The Influence of Antibiotic Resistance on Innate Immune Responses to Staphylococcus aureus Infection
Abstract
:1. Introduction
2. Innate Sensing of S. aureus
2.1. TLRs
2.2. C-Type Lectin Receptor
2.3. Cytosolic Sensors
2.3.1. NOD-like Receptors (NLR)
2.3.2. RIG-I-like Receptors Family
2.3.3. cGAS–STING Pathway
3. Key Cells of the Innate Immune Response to S. aureus Infection
3.1. Keratinocytes (KCs)
3.2. Macrophages
3.3. Neutrophils
3.4. Dendritic Cells
4. Strategies of S. aureus to Evade Immune Defence Mechanisms
4.1. Inhibition of Pattern Recognition Pathways
4.2. Altering Secretion of Pro-Inflammatory Cytokines
4.3. Inhibition of Intracellular Degradation
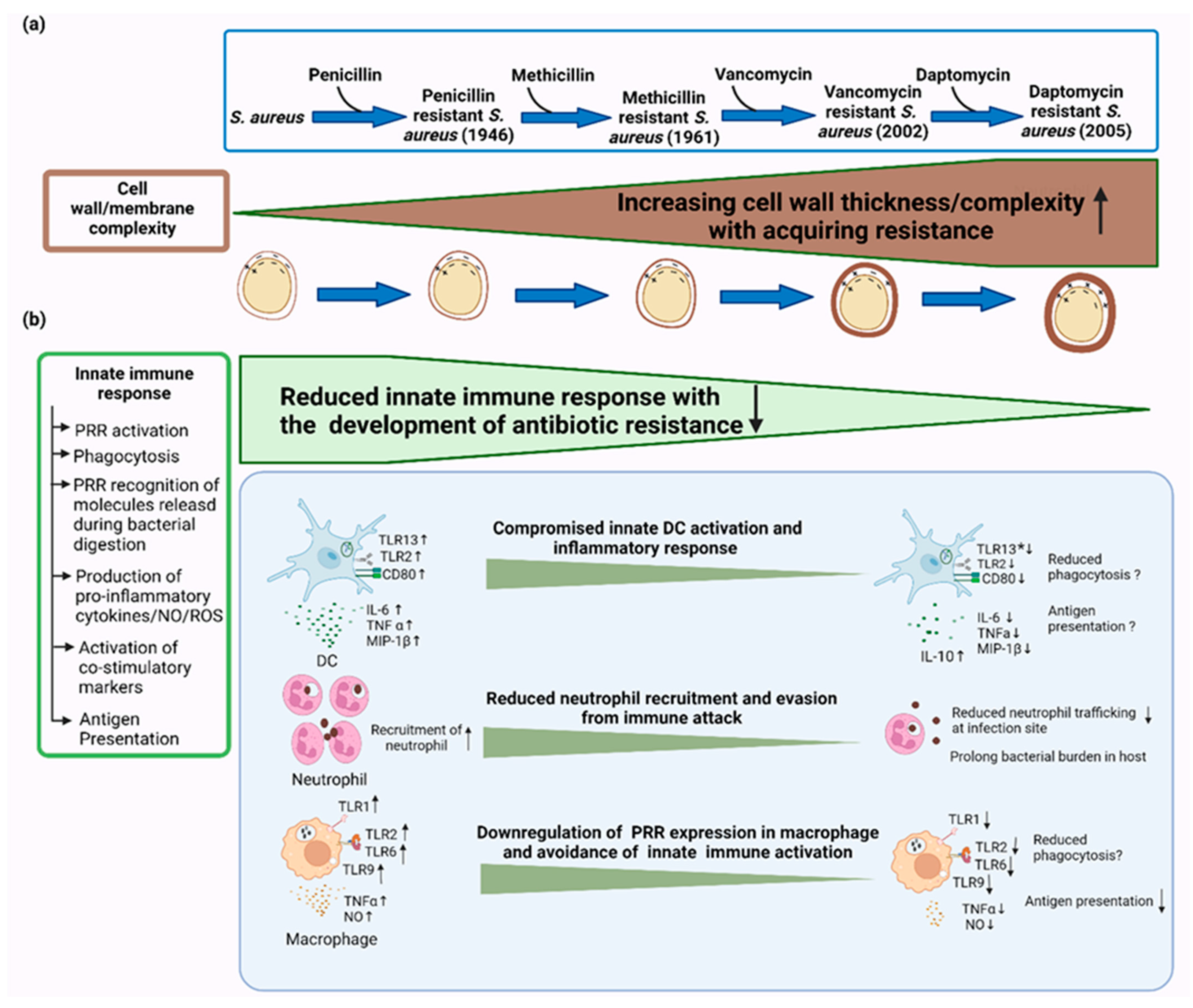
5. Development of Antibiotic Resistance and Evasion of Immune Recognition
6. Interplay between Antibiotic Resistance and Innate Immunity
7. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brown, A.F.; Leech, J.M.; Rogers, T.R.; McLoughlin, R.M. Staphylococcus aureus colonization: Modulation of host immune response and impact on human vaccine design. Front. Immunol. 2014, 4, 507. [Google Scholar] [CrossRef] [PubMed]
- Wertheim, H.F.; Melles, D.C.; Vos, M.C.; van Leeuwen, W.; van Belkum, A.; Verbrugh, H.A.; Nouwen, J.L. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 2005, 5, 751–762. [Google Scholar] [CrossRef]
- Kluytmans, J.; van Belkum, A.; Verbrugh, H. Nasal carriage of Staphylococcus aureus: Epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 1997, 10, 505–520. [Google Scholar] [CrossRef] [PubMed]
- Fowler, V.G., Jr.; Boucher, H.W.; Corey, G.R.; Abrutyn, E.; Karchmer, A.W.; Rupp, M.E.; Levine, D.P.; Chambers, H.F.; Tally, F.P.; Vigliani, G.A.; et al. Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N. Engl. J. Med. 2006, 355, 653–665. [Google Scholar] [CrossRef] [Green Version]
- Howden, B.P.; Davies, J.K.; Johnson, P.D.; Stinear, T.P.; Grayson, M.L. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: Resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 2010, 23, 99–139. [Google Scholar] [CrossRef] [Green Version]
- Parker, D.; Prince, A. Immunopathogenesis of Staphylococcus aureus pulmonary infection. Semin. Immunopathol. 2012, 34, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Gillet, Y.; Issartel, B.; Vanhems, P.; Fournet, J.-C.; Lina, G.; Bes, M.; Vandenesch, F.; Piémont, Y.; Brousse, N.; Floret, D.; et al. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet 2002, 359, 753–759. [Google Scholar] [CrossRef]
- Hayashida, A.; Bartlett, A.H.; Foster, T.J.; Park, P.W. Staphylococcus aureus beta-toxin induces lung injury through syndecan-1. Am. J. Pathol. 2009, 174, 509–518. [Google Scholar] [CrossRef] [Green Version]
- Rastogi, D.; Ratner, A.J.; Prince, A. Host–bacterial interactions in the initiation of inflammation. Paediatr. Respir. Rev. 2001, 2, 245–252. [Google Scholar] [CrossRef]
- Balasubramanian, D.; Harper, L.; Shopsin, B.; Torres, V.J. Staphylococcus aureus pathogenesis in diverse host environments. Pathog. Dis. 2017, 75, ftx005. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.H.; Bhuiyan, M.S.; Shen, H.H.; Cameron, D.R.; Rupasinghe, T.W.T.; Wu, C.M.; Le Brun, A.P.; Kostoulias, X.; Domene, C.; Fulcher, A.J.; et al. Antibiotic resistance and host immune evasion in Staphylococcus aureus mediated by a metabolic adaptation. Proc. Natl. Acad. Sci. USA 2019, 116, 3722–3727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaplin, D.D. Overview of the immune response. J. Allergy Clin. Immunol. 2003, 111, S442–S459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fournier, B.; Philpott, D.J. Recognition of Staphylococcus aureus by the innate immune system. Clin. Microbiol. Rev. 2005, 18, 521–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinman, R.M. The dendritic cell system and its role in immunogenicity. Annu. Rev. Immunol. 1991, 9, 271–296. [Google Scholar] [CrossRef] [PubMed]
- Mogensen, T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishnaswamy, J.K.; Chu, T.; Eisenbarth, S.C. Beyond pattern recognition: NOD-like receptors in dendritic cells. Trends Immunol. 2013, 34, 224–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palm, N.W.; Medzhitov, R. Pattern recognition receptors and control of adaptive immunity. Immunol. Rev. 2009, 227, 221–233. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef]
- Takeuchi, O.; Hoshino, K.; Akira, S. Cutting edge: TLR2-deficient and MyD88-deficient mice are highly susceptible to Staphylococcus aureus infection. J. Immunol. 2000, 165, 5392–5396. [Google Scholar] [CrossRef] [Green Version]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [Green Version]
- Brandt, S.L.; Putnam, N.E.; Cassat, J.E.; Serezani, C.H. Innate immunity to Staphylococcus aureus: Evolving paradigms in soft tissue and invasive infections. J. Immunol. 2018, 200, 3871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stenzel, W.; Soltek, S.; Sanchez-Ruiz, M.; Akira, S.; Miletic, H.; Schlüter, D.; Deckert, M. Both TLR2 and TLR4 are required for the effective immune response in Staphylococcus aureus-induced experimental murine brain abscess. Am. J. Pathol. 2008, 172, 132–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lentini, G.; Famà, A.; De Gaetano, G.V.; Galbo, R.; Coppolino, F.; Venza, M.; Teti, G.; Beninati, C. Role of endosomal TLRs in Staphylococcus aureus infection. J. Immunol. 2021, ji2100389. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, B.V.; Kuehn, M.J. Staphylococcus aureus secretes immunomodulatory RNA and DNA via membrane vesicles. Sci. Rep. 2020, 10, 18293. [Google Scholar] [CrossRef] [PubMed]
- Moen, S.H.; Ehrnström, B.; Kojen, J.F.; Yurchenko, M.; Beckwith, K.S.; Afset, J.E.; Damås, J.K.; Hu, Z.; Yin, H.; Espevik, T.; et al. Human Toll-like Receptor 8 (TLR8) is an important sensor of pyogenic bacteria, and is attenuated by cell surface TLR signaling. Front. Immunol. 2019, 10, 1209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jöckel, S.; Nees, G.; Sommer, R.; Zhao, Y.; Cherkasov, D.; Hori, H.; Ehm, G.; Schnare, M.; Nain, M.; Kaufmann, A.; et al. The 2′-O-methylation status of a single guanosine controls transfer RNA-mediated Toll-like receptor 7 activation or inhibition. J. Exp. Med. 2012, 209, 235–241. [Google Scholar] [CrossRef]
- Parker, D.; Prince, A. Staphylococcus aureus induces type I IFN signaling in dendritic cells via TLR9. J. Immunol. 2012, 189, 4040–4046. [Google Scholar] [CrossRef] [Green Version]
- Zizzari, I.G.; Napoletano, C.; Battisti, F.; Rahimi, H.; Caponnetto, S.; Pierelli, L.; Nuti, M.; Rughetti, A. MGL receptor and immunity: When the ligand can make the difference. J. Immunol. Res. 2015, 2015, 450695. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.; Takahashi, K.; Dundee, J.; Shahroor-Karni, S.; Thiel, S.; Jensenius, J.C.; Gad, F.; Hamblin, M.R.; Sastry, K.N.; Ezekowitz, R.A.B. Mannose-binding lectin-deficient mice are susceptible to infection with Staphylococcus aureus. J. Exp. Med. 2004, 199, 1379–1390. [Google Scholar] [CrossRef] [Green Version]
- Ip, W.K.E.; Takahashi, K.; Moore, K.J.; Stuart, L.M.; Ezekowitz, R.A.B. Mannose-binding lectin enhances Toll-like receptors 2 and 6 signaling from the phagosome. J. Exp. Med. 2008, 205, 169–181. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.K.; Shin, J.S.; Nahm, M.H. NOD-Like receptors in infection, immunity, and diseases. Yonsei Med. J. 2016, 57, 5–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franchi, L.; Warner, N.; Viani, K.; Nuñez, G. Function of Nod-like receptors in microbial recognition and host defense. Immunol. Rev. 2009, 227, 106–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deshmukh, H.S.; Hamburger, J.B.; Ahn, S.H.; McCafferty, D.G.; Yang, S.R.; Fowler, V.G., Jr. Critical role of NOD2 in regulating the immune response to Staphylococcus aureus. Infect. Immun. 2009, 77, 1376–1382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, M.B.; Halman, J.R.; Burmeister, A.R.; Currin, S.; Khisamutdinov, E.F.; Afonin, K.A.; Marriott, I. Retinoic acid inducible gene-I mediated detection of bacterial nucleic acids in human microglial cells. J. Neuroinflamm. 2020, 17, 139. [Google Scholar] [CrossRef]
- Sun, Q.; Sun, L.; Liu, H.H.; Chen, X.; Seth, R.B.; Forman, J.; Chen, Z.J. The specific and essential role of MAVS in antiviral innate immune responses. Immunity 2006, 24, 633–642. [Google Scholar] [CrossRef] [Green Version]
- Marinho, F.V.; Benmerzoug, S.; Oliveira, S.C.; Ryffel, B.; Quesniaux, V.F.J. The emerging roles of STING in bacterial infections. Trends Microbiol. 2017, 25, 906–918. [Google Scholar] [CrossRef]
- Dobbs, N.; Burnaevskiy, N.; Chen, D.; Gonugunta, V.K.; Alto, N.M.; Yan, N. STING activation by translocation from the ER is associated with infection and autoinflammatory disease. Cell Host Microbe 2015, 18, 157–168. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Cai, X.; Wu, J.; Cong, Q.; Chen, X.; Li, T.; Du, F.; Ren, J.; Wu, Y.T.; Grishin, N.V.; et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 2015, 347, aaa2630. [Google Scholar] [CrossRef] [Green Version]
- Scumpia, P.O.; Botten, G.A.; Norman, J.S.; Kelly-Scumpia, K.M.; Spreafico, R.; Ruccia, A.R.; Purbey, P.K.; Thomas, B.J.; Modlin, R.L.; Smale, S.T. Opposing roles of Toll-like receptor and cytosolic DNA-STING signaling pathways for Staphylococcus aureus cutaneous host defense. PLoS Pathog. 2017, 13, e1006496. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhao, S.; Gao, X.; Jiang, S.; Ma, J.; Wang, R.; Li, Q.; Qin, L.; Tong, Z.; Wu, J.; et al. Staphylococcus aureus induces IFN-β production via a CARMA3-independent mechanism. Pathogens 2021, 10, 300. [Google Scholar] [CrossRef]
- Picard, C.; von Bernuth, H.; Ghandil, P.; Chrabieh, M.; Levy, O.; Arkwright, P.D.; McDonald, D.; Geha, R.S.; Takada, H.; Krause, J.C.; et al. Clinical features and outcome of patients with IRAK-4 and MyD88 deficiency. Medicine 2010, 89, 403–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, M.; Zhou, M.; Jiang, C.; Chen, X.; Guo, L.; Zhang, M.; Chu, Z.; Wang, Y. Staphylococcus aureus Phenol-Soluble Modulins α1-α3 act as novel Toll-Like Receptor (TLR) 4 antagonists to inhibit HMGB1/TLR4/NF-κB signaling pathway. Front. Immunol. 2018, 9, 862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mae, M.; Iyori, M.; Yasuda, M.; Shamsul, H.M.; Kataoka, H.; Kiura, K.; Hasebe, A.; Totsuka, Y.; Shibata, K. The diacylated lipopeptide FSL-1 enhances phagocytosis of bacteria by macrophages through a Toll-like receptor 2-mediated signalling pathway. FEMS Immunol. Med. Microbiol. 2007, 49, 398–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jann, N.J.; Schmaler, M.; Ferracin, F.; Landmann, R. TLR2 enhances NADPH oxidase activity and killing of Staphylococcus aureus by PMN. Immunol. Lett. 2011, 135, 17–23. [Google Scholar] [CrossRef]
- Fang, L.; Wu, H.-M.; Ding, P.-S.; Liu, R.-Y. TLR2 mediates phagocytosis and autophagy through JNK signaling pathway in Staphylococcus aureus-stimulated RAW264.7 cells. Cell Signal. 2014, 26, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Ip, W.K.E.; Sokolovska, A.; Charriere, G.M.; Boyer, L.; Dejardin, S.; Cappillino, M.P.; Yantosca, L.M.; Takahashi, K.; Moore, K.J.; Lacy-Hulbert, A.; et al. Phagocytosis and phagosome acidification are required for pathogen processing and MyD88-dependent responses to Staphylococcus aureus. J. Immunol. 2010, 184, 7071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jimenez-Dalmaroni, M.J.; Xiao, N.; Corper, A.L.; Verdino, P.; Ainge, G.D.; Larsen, D.S.; Painter, G.F.; Rudd, P.M.; Dwek, R.A.; Hoebe, K.; et al. Soluble CD36 ectodomain binds negatively charged diacylglycerol ligands and acts as a co-receptor for TLR2. PLoS ONE 2009, 4, e7411. [Google Scholar] [CrossRef]
- Israel, L.; Wang, Y.; Bulek, K.; Della Mina, E.; Zhang, Z.; Pedergnana, V.; Chrabieh, M.; Lemmens, N.A.; Sancho-Shimizu, V.; Descatoire, M.; et al. Human Adaptive Immunity Rescues an Inborn Error of Innate Immunity. Cell 2017, 168, 789–800. [Google Scholar] [CrossRef] [Green Version]
- Kanazawa, N. C-Type Lectin Receptors. In Immunology of the Skin: Basic and Clinical Sciences in Skin Immune Responses; Kabashima, K., Ed.; Springer: Tokyo, Japan, 2016; pp. 255–274. [Google Scholar] [CrossRef]
- De Monte, A.; Olivieri, C.-V.; Vitale, S.; Bailleux, S.; Castillo, L.; Giordanengo, V.; Maryanski, J.L.; Segura, E.; Doglio, A. CD1c-related DCs that express CD207/Langerin, but are distinguishable from langerhans cells, are consistently present in human tonsils. Front. Immunol. 2016, 7, 197. [Google Scholar] [CrossRef] [Green Version]
- van Dalen, R.; De La Cruz Diaz Jacinto, S.; Rumpret, M.; Fuchsberger Felix, F.; van Teijlingen Nienke, H.; Hanske, J.; Rademacher, C.; Geijtenbeek Teunis, B.H.; van Strijp Jos, A.G.; Weidenmaier, C.; et al. Langerhans cells sense Staphylococcus aureus wall teichoic acid through langerin to induce inflammatory responses. mBio 2019, 10, e00330-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mnich, M.E.; van Dalen, R.; Gerlach, D.; Hendriks, A.; Xia, G.; Peschel, A.; van Strijp, J.A.G.; van Sorge, N.M. The C-type lectin receptor MGL senses N-acetylgalactosamine on the unique Staphylococcus aureus ST395 wall teichoic acid. Cell. Microbiol. 2019, 21, e13072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurokawa, K.; Jung, D.-J.; An, J.-H.; Fuchs, K.; Jeon, Y.-J.; Kim, N.-H.; Li, X.; Tateishi, K.; Park, J.A.; Xia, G.; et al. Glycoepitopes of Staphylococcal wall teichoic acid govern complement-mediated opsonophagocytosis via human serum antibody and mannose-binding lectin. J. Biol. Chem. 2013, 288, 30956–30968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Dalen, R.; Peschel, A.; van Sorge, N.M. Wall teichoic acid in Staphylococcus aureus host interaction. Trends Microbiol. 2020, 28, 985–998. [Google Scholar] [CrossRef] [PubMed]
- Chong, Y.P.; Park, K.H.; Kim, E.S.; Kim, M.N.; Kim, S.H.; Lee, S.O.; Choi, S.H.; Jeong, J.Y.; Woo, J.H.; Kim, Y.S. Association of mannose-binding lectin 2 gene polymorphisms with persistent Staphylococcus aureus bacteremia. PLoS ONE 2014, 9, e89139. [Google Scholar] [CrossRef] [PubMed]
- Park, K.-H.; Kurokawa, K.; Zheng, L.; Jung, D.-J.; Tateishi, K.; Jin, J.-O.; Ha, N.-C.; Kang, H.J.; Matsushita, M.; Kwak, J.-Y.; et al. Human serum mannose-binding lectin senses wall teichoic acid glycopolymer of Staphylococcus aureus, which is restricted in infancy. J. Biol. Chem. 2010, 285, 27167–27175. [Google Scholar] [CrossRef] [Green Version]
- Mnich, M.E.; van Dalen, R.; van Sorge, N.M. C-Type lectin receptors in host defense against bacterial pathogens. Front. Cell. Infect. Microbiol. 2020, 10, 309. [Google Scholar] [CrossRef]
- Motta, V.; Soares, F.; Sun, T.; Philpott, D.J. NOD-like receptors: Versatile cytosolic sentinels. Physiol. Rev. 2015, 95, 149–178. [Google Scholar] [CrossRef] [Green Version]
- Holzinger, D.; Gieldon, L.; Mysore, V.; Nippe, N.; Taxman, D.J.; Duncan, J.A.; Broglie, P.M.; Marketon, K.; Austermann, J.; Vogl, T.; et al. Staphylococcus aureus panton-valentine leukocidin induces an inflammatory response in human phagocytes via the NLRP3 inflammasome. J. Leukoc. Biol. 2012, 92, 1069–1081. [Google Scholar] [CrossRef] [Green Version]
- Melehani, J.H.; James, D.B.; DuMont, A.L.; Torres, V.J.; Duncan, J.A. Staphylococcus aureus Leukocidin A/B (LukAB) kills human monocytes via host NLRP3 and ASC when extracellular, but not intracellular. PLoS Pathog. 2015, 11, e1004970. [Google Scholar] [CrossRef]
- Shimada, T.; Park, B.G.; Wolf, A.J.; Brikos, C.; Goodridge, H.S.; Becker, C.A.; Reyes, C.N.; Miao, E.A.; Aderem, A.; Götz, F.; et al. Staphylococcus aureus evades lysozyme-based peptidoglycan digestion that links phagocytosis, inflammasome activation, and IL-1beta secretion. Cell Host Microbe 2010, 7, 38–49. [Google Scholar] [CrossRef] [Green Version]
- Muñoz-Planillo, R.; Franchi, L.; Miller, L.S.; Núñez, G. A critical role for hemolysins and bacterial lipoproteins in Staphylococcus aureus-induced activation of the Nlrp3 inflammasome. J. Immunol. 2009, 183, 3942–3948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Craven, R.R.; Gao, X.; Allen, I.C.; Gris, D.; Bubeck Wardenburg, J.; McElvania-Tekippe, E.; Ting, J.P.; Duncan, J.A. Staphylococcus aureus alpha-hemolysin activates the NLRP3-inflammasome in human and mouse monocytic cells. PLoS ONE 2009, 4, e7446. [Google Scholar] [CrossRef] [PubMed]
- Khare, S.; Dorfleutner, A.; Bryan, N.B.; Yun, C.; Radian, A.D.; de Almeida, L.; Rojanasakul, Y.; Stehlik, C. An NLRP7-containing inflammasome mediates recognition of microbial lipopeptides in human macrophages. Immunity 2012, 36, 464–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, M.R.; Kaminski, J.J.; Kurt-Jones, E.A.; Fitzgerald, K.A. Pattern recognition receptors and the innate immune response to viral infection. Viruses 2011, 3, 920. [Google Scholar] [CrossRef] [Green Version]
- Yoneyama, M.; Kikuchi, M.; Natsukawa, T.; Shinobu, N.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Akira, S.; Fujita, T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004, 5, 730–737. [Google Scholar] [CrossRef]
- Saito, T.; Hirai, R.; Loo, Y.M.; Owen, D.; Johnson, C.L.; Sinha, S.C.; Akira, S.; Fujita, T.; Gale, M., Jr. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. USA 2007, 104, 582–587. [Google Scholar] [CrossRef] [Green Version]
- Yoneyama, M.; Kikuchi, M.; Matsumoto, K.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Foy, E.; Loo, Y.M.; Gale, M., Jr.; Akira, S.; et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 2005, 175, 2851–2858. [Google Scholar] [CrossRef] [Green Version]
- Komuro, A.; Horvath, C.M. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J. Virol. 2006, 80, 12332–12342. [Google Scholar] [CrossRef] [Green Version]
- Venkataraman, T.; Valdes, M.; Elsby, R.; Kakuta, S.; Caceres, G.; Saijo, S.; Iwakura, Y.; Barber, G.N. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J. Immunol. 2007, 178, 6444–6455. [Google Scholar] [CrossRef]
- Rothenfusser, S.; Goutagny, N.; DiPerna, G.; Gong, M.; Monks, B.G.; Schoenemeyer, A.; Yamamoto, M.; Akira, S.; Fitzgerald, K.A. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 2005, 175, 5260–5268. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, O.; Akira, S. Innate immunity to virus infection. Immunol. Rev. 2009, 227, 75–86. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A.; Bowie, A.G. Sensing and signaling in antiviral innate immunity. Curr. Biol. 2010, 20, R328–R333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schroder, M.; Baran, M.; Bowie, A.G. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J. 2008, 27, 2147–2157. [Google Scholar] [CrossRef] [PubMed]
- Michallet, M.C.; Meylan, E.; Ermolaeva, M.A.; Vazquez, J.; Rebsamen, M.; Curran, J.; Poeck, H.; Bscheider, M.; Hartmann, G.; Konig, M.; et al. TRADD protein is an essential component of the RIG-like helicase antiviral pathway. Immunity 2008, 28, 651–661. [Google Scholar] [CrossRef]
- Matsumiya, T.; Stafforini, D.M. Function and regulation of retinoic acid-inducible gene-I. Crit. Rev. Immunol. 2010, 30, 489–513. [Google Scholar] [CrossRef] [Green Version]
- Rehwinkel, J.; Gack, M.U. RIG-I-like receptors: Their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020, 20, 537–551. [Google Scholar] [CrossRef]
- Chiu, Y.-H.; MacMillan, J.B.; Chen, Z.J. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 2009, 138, 576–591. [Google Scholar] [CrossRef] [Green Version]
- Monroe, K.M.; McWhirter, S.M.; Vance, R.E. Identification of host cytosolic sensors and bacterial factors regulating the Type I interferon response to Legionella pneumophila. PLoS Pathog. 2009, 5, e1000665. [Google Scholar] [CrossRef]
- Schmolke, M.; Patel, J.R.; Castro, E.d.; Sánchez-Aparicio, M.T.; Uccellini, M.B.; Miller, J.C.; Manicassamy, B.; Satoh, T.; Kawai, T.; Akira, S.; et al. RIG-I detects mRNA of intracellular Salmonella enterica serovar Typhimurium during bacterial infection. mBio 2014, 5, e01006-14. [Google Scholar] [CrossRef] [Green Version]
- Abdullah, Z.; Schlee, M.; Roth, S.; Mraheil, M.A.; Barchet, W.; Böttcher, J.; Hain, T.; Geiger, S.; Hayakawa, Y.; Fritz, J.H. RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids. EMBO J. 2012, 31, 4153–4164. [Google Scholar] [CrossRef] [Green Version]
- Hagmann, C.A.; Herzner, A.M.; Abdullah, Z.; Zillinger, T.; Jakobs, C.; Schuberth, C.; Coch, C.; Higgins, P.G.; Wisplinghoff, H.; Barchet, W.; et al. RIG-I detects triphosphorylated RNA of Listeria monocytogenes during infection in non-immune cells. PLoS ONE 2013, 8, e62872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asadpour-Behzadi, A.; Kariminik, A. RIG-1 and MDA5 are the important intracellular sensors against bacteria in septicemia suffering patients. J. Appl. Biomed. 2018, 16, 358–361. [Google Scholar] [CrossRef]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013, 339, 786–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.D.; Wu, J.; Gao, D.; Wang, H.; Sun, L.; Chen, Z.J. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 2013, 341, 1390–1394. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Sun, L.; Chen, Z.J. Regulation and function of the cGAS–STING pathway of cytosolic DNA sensing. Nat. Immunol. 2016, 17, 1142. [Google Scholar] [CrossRef]
- Wu, J.; Sun, L.; Chen, X.; Du, F.; Shi, H.; Chen, C.; Chen, Z.J. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 2013, 339, 826–830. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.-Z.; Yang, Y.-J.; Zhou, C.-K.; Yan, S.-Q.; Ma, K.; Gao, Y.; Chen, W. STING contributes to host defense against Staphylococcus aureus pneumonia through suppressing necroptosis. Front. Immunol. 2021, 12, 2076. [Google Scholar] [CrossRef]
- Kaplan, A.; Ma, J.; Kyme, P.; Wolf, A.J.; Becker, C.A.; Tseng, C.W.; Liu, G.Y.; Underhill, D.M. Failure to induce IFN-β production during Staphylococcus aureus infection contributes to pathogenicity. J. Immunol. 2012, 189, 4537. [Google Scholar] [CrossRef] [Green Version]
- Mistry, R.D. Skin and soft tissue infections. Pediatr. Clin. N. Am. 2013, 60, 1063–1082. [Google Scholar] [CrossRef]
- Pasparakis, M.; Haase, I.; Nestle, F.O. Mechanisms regulating skin immunity and inflammation. Nat. Rev. Immunol. 2014, 14, 289–301. [Google Scholar] [CrossRef]
- Miller, L.S. Toll-like receptors in skin. Adv. Dermatol. 2008, 24, 71–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soong, G.; Paulino, F.; Wachtel, S.; Parker, D.; Wickersham, M.; Zhang, D.; Brown, A.; Lauren, C.; Dowd, M.; West, E.; et al. Methicillin-resistant Staphylococcus aureus adaptation to human keratinocytes. mBio 2015, 6, e00289-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, M.T.; Kraft, B.; Yu, W.; Demicrioglu, D.D.; Hertlein, T.; Burian, M.; Schmaler, M.; Boller, K.; Bekeredjian-Ding, I.; Ohlsen, K.; et al. The νSaα specific Lipoprotein Like Cluster (lpl) of S. aureus USA300 contributes to immune stimulation and invasion in human ells. PLoS Pathog. 2015, 11, e1004984. [Google Scholar] [CrossRef] [Green Version]
- Kim, N.J.; Ahn, K.B.; Jeon, J.H.; Yun, C.-H.; Finlay, B.B.; Han, S.H. Lipoprotein in the cell wall of Staphylococcus aureus is a major inducer of nitric oxide production in murine macrophages. Mol. Immunol. 2015, 65, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Nandi, A.; Dey, S.; Biswas, J.; Jaiswal, P.; Naaz, S.; Yasmin, T.; Bishayi, B. Differential induction of inflammatory cytokines and reactive oxygen species in murine peritoneal macrophages and resident fresh bone marrow cells by acute staphylococcus aureus infection: Contribution of toll-like receptor 2 (TLR2). Inflammation 2015, 38, 224–244. [Google Scholar] [CrossRef]
- Bekeredjian-Ding, I.; Stein, C.; Uebele, J. The innate immune response against Staphylococcus aureus. In Staphylococcus aureus: Microbiology, Pathology, Immunology, Therapy and Prophylaxis; Bagnoli, F., Rappuoli, R., Grandi, G., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 385–418. [Google Scholar] [CrossRef]
- Kim, B.; Jiang, T.; Bae, J.H.; Yun, H.S.; Jang, S.H.; Kim, J.H.; Kim, J.D.; Hur, J.H.; Shibata, K.; Kurokawa, K.; et al. In Staphylococcus aureus, the particulate state of the cell envelope is required for the efficient induction of host defense responses. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef] [PubMed]
- Surewaard, B.G.; Deniset, J.F.; Zemp, F.J.; Amrein, M.; Otto, M.; Conly, J.; Omri, A.; Yates, R.M.; Kubes, P. Identification and treatment of the Staphylococcus aureus reservoir in vivo. J. Exp. Med. 2016, 213, 1141–1151. [Google Scholar] [CrossRef] [Green Version]
- Kitur, K.; Parker, D.; Nieto, P.; Ahn, D.S.; Cohen, T.S.; Chung, S.; Wachtel, S.; Bueno, S.; Prince, A. Toxin-induced necroptosis is a major mechanism of Staphylococcus aureus lung damage. PLoS Pathog. 2015, 11, e1004820. [Google Scholar] [CrossRef]
- Martin, F.J.; Parker, D.; Harfenist, B.S.; Soong, G.; Prince, A. Participation of CD11c(+) leukocytes in methicillin-resistant Staphylococcus aureus clearance from the lung. Infect. Immun. 2011, 79, 1898–1904. [Google Scholar] [CrossRef] [Green Version]
- Prajsnar, T.K.; Cunliffe, V.T.; Foster, S.J.; Renshaw, S.A. A novel vertebrate model of Staphylococcus aureus infection reveals phagocyte-dependent resistance of zebrafish to non-host specialized pathogens. Cell. Microbiol. 2008, 10, 2312–2325. [Google Scholar] [CrossRef]
- Nacken, W.; Roth, J.; Sorg, C.; Kerkhoff, C. S100A9/S100A8: Myeloid representatives of the S100 protein family as prominent players in innate immunity. Microsc. Res. Tech. 2003, 60, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Corbin, B.D.; Seeley, E.H.; Raab, A.; Feldmann, J.; Miller, M.R.; Torres, V.J.; Anderson, K.L.; Dattilo, B.M.; Dunman, P.M.; Gerads, R.; et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 2008, 319, 962–965. [Google Scholar] [CrossRef] [PubMed]
- Damo, S.M.; Kehl-Fie, T.E.; Sugitani, N.; Holt, M.E.; Rathi, S.; Murphy, W.J.; Zhang, Y.; Betz, C.; Hench, L.; Fritz, G.; et al. Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc. Natl. Acad. Sci. USA 2013, 110, 3841–3846. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Eagen, W.J.; Lee, J.C. Orchestration of human macrophage NLRP3 inflammasome activation by Staphylococcus aureus extracellular vesicles. Proc. Natl. Acad. Sci. USA 2020, 117, 3174. [Google Scholar] [CrossRef]
- Yao, R.; Chen, Y.; Hao, H.; Guo, Z.; Cheng, X.; Ma, Y.; Ji, Q.; Yang, X.; Wang, Y.; Li, X.; et al. Pathogenic effects of inhibition of mTORC1/STAT3 axis facilitates Staphylococcus aureus-induced pyroptosis in human macrophages. Cell Commun. Signal. 2020, 18, 187. [Google Scholar] [CrossRef]
- Kubica, M.; Guzik, K.; Koziel, J.; Zarebski, M.; Richter, W.; Gajkowska, B.; Golda, A.; Maciag-Gudowska, A.; Brix, K.; Shaw, L.; et al. A potential new pathway for Staphylococcus aureus dissemination: The silent survival of S. aureus phagocytosed by human monocyte-derived macrophages. PLoS ONE 2008, 3, e1409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Barca, E.; Carratalà, J.; Mykietiuk, A.; Fernández-Sevilla, A.; Gudiol, F. Predisposing factors and outcome of Staphylococcus aureus bacteremia in neutropenic patients with cancer. Eur. J. Clin. Microbiol. Infect. Dis 2001, 20, 117–119. [Google Scholar] [CrossRef]
- Bouma, G.; Ancliff, P.J.; Thrasher, A.J.; Burns, S.O. Recent advances in the understanding of genetic defects of neutrophil number and function. Br. J. Haematol. 2010, 151, 312–326. [Google Scholar] [CrossRef]
- Lakshman, R.; Finn, A. Neutrophil disorders and their management. J. Clin. Pathol. 2001, 54, 7–19. [Google Scholar] [CrossRef]
- Miller, L.S.; Cho, J.S. Immunity against Staphylococcus aureus cutaneous infections. Nat. Rev. Immunol. 2011, 11, 505–518. [Google Scholar] [CrossRef] [Green Version]
- El-Benna, J.; Dang, P.M.; Gougerot-Pocidalo, M.A. Priming of the neutrophil NADPH oxidase activation: Role of p47phox phosphorylation and NOX2 mobilization to the plasma membrane. Semin. Immunopathol. 2008, 30, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Clarke, T.B.; Davis, K.M.; Lysenko, E.S.; Zhou, A.Y.; Yu, Y.; Weiser, J.N. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 2010, 16, 228–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellis, T.N.; Beaman, B.L. Interferon-gamma activation of polymorphonuclear neutrophil function. Immunology 2004, 112, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Teng, T.-S.; Ji, A.-l.; Ji, X.-Y.; Li, Y.-Z. Neutrophils and immunity: From bactericidal action to being conquered. J. Immunol. Res. 2017, 2017, 9671604. [Google Scholar] [CrossRef]
- Urban, C.F.; Lourido, S.; Zychlinsky, A. How do microbes evade neutrophil killing? Cell. Microbiol. 2006, 8, 1687–1696. [Google Scholar] [CrossRef]
- Pilsczek, F.H.; Salina, D.; Poon, K.K.H.; Fahey, C.; Yipp, B.G.; Sibley, C.D.; Robbins, S.M.; Green, F.H.Y.; Surette, M.G.; Sugai, M.; et al. A novel mechanism of rapid nuclear neutrophil extracellular trap formation in response to Staphylococcus aureus. J. Immunol. 2010, 185, 7413. [Google Scholar] [CrossRef] [Green Version]
- van Kessel, K.P.M.; Bestebroer, J.; van Strijp, J.A.G. Neutrophil-mediated phagocytosis of Staphylococcus aureus. Front. Immunol. 2014, 5, 467. [Google Scholar] [CrossRef] [Green Version]
- Günther, F.; Wabnitz, G.H.; Stroh, P.; Prior, B.; Obst, U.; Samstag, Y.; Wagner, C.; Hänsch, G.M. Host defence against Staphylococcus aureus biofilms infection: Phagocytosis of biofilms by polymorphonuclear neutrophils (PMN). Mol. Immunol. 2009, 46, 1805–1813. [Google Scholar] [CrossRef]
- Rowley, D.A.; Fitch, F.W. The road to the discovery of dendritic cells, a tribute to Ralph Steinman. Cell. Immunol. 2012, 273, 95–98. [Google Scholar] [CrossRef]
- Liu, K. Dendritic cells. Encyclopedia Cell Biol. 2016, 3, 741–749. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, X.; Cheng, Y.; Cao, X. Dendritic cell migration in inflammation and immunity. Cell. Mol. Immunol. 2021, 18, 2461–2471. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, D.; Vollmann, E.H.; von Andrian, U.H. Mechanisms and consequences of dendritic cell migration. Immunity 2008, 29, 325–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solano-Gálvez, G.S.; Tovar-Torres, M.S.; Tron-Gómez, S.M.; Weiser-Smeke, E.A.; Álvarez-Hernández, A.D.; Franyuti-Kelly, A.G.; Tapia-Moreno, M.; Ibarra, A.; Gutiérrez-Kobeh, L.; Vázquez-López, R. Human dendritic cells: Ontogeny and their subsets in health and disease. Med. Sci. 2018, 6, 88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Zhu, Y.; Zhang, G.; Gao, C.; Zhong, W.; Zhang, X. CD83-stimulated monocytes suppress T-cell immune responses through production of prostaglandin E2. Proc. Natl. Acad. Sci. USA 2011, 108, 18778–18783. [Google Scholar] [CrossRef] [Green Version]
- Banchereau, J.; Briere, F.; Caux, C.; Davoust, J.; Lebecque, S.; Liu, Y.J.; Pulendran, B.; Palucka, K. Immunobiology of dendritic cells. Annu. Rev. Immunol. 2000, 18, 767–811. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, K.; Iyonaga, K.; Ichiyasu, H.; Nagano, J.; Suga, M.; Sasaki, Y. Differentiation, maturation, and survival of dendritic cells by osteopontin regulation. Clin. Diagn. Lab. Immunol. 2005, 12, 206–212. [Google Scholar] [CrossRef] [Green Version]
- de Saint-Vis, B.; Vincent, J.; Vandenabeele, S.; Vanbervliet, B.; Pin, J.J.; Aït-Yahia, S.; Patel, S.; Mattei, M.G.; Banchereau, J.; Zurawski, S.; et al. A novel lysosome-associated membrane glycoprotein, DC-LAMP, induced upon DC maturation, is transiently expressed in MHC Class II compartment. Immunity 1998, 9, 325–336. [Google Scholar] [CrossRef] [Green Version]
- Heath, W.R.; Carbone, F.R. Dendritic cell subsets in primary and secondary T cell responses at body surfaces. Nat. Immunol. 2009, 10, 1237–1244. [Google Scholar] [CrossRef]
- Macri, C.; Pang, E.S.; Patton, T.; O’Keeffe, M. Dendritic cell subsets. Semin. Cell Dev. Biol. 2018, 84, 11–21. [Google Scholar] [CrossRef]
- Hespel, C.; Moser, M. Role of inflammatory dendritic cells in innate and adaptive immunity. Eur. J. Immunol. 2012, 42, 2535–2543. [Google Scholar] [CrossRef]
- McKenna, K.; Beignon, A.-S.; Bhardwaj, N. Plasmacytoid dendritic cells: Linking innate and adaptive immunity. J. Virol. 2005, 79, 17–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darisipudi, M.N.; Nordengrün, M.; Bröker, B.M.; Péton, V. Messing with the sentinels-The interaction of Staphylococcus aureus with dendritic Cells. Microorganisms 2018, 6, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pidwill, G.R.; Gibson, J.F.; Cole, J.; Renshaw, S.A.; Foster, S.J. The Role of Macrophages in Staphylococcus aureus Infection. Front. Immunol. 2021, 11, 339. [Google Scholar] [CrossRef] [PubMed]
- Nishat, S.; Wuescher, L.M.; Worth, R.G. Platelets enhance dendritic cell responses against Staphylococcus aureus through CD40-CD40L. Infect. Immun. 2018, 86, e00186-18. [Google Scholar] [CrossRef] [Green Version]
- O’Keeffe, K.M.; Wilk, M.M.; Leech, J.M.; Murphy, A.G.; Laabei, M.; Monk, I.R.; Massey, R.C.; Lindsay, J.A.; Foster, T.J.; Geoghegan, J.A.; et al. Manipulation of autophagy in phagocytes facilitates Staphylococcus aureus bloodstream infection. Infect. Immun. 2015, 83, 3445–3457. [Google Scholar] [CrossRef] [Green Version]
- Uebele, J.; Stein, C.; Nguyen, M.-T.; Schneider, A.; Kleinert, F.; Tichá, O.; Bierbaum, G.; Götz, F.; Bekeredjian-Ding, I. Antigen delivery to dendritic cells shapes human CD4+ and CD8+ T cell memory responses to Staphylococcus aureus. PLoS Pathog. 2017, 13, e1006387. [Google Scholar] [CrossRef]
- Jin, J.O.; Zhang, W.; Du, J.Y.; Yu, Q. BDCA1-positive dendritic cells (DCs) represent a unique human myeloid DC subset that induces innate and adaptive immune responses to Staphylococcus aureus Infection. Infect. Immun. 2014, 82, 4466–4476. [Google Scholar] [CrossRef] [Green Version]
- Parcina, M.; Wendt, C.; Goetz, F.; Zawatzky, R.; Zähringer, U.; Heeg, K.; Bekeredjian-Ding, I. Staphylococcus aureus induced plasmacytoid dendritic cell activation is based on an IgG-mediated memory response. J. Immunol. 2008, 181, 3823. [Google Scholar] [CrossRef] [Green Version]
- Michea, P.; Vargas, P.; Donnadieu, M.-H.; Rosemblatt, M.; Bono, M.; Dumenil, G.; Soumelis, V. Epithelial control of the human pDC response to extracellular bacteria. Eur. J. Immunol. 2013, 43. [Google Scholar] [CrossRef]
- Schindler, D.; Gutierrez, M.G.; Beineke, A.; Rauter, Y.; Rohde, M.; Foster, S.; Goldmann, O.; Medina, E. Dendritic cells are central coordinators of the host immune response to Staphylococcus aureus bloodstream infection. Am. J. Pathol. 2012, 181, 1327–1337. [Google Scholar] [CrossRef]
- Compton, L.A.; Murphy, G.F.; Lian, C.G. Diagnostic immunohistochemistry in cutaneous neoplasia: An update. Dermatopathology 2015, 2, 15–42. [Google Scholar] [CrossRef] [PubMed]
- Richardson, J.R.; Armbruster, N.S.; Gunter, M.; Biljecki, M.; Klenk, J.; Heumos, S.; Autenrieth, S.E. PSM Peptides From Community-Associated Methicillin-Resistant Staphylococcus aureus Impair the Adaptive Immune Response via Modulation of Dendritic Cell Subsets in vivo. Front. Immunol. 2019, 10, 995. [Google Scholar] [CrossRef] [PubMed]
- Berends Evelien, T.M.; Zheng, X.; Zwack Erin, E.; Ménager Mickaël, M.; Cammer, M.; Shopsin, B.; Torres Victor, J.; Freitag Nancy, E. Staphylococcus aureus impairs the function of and kills human dendritic cells via the LukAB toxin. mBio 2019, 10, e01918-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldmann, O.; Medina, E. Staphylococcus aureus strategies to evade the host acquired immune response. Int. J. Med. Microbiol. 2018, 308, 625–630. [Google Scholar] [CrossRef]
- Zhao, Y.; van Kessel, K.P.M.; de Haas, C.J.C.; Rogers, M.R.C.; van Strijp, J.A.G.; Haas, P.-J.A. Staphylococcal superantigen-like protein 13 activates neutrophils via formyl peptide receptor 2. Cell microbiol 2018, 20, e12941. [Google Scholar] [CrossRef] [Green Version]
- Bestebroer, J.; Poppelier, M.J.; Ulfman, L.H.; Lenting, P.J.; Denis, C.V.; van Kessel, K.P.; van Strijp, J.A.; de Haas, C.J. Staphylococcal superantigen-like 5 binds PSGL-1 and inhibits P-selectin-mediated neutrophil rolling. Blood 2007, 109, 2936–2943. [Google Scholar] [CrossRef]
- Koymans, K.J.; Goldmann, O.; Karlsson, C.A.Q.; Sital, W.; Thanert, R.; Bisschop, A.; Vrieling, M.; Malmstrom, J.; van Kessel, K.P.M.; de Haas, C.J.C.; et al. The TLR2 Antagonist Staphylococcal Superantigen-Like Protein 3 Acts as a Virulence Factor to Promote Bacterial Pathogenicity in vivo. J. Innate Immun. 2017, 9, 561–573. [Google Scholar] [CrossRef]
- Walenkamp, A.M.; Boer, I.G.; Bestebroer, J.; Rozeveld, D.; Timmer-Bosscha, H.; Hemrika, W.; van Strijp, J.A.; de Haas, C.J. Staphylococcal superantigen-like 10 inhibits CXCL12-induced human tumor cell migration. Neoplasia 2009, 11, 333–344. [Google Scholar] [CrossRef] [Green Version]
- Bardoel, B.W.; Vos, R.; Bouman, T.; Aerts, P.C.; Bestebroer, J.; Huizinga, E.G.; Brondijk, T.H.; van Strijp, J.A.; de Haas, C.J. Evasion of Toll-like receptor 2 activation by staphylococcal superantigen-like protein 3. J. Mol. Med. 2012, 90, 1109–1120. [Google Scholar] [CrossRef]
- Koymans, K.J.; Feitsma, L.J.; Brondijk, T.H.; Aerts, P.C.; Lukkien, E.; Lossl, P.; van Kessel, K.P.; de Haas, C.J.; van Strijp, J.A.; Huizinga, E.G. Structural basis for inhibition of TLR2 by staphylococcal superantigen-like protein 3 (SSL3). Proc. Natl. Acad. Sci. USA 2015, 112, 11018–11023. [Google Scholar] [CrossRef] [Green Version]
- Yokoyama, R.; Itoh, S.; Kamoshida, G.; Takii, T.; Fujii, S.; Tsuji, T.; Onozaki, K. Staphylococcal superantigen-like protein 3 binds to the Toll-like receptor 2 extracellular domain and inhibits cytokine production induced by Staphylococcus aureus, cell wall component, or lipopeptides in murine macrophages. Infect. Immun. 2012, 80, 2816–2825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, K.; Akira, S. TLR signaling pathways. Semin. Immunol. 2004, 16, 3–9. [Google Scholar] [CrossRef]
- Xu, Y.; Tao, X.; Shen, B.; Horng, T.; Medzhitov, R.; Manley, J.L.; Tong, L. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature 2000, 408, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Askarian, F.; van Sorge, N.M.; Sangvik, M.; Beasley, F.C.; Henriksen, J.R.; Sollid, J.U.; van Strijp, J.A.; Nizet, V.; Johannessen, M. A Staphylococcus aureus TIR domain protein virulence factor blocks TLR2-mediated NF-κB signaling. J. Innate Immun. 2014, 6, 485–498. [Google Scholar] [CrossRef]
- Schiller, M.; Metze, D.; Luger, T.A.; Grabbe, S.; Gunzer, M. Immune response modifiers—Mode of action. Exp. Dermatol. 2006, 15, 331–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Askarian, F.; Wagner, T.; Johannessen, M.; Nizet, V. Staphylococcus aureus modulation of innate immune responses through Toll-like (TLR), (NOD)-like (NLR) and C-type lectin (CLR) receptors. FEMS Microbiol. Rev. 2018, 42, 656–671. [Google Scholar] [CrossRef] [PubMed]
- Patot, S.; Imbert, P.R.C.; Baude, J.; Martins Simões, P.; Campergue, J.-B.; Louche, A.; Nijland, R.; Bès, M.; Tristan, A.; Laurent, F.; et al. Correction: The TIR homologue lies near resistance genes in Staphylococcus aureus, coupling modulation of virulence and antimicrobial susceptibility. PLoS Pathog. 2017, 13, e1006291. [Google Scholar] [CrossRef]
- Kitakaze, M.; Hori, M.; Kamada, T. Role of adenosine and its interaction with alpha adrenoceptor activity in ischaemic and reperfusion injury of the myocardium. Cardiovasc. Res. 1993, 27, 18–27. [Google Scholar] [CrossRef]
- Kaufmann, I.; Hoelzl, A.; Schliephake, F.; Hummel, T.; Chouker, A.; Lysenko, L.; Peter, K.; Thiel, M. Effects of adenosine on functions of polymorphonuclear leukocytes from patients with septic shock. Shock 2007, 27, 25–31. [Google Scholar] [CrossRef]
- Csoka, B.; Nemeth, Z.H.; Virag, L.; Gergely, P.; Leibovich, S.J.; Pacher, P.; Sun, C.X.; Blackburn, M.R.; Vizi, E.S.; Deitch, E.A.; et al. A2A adenosine receptors and C/EBPbeta are crucially required for IL-10 production by macrophages exposed to Escherichia coli. Blood 2007, 110, 2685–2695. [Google Scholar] [CrossRef] [Green Version]
- Edwards, C.K., 3rd; Watts, L.M.; Parmely, M.J.; Linnik, M.D.; Long, R.E.; Borcherding, D.R. Effect of the carbocyclic nucleoside analogue MDL 201,112 on inhibition of interferon-gamma-induced priming of Lewis (LEW/N) rat macrophages for enhanced respiratory burst and MHC class II Ia+ antigen expression. J. Leukoc. Biol. 1994, 56, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Panther, E.; Idzko, M.; Herouy, Y.; Rheinen, H.; Gebicke-Haerter, P.J.; Mrowietz, U.; Dichmann, S.; Norgauer, J. Expression and function of adenosine receptors in human dendritic cells. FASEB J. 2001, 15, 1963–1970. [Google Scholar] [CrossRef] [PubMed]
- HaskÓ, G.; Kuhel, D.G.; Chen, J.-F.; Schwarzschild, M.A.; Deitch, E.A.; Mabley, J.G.; Marton, A.; SzabÓ, C. Adenosine inhibits IL-12 and TNF-α production via adenosine A2a receptor-dependent and independent mechanisms. FASEB J. 2000, 14, 2065–2074. [Google Scholar] [CrossRef] [Green Version]
- Savina, A.; Amigorena, S. Phagocytosis and antigen presentation in dendritic cells. Immunol. Rev. 2007, 219, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Bhinderwala, F.; Lehman, M.K.; Thomas, V.C.; Chaudhari, S.S.; Yamada, K.J.; Foster, K.W.; Powers, R.; Kielian, T.; Fey, P.D. Urease is an essential component of the acid response network of Staphylococcus aureus and is required for a persistent murine kidney infection. PLoS Pathog. 2019, 15, e1007538. [Google Scholar] [CrossRef] [PubMed]
- Collins, L.V.; Kristian, S.A.; Weidenmaier, C.; Faigle, M.; Van Kessel, K.P.; Van Strijp, J.A.; Götz, F.; Neumeister, B.; Peschel, A. Staphylococcus aureus strains lacking D-alanine modifications of teichoic acids are highly susceptible to human neutrophil killing and are virulence attenuated in mice. J. Infect. Dis. 2002, 186, 214–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hampton, M.B.; Winterbourn, C.C. Modification of neutrophil oxidant production with diphenyleneiodonium and its effect on bacterial killing. Free Radic. Biol. Med. 1995, 18, 633–639. [Google Scholar] [CrossRef]
- Monteith, A.J.; Miller, J.M.; Maxwell, C.N.; Chazin, W.J.; Skaar, E.P. Neutrophil extracellular traps enhance macrophage killing of bacterial pathogens. Sci. Adv. 2021, 7, eabj2101. [Google Scholar] [CrossRef]
- Ballal, A.; Manna, A.C. Regulation of superoxide dismutase (sod) genes by SarA in Staphylococcus aureus. J. Bacteriol. 2009, 191, 3301–3310. [Google Scholar] [CrossRef] [Green Version]
- Cosgrove, K.; Coutts, G.; Jonsson, I.M.; Tarkowski, A.; Kokai-Kun, J.F.; Mond, J.J.; Foster, S.J. Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J. Bacteriol. 2007, 189, 1025–1035. [Google Scholar] [CrossRef] [Green Version]
- Schnaith, A.; Kashkar, H.; Leggio, S.A.; Addicks, K.; Krönke, M.; Krut, O. Staphylococcus aureus subvert autophagy for induction of caspase-independent host cell death. J. Biol. Chem. 2007, 282, 2695–2706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neumann, Y.; Bruns, S.A.; Rohde, M.; Prajsnar, T.K.; Foster, S.J.; Schmitz, I. Intracellular Staphylococcus aureus eludes selective autophagy by activating a host cell kinase. Autophagy 2016, 12, 2069–2084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Fan, Z.; Han, H. Autophagy in Staphylococcus aureus infection. Front. Cell. Infect. Microbiol. 2021, 11, 222. [Google Scholar] [CrossRef] [PubMed]
- Jondle, C.N.; Gupta, K.; Mishra, B.B.; Sharma, J. Klebsiella pneumoniae infection of murine neutrophils impairs their efferocytic clearance by modulating cell death machinery. PLoS Pathog. 2018, 14, e1007338. [Google Scholar] [CrossRef] [Green Version]
- Greenlee-Wacker, M.C.; Rigby, K.M.; Kobayashi, S.D.; Porter, A.R.; DeLeo, F.R.; Nauseef, W.M. Phagocytosis of Staphylococcus aureus by human neutrophils prevents macrophage efferocytosis and induces programmed necrosis. J. Immunol. 2014, 192, 4709. [Google Scholar] [CrossRef] [Green Version]
- Cohen, T.S.; Jones-Nelson, O.; Hotz, M.; Cheng, L.; Miller, L.S.; Suzich, J.; Stover, C.K.; Sellman, B.R.S. aureus blocks efferocytosis of neutrophils by macrophages through the activity of its virulence factor alpha toxin. Sci. Rep. 2016, 6, 35466. [Google Scholar] [CrossRef]
- Huitema, L.; Phillips, T.; Alexeev, V.; Tomic-Canic, M.; Pastar, I.; Igoucheva, O. Intracellular escape strategies of Staphylococcus aureus in persistent cutaneous infections. Exp. Dermatol. 2021, 30, 1428–1439. [Google Scholar] [CrossRef]
- Nowicka, D.; Grywalska, E. Staphylococcus aureus and host immunity in recurrent furunculosis. Dermatology 2019, 235, 295–305. [Google Scholar] [CrossRef]
- Bera, A.; Herbert, S.; Jakob, A.; Vollmer, W.; Gotz, F. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 2005, 55, 778–787. [Google Scholar] [CrossRef]
- Pushkaran, A.C.; Nataraj, N.; Nair, N.; Götz, F.; Biswas, R.; Mohan, C.G. Understanding the Structure–Function Relationship of Lysozyme Resistance in Staphylococcus aureus by Peptidoglycan O-Acetylation Using Molecular Docking, Dynamics, and Lysis Assay. J. Chem. Inf. Model. 2015, 55, 760–770. [Google Scholar] [CrossRef]
- Giese, B.; Glowinski, F.; Paprotka, K.; Dittmann, S.; Steiner, T.; Sinha, B.; Fraunholz, M.J. Expression of delta-toxin by Staphylococcus aureus mediates escape from phago-endosomes of human epithelial and endothelial cells in the presence of beta-toxin. Cell. Microbiol. 2011, 13, 316–329. [Google Scholar] [CrossRef] [PubMed]
- Grosz, M.; Kolter, J.; Paprotka, K.; Winkler, A.C.; Schafer, D.; Chatterjee, S.S.; Geiger, T.; Wolz, C.; Ohlsen, K.; Otto, M.; et al. Cytoplasmic replication of Staphylococcus aureus upon phagosomal escape triggered by phenol-soluble modulin alpha. Cell. Microbiol. 2014, 16, 451–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Surewaard, B.G.J.; de Haas, C.J.C.; Vervoort, F.; Rigby, K.M.; DeLeo, F.R.; Otto, M.; van Strijp, J.A.G.; Nijland, R. Staphylococcal alpha-phenol soluble modulins contribute to neutrophil lysis after phagocytosis. Cell. Microbiol. 2013, 15, 1427–1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armbruster, N.S.; Richardson, J.R.; Schreiner, J.; Klenk, J.; Gunter, M.; Kretschmer, D.; Poschel, S.; Schenke-Layland, K.; Kalbacher, H.; Clark, K.; et al. PSM peptides of Staphylococcus aureus activate the p38-CREB pathway in dendritic Cells, thereby modulating cytokine production and T cell priming. J. Immunol. 2016, 196, 1284–1292. [Google Scholar] [CrossRef] [Green Version]
- Armbruster, N.S.; Richardson, J.R.; Schreiner, J.; Klenk, J.; Gunter, M.; Autenrieth, S.E. Staphylococcus aureus PSM peptides induce tolerogenic dendritic cells upon treatment with ligands of extracellular and intracellular TLRs. Int. J. Med. Microbiol. 2016, 306, 666–674. [Google Scholar] [CrossRef]
- Cruciani, M.; Etna, M.P.; Camilli, R.; Giacomini, E.; Percario, Z.A.; Severa, M.; Sandini, S.; Rizzo, F.; Brandi, V.; Balsamo, G.; et al. Staphylococcus aureus Esx factors control human dendritic cell functions conditioning Th1/Th17 response. Front. Cell Infect Microbiol. 2017, 7, 330. [Google Scholar] [CrossRef]
- Miao, E.A.; Leaf, I.A.; Treuting, P.M.; Mao, D.P.; Dors, M.; Sarkar, A.; Warren, S.E.; Wewers, M.D.; Aderem, A. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat. Immunol. 2010, 11, 1136–1142. [Google Scholar] [CrossRef]
- Soe, Y.M.; Bedoui, S.; Stinear, T.P.; Hachani, A. Intracellular Staphylococcus aureus and host cell death pathways. Cell. Microbiol. 2021, 23, e13317. [Google Scholar] [CrossRef]
- Proctor, R.A.; van Langevelde, P.; Kristjansson, M.; Maslow, J.N.; Arbeit, R.D. Persistent and relapsing infections associated with small-colony variants of Staphylococcus aureus. Clin. Infect. Dis. 1995, 20, 95–102. [Google Scholar] [CrossRef]
- Kipp, F.; Ziebuhr, W.; Becker, K.; Krimmer, V.; Höβ, N.; Peters, G.; von Eiff, C. Detection of Staphylococcus aureus by 16S rRNA directed in situ hybridisation in a patient with a brain abscess caused by small colony variants. J. Neurol. Neurosurg Psychiatry 2003, 74, 1000–1002. [Google Scholar] [CrossRef]
- Schröder, A.; Kland, R.; Peschel, A.; von Eiff, C.; Aepfelbacher, M. Live cell imaging of phagosome maturation in Staphylococcus aureus infected human endothelial cells: Small colony variants are able to survive in lysosomes. Med. Microbiol. Immunol. 2006, 195, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Garzoni, C.; Kelley, W.L. Staphylococcus aureus: New evidence for intracellular persistence. Trends Microbiol. 2009, 17, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Fraunholz, M.; Sinha, B. Intracellular Staphylococcus aureus: Live-in and let die. Front. Cell. Infect. Microbiol. 2012, 2, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, M.L. Epidemiology of drug resistance: Implications for a post-antimicrobial era. Science 1992, 257, 1050–1055. [Google Scholar] [CrossRef] [PubMed]
- Dantes, R.; Mu, Y.; Belflower, R.; Aragon, D.; Dumyati, G.; Harrison, L.H.; Lessa, F.C.; Lynfield, R.; Nadle, J.; Petit, S.; et al. National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA Intern. Med. 2013, 173, 1970–1978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeLeo, F.R.; Diep, B.A.; Otto, M. Host defense and pathogenesis in Staphylococcus aureus infections. Infect. Dis. Clin. N. Am. 2009, 23, 17–34. [Google Scholar] [CrossRef] [Green Version]
- Guillemot, D. Antibiotic use in humans and bacterial resistance. Curr. Opin. Microbiol. 1999, 2, 494–498. [Google Scholar] [CrossRef]
- Ito, T.; Okuma, K.; Ma, X.X.; Yuzawa, H.; Hiramatsu, K. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: Genomic island SCC. Drug Resist. Updat. 2003, 6, 41–52. [Google Scholar] [CrossRef]
- Chambers, H.F.; Deleo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef]
- Theuretzbacher, U. Resistance drives antibacterial drug development. Curr. Opin. Pharmacol. 2011, 11, 433–438. [Google Scholar] [CrossRef]
- Peacock, S.J.; Paterson, G.K. Mechanisms of Methicillin Resistance in Staphylococcus aureus. Annu. Rev. Biochem. 2015, 84, 577–601. [Google Scholar] [CrossRef] [PubMed]
- Fuda, C.C.; Fisher, J.F.; Mobashery, S. Beta-lactam resistance in Staphylococcus aureus: The adaptive resistance of a plastic genome. Cell. Mol. Life Sci. 2005, 62, 2617–2633. [Google Scholar] [CrossRef] [PubMed]
- Llarrull, L.I.; Fisher, J.F.; Mobashery, S. Molecular basis and phenotype of methicillin resistance in Staphylococcus aureus and insights into new beta-lactams that meet the challenge. Antimicrob. Agents Chemother. 2009, 53, 4051–4063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stryjewski, M.E.; Corey, G.R. Methicillin-resistant Staphylococcus aureus: An evolving pathogen. Clin. Infect. Dis. 2014, 58, S10–S19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangili, A.; Bica, I.; Snydman, D.R.; Hamer, D.H. Daptomycin-resistant, methicillin-resistant Staphylococcus aureus bacteremia. Clin. Infect. Dis. 2005, 40, 1058–1060. [Google Scholar] [CrossRef] [Green Version]
- Vikram, H.R.; Havill, N.L.; Koeth, L.M.; Boyce, J.M. Clinical progression of methicillin-resistant Staphylococcus aureus vertebral osteomyelitis associated with reduced susceptibility to daptomycin. J. Clin. Microbiol. 2005, 43, 5384–5387. [Google Scholar] [CrossRef] [Green Version]
- Hayden, M.K.; Rezai, K.; Hayes, R.A.; Lolans, K.; Quinn, J.P.; Weinstein, R.A. Development of Daptomycin resistance in vivo in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2005, 43, 5285–5287. [Google Scholar] [CrossRef] [Green Version]
- Tran, T.T.; Munita, J.M.; Arias, C.A. Mechanisms of drug resistance: Daptomycin resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 32–53. [Google Scholar] [CrossRef] [Green Version]
- Peleg, A.Y.; Miyakis, S.; Ward, D.V.; Earl, A.M.; Rubio, A.; Cameron, D.R.; Pillai, S.; Moellering, R.C., Jr.; Eliopoulos, G.M. Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. PLoS ONE 2012, 7, e28316. [Google Scholar] [CrossRef]
- Thitiananpakorn, K.; Aiba, Y.; Tan, X.-E.; Watanabe, S.; Kiga, K.; Sato’o, Y.; Boonsiri, T.; Li, F.-Y.; Sasahara, T.; Taki, Y.; et al. Association of mprF mutations with cross-resistance to daptomycin and vancomycin in methicillin-resistant Staphylococcus aureus (MRSA). Sci. Rep. 2020, 10. [Google Scholar] [CrossRef]
- Bhuiyan, M.S.; Jiang, J.H.; Kostoulias, X.; Theegala, R.; Lieschke, G.J.; Peleg, A.Y. The resistance to host antimicrobial peptides in infections caused by daptomycin-resistant Staphylococcus aureus. Antibiotics 2021, 10, 96. [Google Scholar] [CrossRef] [PubMed]
- Sabat, A.J.; Tinelli, M.; Grundmann, H.; Akkerboom, V.; Monaco, M.; Del Grosso, M.; Errico, G.; Pantosti, A.; Friedrich, A.W. Daptomycin resistant Staphylococcus aureus clinical strain with novel non-synonymous mutations in the mprF and vraS genes: A new insight into daptomycin resistance. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, N.N.; Yang, S.J.; Sawa, A.; Rubio, A.; Nast, C.C.; Yeaman, M.R.; Bayer, A.S. Analysis of cell membrane characteristics of in vitro-selected daptomycin-resistant strains of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2009, 53, 2312–2318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camargo, I.L.; Neoh, H.M.; Cui, L.; Hiramatsu, K. Serial daptomycin selection generates daptomycin-nonsusceptible Staphylococcus aureus strains with a heterogeneous vancomycin-intermediate phenotype. Antimicrob. Agents Chemother. 2008, 52, 4289–4299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, F.-J.; Lauderdale, T.-L.; Lee, C.-H.; Hsu, Y.-C.; Huang, I.W.; Hsu, P.-C.; Yang, C.-S. Effect of a Point Mutation in mprF on Susceptibility to Daptomycin, Vancomycin, and Oxacillin in an MRSA Clinical Strain. Front. Microbiol. 2018, 9, 1086. [Google Scholar] [CrossRef] [PubMed]
- Ohniwa, R.L.; Kitabayashi, K.; Morikawa, K. Alternative cardiolipin synthase Cls1 compensates for stalled Cls2 function in Staphylococcus aureus under conditions of acute acid stress. FEMS Microbiol. Lett. 2013, 338, 141–146. [Google Scholar] [CrossRef] [Green Version]
- Koprivnjak, T.; Zhang, D.; Ernst, C.M.; Peschel, A.; Nauseef, W.M.; Weiss, J.P. Characterization of Staphylococcus aureus cardiolipin synthases 1 and 2 and their contribution to accumulation of cardiolipin in stationary phase and within phagocytes. J. Bacteriol. 2011, 193, 4134–4142. [Google Scholar] [CrossRef] [Green Version]
- Cameron, D.R.; Mortin, L.I.; Rubio, A.; Mylonakis, E.; Moellering, R.C., Jr.; Eliopoulos, G.M.; Peleg, A.Y. Impact of daptomycin resistance on Staphylococcus aureus virulence. Virulence 2015, 6, 127–131. [Google Scholar] [CrossRef] [Green Version]
- Ernst, C.M.; Staubitz, P.; Mishra, N.N.; Yang, S.J.; Hornig, G.; Kalbacher, H.; Bayer, A.S.; Kraus, D.; Peschel, A. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 2009, 5, e1000660. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Xu, F. Dendritic cells during Staphylococcus aureus infection: Subsets and roles. J. Transl. Med. 2014, 12, 358. [Google Scholar] [CrossRef] [Green Version]
- Balraadjsing, P.P.; de Jong, E.C.; van Wamel, W.J.B.; Zaat, S.A.J. Dendritic cells internalize Staphylococcus aureus more efficiently than Staphylococcus epidermidis, but do not differ in induction of antigen-specific T Cell proliferation. Microorganisms 2019, 8, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oldenburg, M.; Krüger, A.; Ferstl, R.; Kaufmann, A.; Nees, G.; Sigmund, A.; Bathke, B.; Lauterbach, H.; Suter, M.; Dreher, S.; et al. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science 2012, 337, 1111–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patton, T.; Jiang, J.-H.; Lundie, R.; Bafit, M.; Gao, W.; Peleg, A.; O’Keeffe, M. Daptomycin resistant Staphylococcus aureus clinical isolates are poorly sensed by dendritic cells. Immunol. Cell. Biol. 2019, 98. [Google Scholar] [CrossRef]
- Cruciani, M.; Sandini, S.; Etna, M.P.; Giacomini, E.; Camilli, R.; Severa, M.; Rizzo, F.; Bagnoli, F.; Hiscott, J.; Coccia, E.M. Differential responses of human dendritic cells to live or inactivated Staphylococcus aureus: Impact on cytokine production and T helper expansion. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef]
- Spaan, A.N.; Surewaard, B.G.; Nijland, R.; van Strijp, J.A. Neutrophils versus Staphylococcus aureus: A biological tug of war. Annu Rev. Microbiol. 2013, 67, 629–650. [Google Scholar] [CrossRef] [Green Version]
- Guerra, F.E.; Borgogna, T.R.; Patel, D.M.; Sward, E.W.; Voyich, J.M. Epic Immune Battles of History: Neutrophils vs. Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2017, 7, 286. [Google Scholar] [CrossRef] [PubMed]
- English, B.K.; Maryniw, E.M.; Talati, A.J.; Meals, E.A. Diminished macrophage inflammatory response to Staphylococcus aureus isolates exposed to daptomycin versus vancomycin or oxacillin. Antimicrob. Agents Chemother. 2006, 50, 2225–2227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelesidis, T. The interplay between daptomycin and the immune system. Front. Immunol. 2014, 5, 52. [Google Scholar] [CrossRef] [Green Version]
- Mishra, N.N.; McKinnell, J.; Yeaman, M.R.; Rubio, A.; Nast, C.C.; Chen, L.; Kreiswirth, B.N.; Bayer, A.S. In vitro cross-resistance to daptomycin and host defense cationic antimicrobial peptides in clinical methicillin-resistant Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 2011, 55, 4012–4018. [Google Scholar] [CrossRef] [Green Version]
- Mishra, N.N.; Yang, S.-J.; Chen, L.; Muller, C.; Saleh-Mghir, A.; Kuhn, S.; Peschel, A.; Yeaman, M.R.; Nast, C.C.; Kreiswirth, B.N.; et al. Emergence of daptomycin resistance in daptomycin-naïve babbits with methicillin-resistant Staphylococcus aureus prosthetic joint Infection is associated with resistance to host defense cationic peptides and mprF polymorphisms. PLoS ONE 2013, 8, e71151. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Piechota, M.; Kot, B.; Frankowska-Maciejewska, A.; Grużewska, A.; Woźniak-Kosek, A. Biofilm formation by Methicillin-Resistant and Methicillin-Sensitive Staphylococcus aureus strains from hospitalized patients in Poland. BioMed Res. Int. 2018, 2018, 4657396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thurlow, L.R.; Hanke, M.L.; Fritz, T.; Angle, A.; Aldrich, A.; Williams, S.H.; Engebretsen, I.L.; Bayles, K.W.; Horswill, A.R.; Kielian, T. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J. Immunol. 2011, 186, 6585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ernst, C.M.; Kuhn, S.; Slavetinsky, C.J.; Krismer, B.; Heilbronner, S.; Gekeler, C.; Kraus, D.; Wagner, S.; Peschel, A. The lipid-modifying multiple peptide resistance factor is an oligomer consisting of distinct interacting synthase and flippase subunits. mBio 2015, 6, e02340-14. [Google Scholar] [CrossRef] [Green Version]
| PRR Family | Receptor | Species | Cellular Location | PAMP | Signal Adapter | References | |
|---|---|---|---|---|---|---|---|
| Human | Mouse | ||||||
| TLR | TLR1/TLR2 heterodimer | + | + | Plasma Membrane | Triacyl lipoproteins | MyD88/TIRAP | [18,19,20,21,22] |
| TLR2/TLR6 heterodimer | + | + | Plasma Membrane | Diacyl lipoprotein, Lipopeptide, Peptidoglycan, Phenol soluble modulin (PSM) | MyD88/TIRAP | [18,19,20] | |
| TLR7 | + | + | Endosome | tRNA | MyD88 | [23,24] | |
| TLR8 | + | − | Endosome | ssRNA | MyD88 | [25,26] | |
| TLR9 | + | + | Endosome | Unmethylated CpG DNA | MyD88 | [23,27] | |
| TLR13 | − | + | Endosome | 23s RNA | MyD88 | [23] | |
| CLR | MBL | + | + | Plasma Membrane | Teichoic acid | [28,29,30] | |
| NLR | NOD2 | + | + | Cytoplasm | Peptidoglycan, α, β-, and γ- hemolysins; Panton–Valentine leukocidin (PVL); Leukocidin A/B; Acylated lipopeptides | RIP2, MAVS | [31,32,33] |
| RLRs | RIG-I | + | + | Cytoplasm | Cytoplasmic RNA | MAVS | [34,35] |
| CDS | cGAS STING | + | + | Cytoplasm | Double stranded DNA, Cyclic dinucleotide | STING | [36,37,38,39,40] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jahan, N.; Patton, T.; O’Keeffe, M. The Influence of Antibiotic Resistance on Innate Immune Responses to Staphylococcus aureus Infection. Antibiotics 2022, 11, 542. https://doi.org/10.3390/antibiotics11050542
Jahan N, Patton T, O’Keeffe M. The Influence of Antibiotic Resistance on Innate Immune Responses to Staphylococcus aureus Infection. Antibiotics. 2022; 11(5):542. https://doi.org/10.3390/antibiotics11050542
Chicago/Turabian StyleJahan, Nazneen, Timothy Patton, and Meredith O’Keeffe. 2022. "The Influence of Antibiotic Resistance on Innate Immune Responses to Staphylococcus aureus Infection" Antibiotics 11, no. 5: 542. https://doi.org/10.3390/antibiotics11050542
APA StyleJahan, N., Patton, T., & O’Keeffe, M. (2022). The Influence of Antibiotic Resistance on Innate Immune Responses to Staphylococcus aureus Infection. Antibiotics, 11(5), 542. https://doi.org/10.3390/antibiotics11050542






