughes, D.; Andersson, D.I. Evolutionary trajectories to antibiotic resistance. Annu. Rev. Microbiol. 2017, 71, 579–596. [Google Scholar] [CrossRef] [Green Version]
- Santos-Lopez, A.; Marshall, C.W.; Scribner, M.R.; Snyder, D.J.; Cooper, V.S. Evolutionary pathways to antibiotic resistance are dependent upon environmental structure and bacterial lifestyle. eLife 2019, 8, e47612. [Google Scholar] [CrossRef]
- Card, K.J.; Jordan, J.A.; Lenski, R.E. Idiosyncratic variation in the fitness costs of tetracycline-resistance mutations in Escherichia coli. Evolution 2021, 75, 1230–1238. [Google Scholar] [CrossRef]
- Vogwill, T.; Kojadinovic, M.; MacLean, R.C. Epistasis between antibiotic resistance mutations and genetic background shape the fitness effect of resistance across species of Pseudomonas. Proc. R. Soc. B 2016, 283, 20160151. [Google Scholar] [CrossRef] [Green Version]
- Knopp, M.; Andersson, D.I. Predictable phenotypes of antibiotic resistance mutations. mBio 2018, 9, e00770-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokoyama, M.; Stevens, E.; Laabei, M.; Bacon, L.; Heesom, K.; Bayliss, S.; Ooi, N.; O’Neill, A.J.; Murray, E.; Williams, P.; et al. Epistasis analysis uncovers hidden antibiotic resistance-associated fitness costs hampering the evolution of MRSA. Genome Biol. 2018, 19, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apjok, G.; Boross, G.; Nyerges, Á.; Fekete, G.; Lázár, V.; Papp, B.; Pál, C.; Csörgo, B. Limited evolutionary conservation of the phenotypic effects of antibiotic resistance mutations. Mol. Biol. Evol. 2019, 36, 1601–1611. [Google Scholar] [CrossRef] [PubMed]
- Castro, R.A.D.; Ross, A.; Kamwela, L.; Reinhard, M.; Loiseau, C.; Feldmann, J.; Borrell, S.; Trauner, A.; Gagneux, S. The genetic background modulates the evolution of fluoroquinolone-resistance in Mycobacterium tuberculosis. Mol. Biol. Evol. 2020, 37, 195–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Card, K.J.; LaBar, T.; Gomez, J.B.; Lenski, R.E. Historical contingency in the evolution of antibiotic resistance after decades of relaxed selection. PLoS Biol. 2019, 17, e3000397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Card, K.J.; Thomas, M.D.; Graves, J.L.; Barrick, J.E.; Lenski, R.E. Genomic evolution of antibiotic resistance is contingent on genetic background following a long-term experiment with Escherichia coli. Proc. Natl. Acad. Sci. USA 2021, 118, e2016886118. [Google Scholar] [CrossRef] [PubMed]
- Lenski, R.E.; Rose, M.R.; Simpson, S.C.; Tadler, S.C. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2000 generations. Am. Nat. 1991, 138, 1315–1341. [Google Scholar] [CrossRef]
- Atlas, R.M. Handbook of Microbiological Media, 3rd ed.; CRC Press: Boca Raton, FL, USA, 2004. [Google Scholar]
- Wiser, M.J.; Ribeck, N.; Lenski, R.E. Long-term dynamics of adaptation in asexual populations. Science 2013, 342, 1364–1367. [Google Scholar] [CrossRef] [Green Version]
- Lenski, R.E.; Wiser, M.J.; Ribeck, N.; Blount, Z.D.; Nahum, J.R.; Morris, J.J.; Zaman, L.; Turner, C.B.; Wade, B.D.; Maddamsetti, R.; et al. Sustained fitness gains and variability in fitness trajectories in the long-term evolution experiment with Escherichia coli. Proc. R. Soc. B 2015, 282, 20152292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melnyk, A.H.; Wong, A.; Kassen, R. The fitness costs of antibiotic resistance mutations. Evol. Appl. 2015, 8, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Kenney, L.J. A new role of OmpR in acid and osmotic stress in Salmonella and E. coli. Front. Microbiol. 2018, 9, 2656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, U.; Lee, C.-R. Distinct roles of outer membrane porins in antibiotic resistance and membrane integrity in Escherichia coli. Front. Microbiol. 2019, 10, 953. [Google Scholar] [CrossRef] [PubMed]
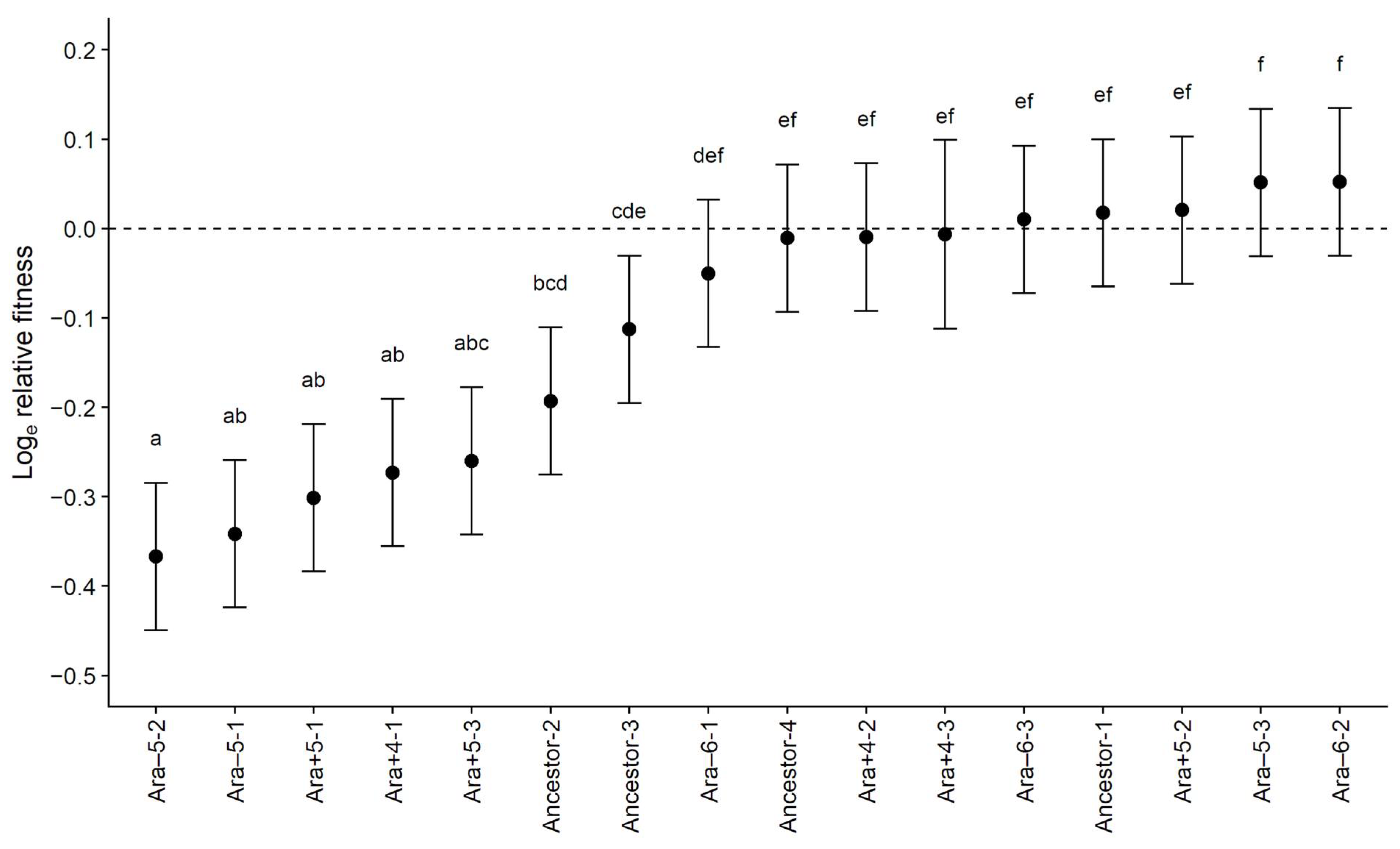
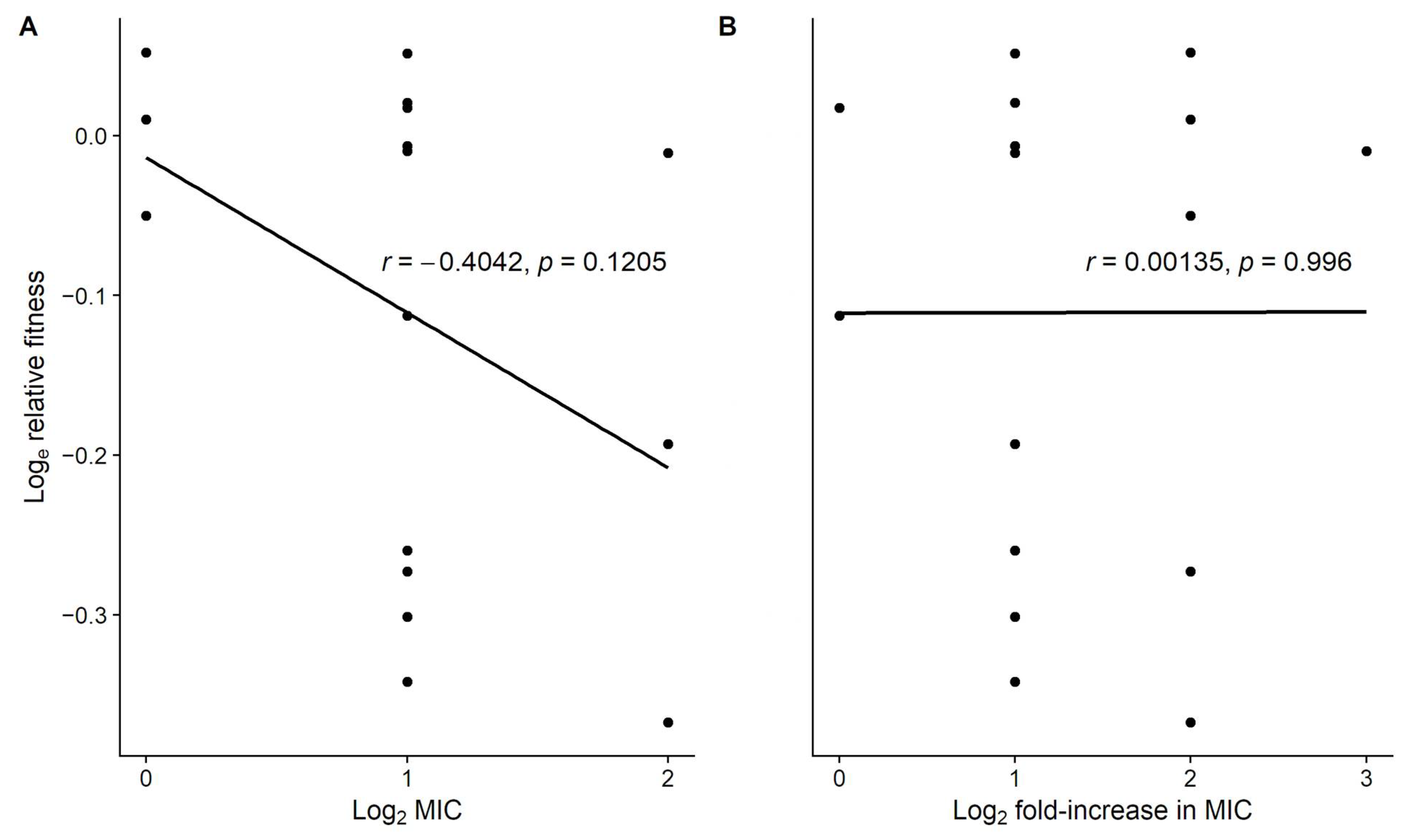
| Source | SS | d.f. | MS | F | p |
|---|---|---|---|---|---|
| Line | 1.7220 | 15 | 0.1148 | 26.04 | <<0.0001 |
| Error | 0.2777 | 63 | 0.0044 | ||
| Total | 1.9997 | 78 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jordan, J.A.; Lenski, R.E.; Card, K.J. Idiosyncratic Fitness Costs of Ampicillin-Resistant Mutants Derived from a Long-Term Experiment with Escherichia coli. Antibiotics 2022, 11, 347. https://doi.org/10.3390/antibiotics11030347
Jordan JA, Lenski RE, Card KJ. Idiosyncratic Fitness Costs of Ampicillin-Resistant Mutants Derived from a Long-Term Experiment with Escherichia coli. Antibiotics. 2022; 11(3):347. https://doi.org/10.3390/antibiotics11030347
Chicago/Turabian StyleJordan, Jalin A., Richard E. Lenski, and Kyle J. Card. 2022. "Idiosyncratic Fitness Costs of Ampicillin-Resistant Mutants Derived from a Long-Term Experiment with Escherichia coli" Antibiotics 11, no. 3: 347. https://doi.org/10.3390/antibiotics11030347
APA StyleJordan, J. A., Lenski, R. E., & Card, K. J. (2022). Idiosyncratic Fitness Costs of Ampicillin-Resistant Mutants Derived from a Long-Term Experiment with Escherichia coli. Antibiotics, 11(3), 347. https://doi.org/10.3390/antibiotics11030347






