The Synthesis of Triazolium Salts as Antifungal Agents: A Biological and In Silico Evaluation
Abstract
1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Biological Evaluation
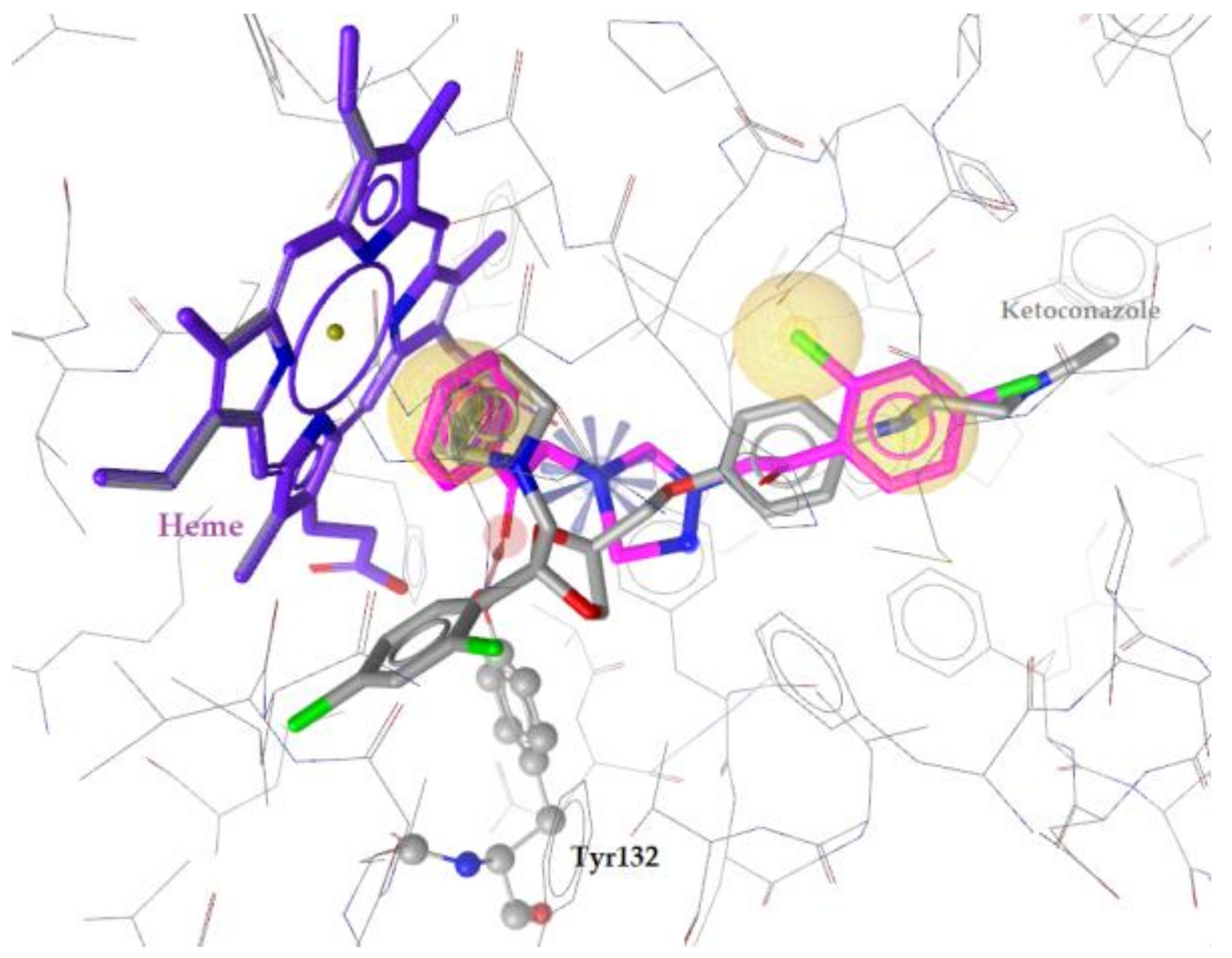
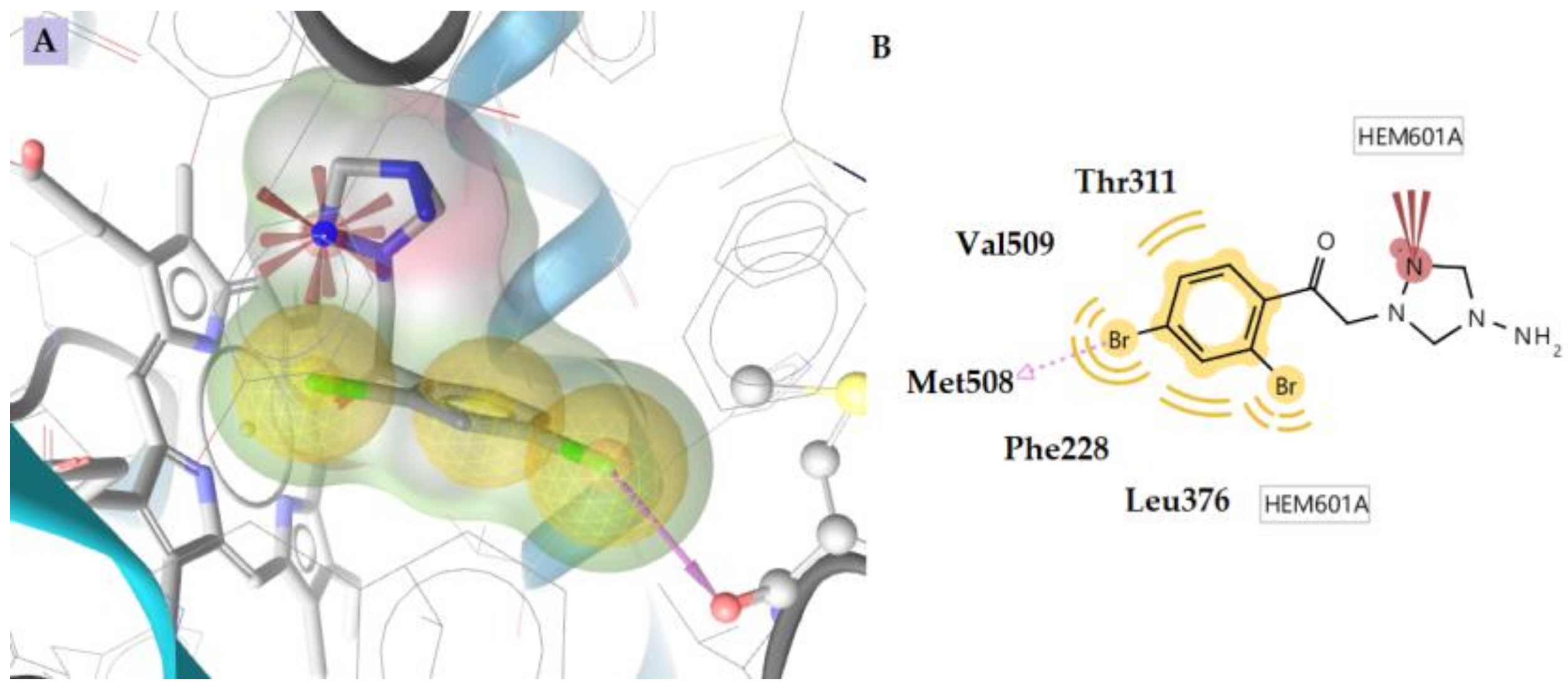
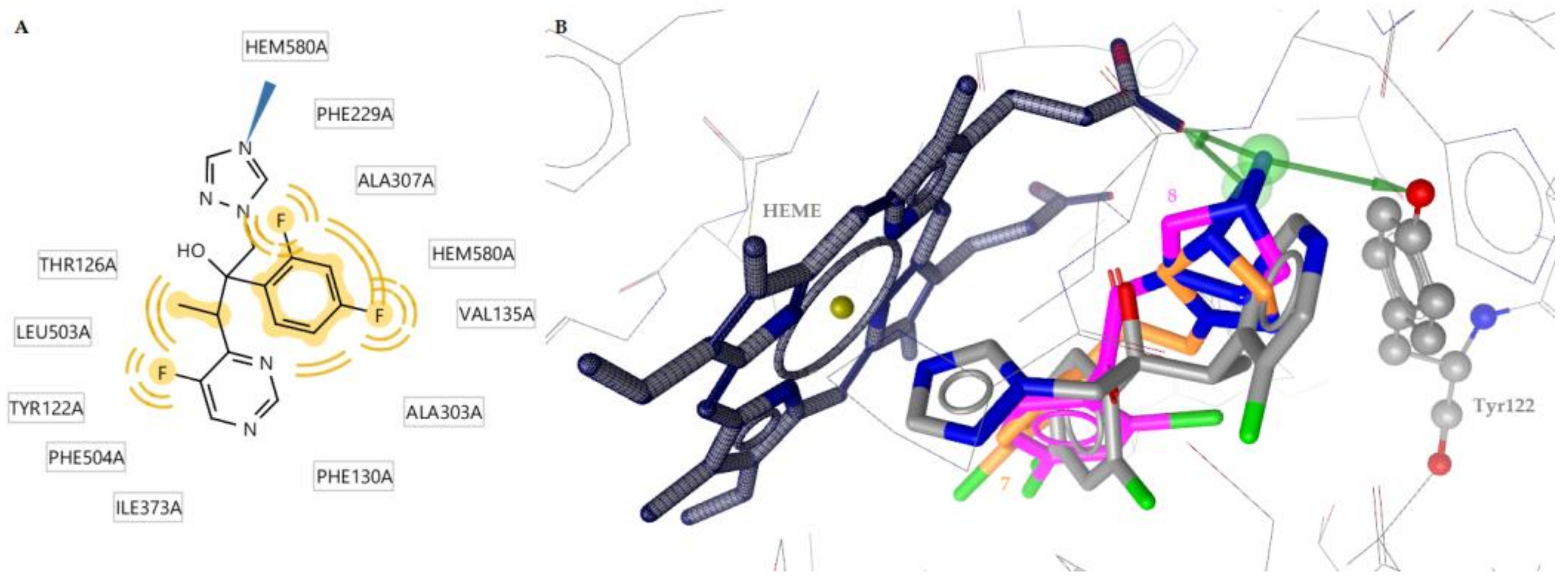
2.3. Docking to Antifungal Targets
2.4. Drug-likeness
3. Materials and Methods
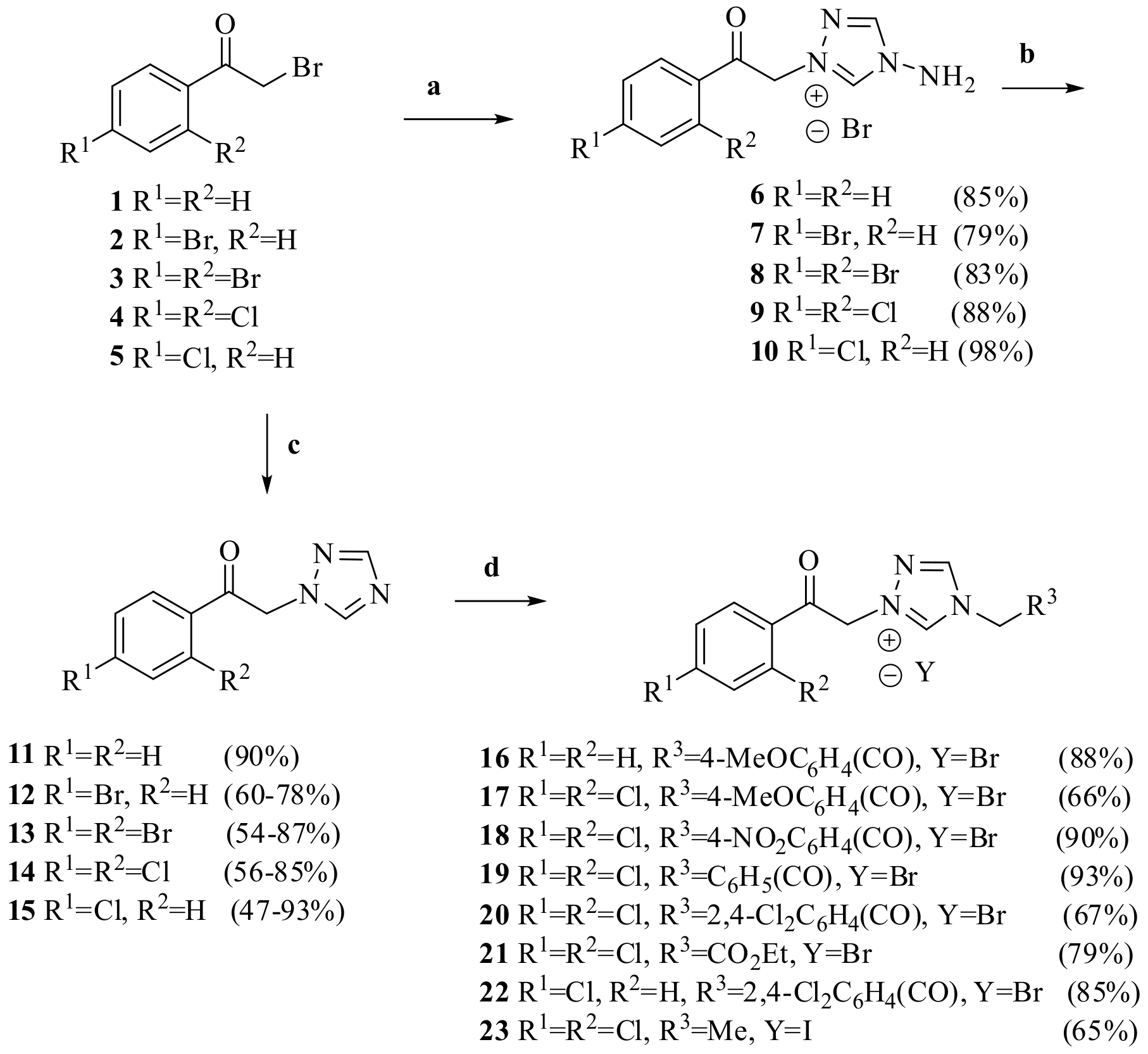
3.1. Chemistry
3.1.1. Typical Experimental Procedure for the Synthesis of 1-substituted-2-(1H-1,2,4-triazol-1-yl) ethanones 11-15
3.1.2. General Experimental Procedure for Synthesis Ammonium Salts 6-10 and 15-23
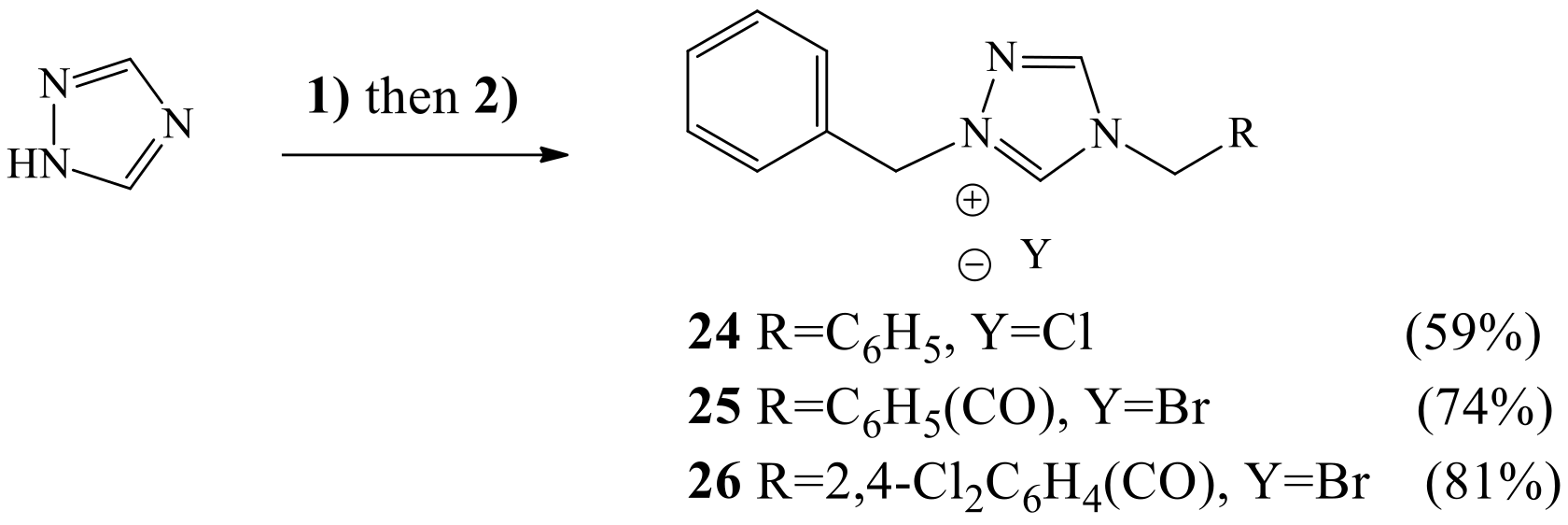
3.1.3. General Experimental Procedure for the Synthesis of 1-benzyl-4-substituted 1H-1,2,4-triazol-4-ium halides 24-26
3.2. Antifungal Action
3.3. In-Silico Molecular Docking Studies
3.4. In Silico Predictive Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Amorim, A.; Guedes-Vaz, L.; Araujo, R. Susceptibility to five antifungals of Aspergillus fumigatus strains isolated from chronically colonised cystic fibrosis patients receiving azole therapy. Int. J. Antimicrob. Agents 2010, 35, 396–399. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jain, K.S.; Khedkar, V.M.; Arya, N.; Rane, P.V.; Chaskar, P.K.; Coutinho, E.C. Design, synthesis & evaluation of condensed 2H-4-arylaminopyrimidines as novel antifungal agents. Eur. J. Med. Chem. 2014, 77, 166–175. [Google Scholar] [PubMed]
- Kniemeyer, O.; Schmidt, A.D.; Vödisch, M.; Wartenberg, D.; Brakhage, A.A. Identification of virulence determinants of the human pathogenic fungi Aspergillus fumigatus and Candida albicans by proteomics. Int. J. Med. Microbiol. 2011, 301, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Cao, Y.; Zhang, J.; Yu, S.; Zou, Y.; Chai, X.; Wu, Q.; Zhang, D.; Jiang, Y.; Sun, Q. Design, synthesis and antifungal activities of novel 1,2,4-triazole derivatives. Eur. J. Med. Chem. 2011, 46, 3142–3148. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.D.; Patel, M.R.; Kocsis, B.; Kocsis, E.; Graham, S.M.; Warren, A.R.; Nicholson, S.M.; Billack, B.; Fronczek, F.R.; Talele, T.T. Design, synthesis and determination of antifungal activity of 5(6)-substituted benzotriazoles. Eur. J. Med. Chem. 2010, 45, 2214–2222. [Google Scholar] [CrossRef]
- Gao, F.; Wang, T.; Xiao, J.; Huang, G. Antibacterial activity study of 1,2,4-triazole derivatives. Eur. J. Med. Chem. 2019, 173, 274–281. [Google Scholar] [CrossRef]
- Stingaci, E.; Zveaghinteva, M.; Pogrebnoi, S.; Lupascu, L.; Valica, V.; Uncu, L.; Smetanscaia, A.; Drumea, M.; Petrou, A.; Ciric, A.; et al. New vinyl-1,2,4-triazole derivatives as antimicrobial agents: Synthesis, biological evaluation and molecular docking studies. Bioorg. Med. Chem. Lett. 2020, 30, 127368. [Google Scholar] [CrossRef]
- Shaikha, M.H.; Subhedara, D.D.; Akolkara, S.V.; Nagargojea, A.A.; Khedkard, V.M.; Sarkare, D.; Shingatea, B.B. Tetrazoloquinoline-1,2,3-Triazole Derivatives as Antimicrobial Agents: Synthesis, Biological Evaluation and Molecular Docking Study. Polycycl. Aromat. Compd. 2020. epub ahead of printing. [Google Scholar] [CrossRef]
- Gupta, D.; Jain, D.K. Synthesis, antifungal and antibacterial activity of novel 1,2,4-triazole derivatives. J. Adv. Pharm. Technol. Res. 2015, 6, 141–146. [Google Scholar] [CrossRef]
- da Silva, N.M.; Gentz, C.B.; Reginatto, P.; Fernandes, T.H.M.; Kaminski, T.F.A.; Lopes, W.; Quatrin, P.M.; Vainstein, M.H.; Abegg, M.A.; Lopes, M.S.; et al. 8-Hydroxyquinoline 1,2,3-triazole derivatives with promising and selective antifungal activity. Med. Mycol. 2021, 59, 431–440. [Google Scholar] [CrossRef]
- Pagniez, F.; Lebouvier, N.; Na, Y.M.; Ourliac-Garnier, I.; Picot, C.; Le Borgne, M.; Le Pape, P. Biological exploration of a novel 1,2,4-triazole-indole hybrid molecule as antifungal agent. J. Enzym. Inhib. Med. Chem. 2020, 35, 398–403. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, Z.; Gao, C.; Ren, Q.C.; Chang, L.; Lv, Z.S.; Feng, L.S. Triazole derivatives and their anti-tubercular activity. Eur. J. Med. Chem. 2017, 138, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Karczmarzyk, Z.; Swatko-Ossor, M.; Wysocki, W.; Drozd, M.; Ginalska, G.; Pachuta-Stec, A.; Pitucha, M. New Application of 1,2,4-Triazole Derivatives as Antitubercular Agents. Structure, In Vitro Screening and Docking Studies. Molecules 2020, 25, 6033. [Google Scholar] [CrossRef] [PubMed]
- Casertano, M.; Menna, M.; Imperatore, C. The Ascidian-Derived Metabolites with Antimicrobial Properties. Antibiotics 2020, 9, 510. [Google Scholar] [CrossRef] [PubMed]
- Zalacain, M.; Lozano, C.; Llanos, A.; Sprynski, N.; Valmont, T.; De Piano, C.; Davies, D.; Leiris, S.; Sable, C.; Ledoux, A.; et al. Novel Specific Metallo-β-Lactamase Inhibitor ANT2681 Restores Meropenem Activity to Clinically Effective Levels against NDM-Positive Enterobacterales. Antimicrob. Agents Chemother. 2021, 65, e00203-21. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y.; Zhou, Y.; Gao, Y.; Wang, X.; Wang, J.; Niu, X. Discovery of a Novel Natural Allosteric Inhibitor That Targets NDM-1 Against Escherichia coli. Front. Pharmacol. 2020, 11, 581001. [Google Scholar] [CrossRef] [PubMed]
- Wade, N.; Tehrani, K.H.; Brüchle, N.C.; van Haren, M.J.; Mashayekhi, V.; Martin, N.I. Mechanistic Investigations of Metallo-β-lactamase Inhibitors: StrongZinc Binding Is Not Required for Potent Enzyme Inhibition. ChemMedChem 2021, 16, 1651–1659. [Google Scholar] [CrossRef]
- Tratrat, C.; Haroun, M.; Paparisva, A.; Kamoutsis, C.; Petrou, A.; Gavalas, A.; Eleftheriou, P.; Geronikaki, A.; Venugopala, N.; Kochkar, H.; et al. New Substituted 5-Benzylideno-2-Adamantylthiazol[3,2-b][1,2,4]Triazol-6(5H)ones as Possible Anti-Inflammatory Agents. Molecules 2021, 26, 659. [Google Scholar] [CrossRef]
- Shafi, S.; Alam, M.M.; Mulakayala, N.; Mulakayala, C.; Vanaja, G.; Kalle, A.M.; Pallu, R.; Alam, M.S. Synthesis of novel 2-mercapto benzothiazole and 1,2,3-triazole based bis-heterocycles: Their anti-inflammatory and anti-nociceptive activities. Eur. J. Med. Chem. 2012, 49, 324–333. [Google Scholar] [CrossRef]
- Simurova, N.V.; Maiboroda, O.I. Antiviral activity of 1,2,4-triazole derivatives (micro review). Chem. Heterocycl. Compd. 2021, 57, 420–422. [Google Scholar] [CrossRef]
- Aouad, M.R.; Khan, D.J.O.; Said, M.A.; Al-Kaff, N.S.; Rezki, N.; Ali, A.A.; Bouqellah, N.; Hagar, M. Novel 1,2,3-Triazole Derivatives as Potential Inhibitors against Covid-19 Main Protease: Synthesis, Characterization, Molecular Docking and DFT Studies. Chem. Select 2021, 6, 3468–3486. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wu, G.; Zalloum, W.A.; Meuser, M.E.; Dick, A.; Sun, L.; Chen, C.H.; Kang, D.; Jing, L.; Jia, R.; et al. Discovery of novel 1,4-disubstituted 1,2,3-triazole phenylalanine derivatives as HIV-1 capsid inhibitors. RSC Adv. 2019, 9, 28961–28986. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.-S.; Zheng, M.-J.; Zhao, F.; Liu, D. 1,2,3-Triazole hybrids with anti-HIV-1 activity. Arch. Pharm. 2021, 354, e2000163. [Google Scholar] [CrossRef] [PubMed]
- Asgari, M.; Tahmasebi, B.; Mojtabavi, S.; Faramarzi, M.A.; Rahimi, R.; Ranjbar, P.; Biglar, M.; Rastegar, H.; Mohammadi-Khanaposhtani, M.; Mahdavi, M. Design, synthesis, biological evaluation, and docking study of new acridine-9-carboxamide linked to 1,2,3-triazole derivatives as antidiabetic agents targeting α-glucosidase. J. Heterocycl. Chem. 2020, 57, 4348–4357. [Google Scholar] [CrossRef]
- Gani, S.; Timanagouda, K.; Madhushree, S.; Joshi, D.; Hiremath, M.; Begum, S.; Kudva, A. Synthesis of novel indole, 1,2,4-triazole derivatives as potential glucosidase inhibitors. J. King Saud Univ.-Sci. 2020, 32, 3388–3399. [Google Scholar] [CrossRef]
- Kumar, S.; Khokra, S.; Yadav, A. Triazole analogues as potential pharmacological agents: A brief review. Future J. Pharm. Sci. 2021, 7, 106. [Google Scholar] [CrossRef]
- Aouad, M.R.; Soliman, M.A.; Alharbi, M.O.; Bardaweel, S.K.; Sahu, P.K.; Ali, A.A.; Messali, M.; Rezki, N.; Al-Soud, Y.A. Design, synthesis and anticancer screening ofnovel benzothiazole-piperazine-1,2,3-triazole hybrids. Molecules 2018, 23, 2788. [Google Scholar] [CrossRef]
- Maddali, N.; Ivaturi, V.; Yellajyosula, M.; Malkhed, V.; Brahman, P.; Pindiprolu, S.; Kondaparthi, V.; Nethinti, S. New 1,2,4-Triazole Scaffolds as Anticancer Agents: Synthesis, Biological Evaluation and Docking Studies. Chem. Select 2021, 6, 6788–6796. [Google Scholar] [CrossRef]
- Mahantia, S.; Sunkaraa, S.; Bhavani, R. Synthesis, biological evaluation and computational studies of fused acridine containing 1,2,4-triazole derivatives as anticancer agents. Synth. Commun. 2019, 49, 1729–1740. [Google Scholar] [CrossRef]
- Song, M.-X.; Deng, X.-Q. Recent developments on triazole nucleus in anticonvulsant compounds: A review. J. Enzym. Inhib. Med. Chem. 2018, 33, 453–478. [Google Scholar] [CrossRef]
- Kaproń, B.; Czarnomysy, R.; Wysokiński, M.; Andrys, R.; Musilek, K.; Angeli, A.; Supuran, C.T.; Plech, T. 1,2,4-Triazole-based anticonvulsant agents with additional ROS scavenging activity are effective in a model of pharmacoresistant epilepsy. J. Enzym. Inhib. Med. Chem. 2020, 35, 993–1002. [Google Scholar] [CrossRef] [PubMed]
- Akin, S.; Ayaloglu, H.; Gultekin, E.; Colak, A.; Bekircan, O.; Akatin, M.Y. Synthesis of 1,2,4-triazole-5-on derivatives and determination of carbonic anhydrase II isoenzyme inhibition effects. Bioorg. Chem. 2019, 83, 170–179. [Google Scholar] [CrossRef] [PubMed]
- El-Gazzar, M.; Nafie, N.; Nocentini, A.; Ghorab, M.; Heiba, H.; Supuran, C.T. Carbonic anhydrase inhibition with a series ofnovel benzenesulfonamide-triazole conjugates. J. Enzym. Inhib. Med. Chem. 2018, 33, 1565–1574. [Google Scholar] [CrossRef] [PubMed]
- Meinel, R.S.; Almeida, A.D.C.; Stroppa, P.H.F.; Glanzmann, N.; Coimbra, E.S.; da Silva, A.D. Novel functionalized 1,2,3-triazole derivatives exhibit antileishmanial activity, increase in total and mitochondrial-ROS and depolarization of mitochondrial membrane potential of Leishmania amazonensis. Chem. Biol. Interact. 2020, 315, 108850. [Google Scholar] [CrossRef] [PubMed]
- Nandikolla, N.; Srinivasarao, S.; Kumar, B.; Murugesan, M.; Aggarwal, H.; Major, L.; Smithc, T.; Sekhar, T. Synthesis, study of antileishmanial and antitrypanosomal activity of imidazo pyridine fused triazole analogues. RSC Adv. 2020, 10, 38328. [Google Scholar] [CrossRef]
- Pertino, M.W.; Vega, C.; Rolón, M.; Coronel, C.; Rojas de Arias, A.; Schmeda-Hirschmann, G. Antiprotozoal Activity of Triazole Derivatives of Dehydroabietic Acid and Oleanolic Acid. Molecules 2017, 22, 369. [Google Scholar] [CrossRef]
- Mabasa, T.F.; Awe, B.; Laming, D.; Kinfe, H.H. Design, Synthesis and Antiplasmodial Evaluation of Sulfoximine-triazole Hybrids as Potential Antimalarial Prototypes. Med. Chem. 2019, 15, 685–692. [Google Scholar] [CrossRef]
- Ibrahim, Z.; Uzairu, A.; Shallangwa, G.; Abechi, S. Molecular modeling and design of someβ-amino alcohol grafted1,4,5-trisubstituted 1,2,3-triazoles derivatives against chloroquine sensitive, 3D7 strain of Plasmodium falciparum. Heliyon 2021, 7, e05924. [Google Scholar] [CrossRef]
- Abuelizz, H.; Taie, H.; Bakheit, A.; Marzouk, M.; Abdellatif, M.; Al-Salahi, M. Biological Evaluation of 4-(1H-triazol-1-yl)benzoic Acid Hybrids as Antioxidant Agents: In Vitro Screening and DFT Study. Appl. Sci. 2021, 11, 11642. [Google Scholar] [CrossRef]
- Perekhoda, L.; Georgiyants, V.; Yeromina, H.; Drapak, I.; Lubenets, V.; Ieromina, Z.; Sych, I.; Severina, H.; Demchenko, A. The synthesis and in silico antihypertensive activity prognosis of new mannich bases containing the 1, 2, 4-triazole moiety. Chem. Chem. Technol. 2020, 2, 214–220. [Google Scholar] [CrossRef]
- Al-Salahi, R.; El-Tahir, K.-E.; Alswaidan, I.; Lolak, N.; Hamidaddin, M.; Marzouk, M. Biological effects of a new set 1,2,4- triazolo[1,5-a]quinazolines on heart rate and blood pressure. Chem. Cent. J. 2014, 8, 3. [Google Scholar] [CrossRef] [PubMed]
- Küçükgüzel, S.G.; Çıkla-Süzgün, P. Recent advances bioactive 1, 2, 4-triazole-3-thiones. Eur. J. Med. Chem. 2015, 97, 830–870. [Google Scholar] [CrossRef] [PubMed]
- Zveaghintseva, M.; Stingaci, E.; Pogrebnoi, S.; Smetanscaia, A.; Valica, V.; Uncu, L.; Kravtsov, V.; Melnic, E.; Petrou, A.; Glamočlija, J.; et al. Chromenol Derivatives as Novel Antifungal Agents: Synthesis, In Silico and In Vitro Evaluation. Molecules 2021, 26, 4304. [Google Scholar] [CrossRef] [PubMed]
- Kamoutsis, C.; Fesatidou, M.; Petrou, A.; Geronikaki, A.; Poroikov, V.; Ivanov, M.; Soković, M.; Cirić, A.; Carazo, A.; Mladenka, M. Triazolo Based-Thiadiazole Derivatives. Synthesis, Biological Evaluation and Molecular Docking Studies. Antibiotics 2021, 10, 804. [Google Scholar] [CrossRef] [PubMed]
- Krimer, M.Z.; Tashchi, V.P.; Roitburd, G.V.; Shtyrkov, I.M.; Manaev, S.A.; Malinovsky, S.T.; Putsykin, Y.G. Regiospecific synthesis of aryl- and alkyl(1,2,4-triazolyl-1-ylmethyl) ketones. Doklady Akademii Nauk USSR 1989, 308, 1155–1158. [Google Scholar]
- Lebouvier, N.; Giraud, F.; Corbin, T.; Na, Y.M.; Le Baut, G.; Marchand, P.; Le Borgne, M. Efficient microwave-assisted synthesis of 1-(1H-indol-1-yl)-2-phenyl-3-(1H-1,2,4-triazol-1-yl)-propan-2-ols as antifungal agents. Tetrahedron Lett. 2006, 47, 6479–6483. [Google Scholar] [CrossRef]
- Kritsi, E.; Matsoukas, M.T.; Potamitis, C.; Detsi, A.; Ivanov, M.; Sokovic, M.; Zoumpoulakis, P. Novel Hit Compounds as Putative Antifungals: The Case of Aspergillus fumigatus. Molecules 2019, 24, 3853. [Google Scholar] [CrossRef]
- Aleksić, M.; Stanisavljević, D.; Smiljković, M.; Vasiljević, P.; Stevanović, M.; Soković, M.; Stojković, D. Pyrimethanil: Between efficient fungicide against Aspergillus rot on cherry tomato and cytotoxic agent on human cell lines. Ann. Appl. Biol. 2019, 175, 228–235. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Autodock4 and AutoDockTools4: Automated docking with selective receptor flexiblity. J. Comput. Chem. 2009, 16, 2785–2791. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Lagorce, D.; Sperandio, O.; Galons, H.; Miteva, M.A.; Villoutreix, B.O. FAF-Drugs2: Free ADME/tox filtering tool to assist drug discovery and chemical biology projects. BMC Bioinform. 2008, 9, 396. [Google Scholar] [CrossRef] [PubMed]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Wars, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Muegge, I.; Heald, S.L.; Brittelli, D. Simple selection criteria for drug-like chemical matter. J. Med. Chem. 2001, 44, 1841–1846. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead- and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef]



| Compounds | A.fum. | A.v. | A.o. | A.n. | T.v. | P.f. | P.o. | P.v.c. | |
|---|---|---|---|---|---|---|---|---|---|
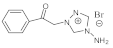 6 | MIC | 0.15 | 0.10 | 0.10 | 0.05 | 0.025 | 0.10 | 0.07 | 0.20 |
| MFC | 0.20 | 0.20 | 0.20 | 0.10 | 0.05 | 0.20 | 0.10 | 0.40 | |
 7 | MIC | 0.025 | 0.025 | 0.0125 | 0.025 | 0.05 | 0.05 | 0.025 | 0.05 |
| MFC | 0.05 | 0.05 | 0.025 | 0.05 | 0.10 | 0.10 | 0.05 | 0.10 | |
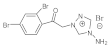 8 | MIC | 0.025 | 0.0125 | 0.07 | 0.0 | 0.009 | 0.0125 | 0.0125 | 0.025 |
| MFC | 0.05 | 0.025 | 0.10 | 0.10 | 0.0125 | 0.025 | 0.025 | 0.05 | |
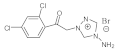 9 | MIC | 0.05 | 0.05 | 0.025 | 0.037 | 0.015 | 0.03 | 0.025 | 0.05 |
| MFC | 0.10 | 0.10 | 0.05 | 0.0 | 0.025 | 0.05 | 0.05 | 0.10 | |
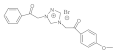 16 | MIC | 0.07 | 0.006 | 0.05 | 0.025 | 0.025 | 0.05 | 0.05 | 0.10 |
| MFC | 0.10 | 0.0125 | 0.10 | 0.05 | 0.05 | 0.10 | 0.10 | 0.20 | |
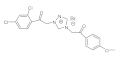 17 | MIC | 0.037 | 0.015 | 0.025 | 0.05 | 0.009 | 0.037 | 0.037 | 0.037 |
| MFC | 0.05 | 0.025 | 0.05 | 0.10 | 0.0125 | 0.05 | 0.05 | 0.05 | |
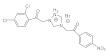 18 | MIC | 0.05 | 0.03 | 0.025 | 0.05 | 0.015 | 0.05 | 0.03 | 0.07 |
| MFC | 0.10 | 0.05 | 0.05 | 0.10 | 0.025 | 0.10 | 0.05 | 0.10 | |
 19 | MIC | 0.037 | 0.0125 | 0.025 | 0.037 | 0.009 | 0.02 | 0.0125 | 0.025 |
| MFC | 0.05 | 0.025 | 0.05 | 0.05 | 0.0125 | 0.025 | 0.025 | 0.05 | |
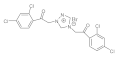 20 | MIC | 0.07 | 0.0003 | 0.05 | 0.05 | 0.006 | 0.037 | 0.025 | 0.025 |
| MFC | 0.10 | 0.0006 | 0.10 | 0.10 | 0.0125 | 0.05 | 0.05 | 0.05 | |
 21 | MIC | 0.05 | 0.05 | 0.025 | 0.025 | 0.012 | 0.05 | 0.037 | 0.025 |
| MFC | 0.10 | 0.10 | 0.05 | 0.05 | 0.025 | 0.10 | 0.05 | 0.05 | |
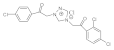 22 | MIC | 0.05 | 0.015 | 0.037 | 0.05 | 0.009 | 0.037 | 0.025 | 0.027 |
| MFC | 0.10 | 0.025 | 0.05 | 0.10 | 0.0125 | 0.05 | 0.05 | 0.05 | |
 23 | MIC | 0.10 | 0.07 | 0.05 | 0.05 | 0.037 | 0.07 | 0.05 | 0.07 |
| MFC | 0.20 | 0.10 | 0.10 | 0.10 | 0.05 | 0.10 | 0.10 | 0.10 | |
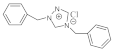 24 | MIC | 0.10 | 0.05 | 0.037 | 0.05 | 0.015 | 0.05 | 0.05 | 0.10 |
| MFC | 0.20 | 0.10 | 0.05 | 0.10 | 0.05 | 0.10 | 0.10 | 0.20 | |
 25 | MIC | 0.10 | 0.07 | 0.037 | 0.05 | 0.025 | 0.20 | 0.025 | 0.20 |
| MFC | 0.20 | 0.10 | 0.05 | 0.10 | 0.05 | 0.40 | 0.05 | 0.40 | |
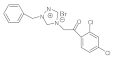 26 | MIC | 0.10 | 0.037 | 0.05 | 0.05 | 0.015 | 0.037 | 0.037 | 0.037 |
| MFC | 0.20 | 0.05 | 0.10 | 0.10 | 0.025 | 0.05 | 0.05 | 0.05 | |
| Ketoconazole | MIC | 0.38 | 0.38 | 0.28 | 0.38 | 1.88 | 0.38 | 1.88 | 0.38 |
| MFC | 0.94 | 0.94 | 0.38 | 0.94 | 2.82 | 0.94 | 2.82 | 0.57 | |
| Bifonazole | MIC | 0.48 | 0.32 | 0.48 | 0.48 | 0.48 | 0.64 | 0.64 | 0.32 |
| MFC | 0.64 | 0.64 | 0.64 | 0.64 | 0.64 | 0.81 | 0.81 | 0.64 | |
| Est. Binding Energy (kcal/mol) | ||||||
|---|---|---|---|---|---|---|
| N/N | DNA TopoIV 1S16 | CYP51 of C. albicans 5V5Z | Residues Involved in H-Bond Interactions | Halogen Interactions | Hydrophobic Interactions | Interactions with HEM601 |
| 6 | - | −6.60 | - | - | Phe233, Leu300, Phe380, Met508, Hem601 | Hydrophobic |
| 7 | −4.11 | −9.27 | Tyr132 | - | Phe233, Leu376, Phe380, Met508, Val509, Hem601 | Hydrophobic, Aromatic |
| 8 | −2.27 | −10.60 | - | Met508 | Phe228, Thr311, Leu376, Met508, Val509 | Negative ionizable |
| 9 | −3.37 | −9.10 | Tyr118 | - | Leu376, Phe380, Met508, Val509, Hem601 | Hydrophobic, Aromatic |
| 16 | −2.82 | −6.95 | - | - | Leu376, Met508, Hem601 | Hydrophobic |
| 17 | −1.86 | −8.80 | Tyr118 | - | Tyr118, Leu376, Met508, Hem601 | Hydrophobic, Aromatic |
| 18 | −4.16 | −8.84 | Tyr64 | - | Tyr118, Leu300, Leu376, Phe380, Met508, Hem601 | Hydrophobic, Aromatic |
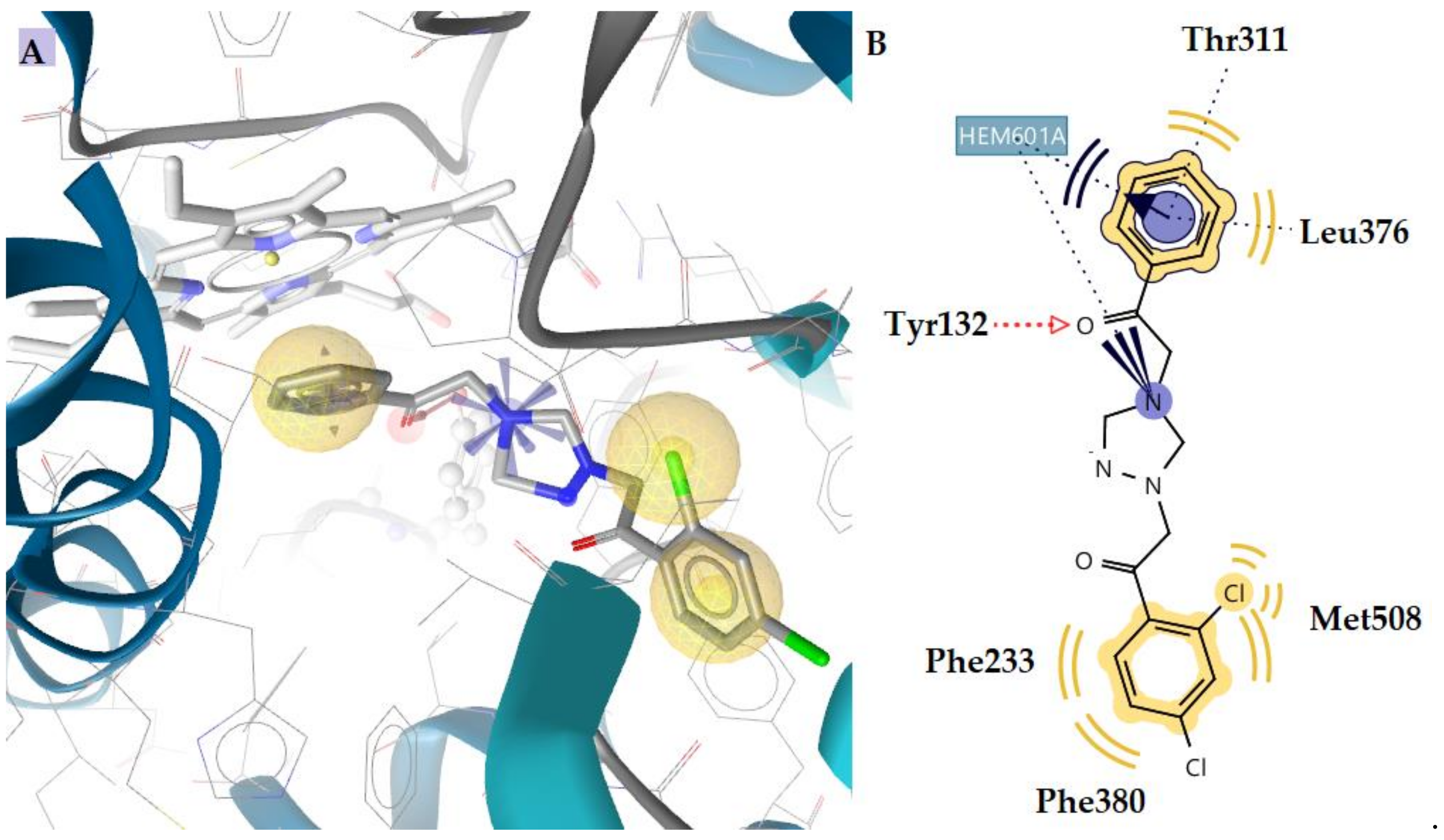
| 19 | −3.30 | −10.83 | Tyr132 | - | Phe233, Thr311, Leu376, Phe380, Met508, Hem601 | Hydrophobic, Aromatic, Positive ionizable |
| 20 | - | −7.11 | - | - | Thr311, Leu376, Met508, Hem601 | Hydrophobic |
| 21 | - | −8.10 | - | Met508 | Thr311, Leu376, Met508, Hem601 | Hydrophobic, Aromatic |
| 22 | −2.52 | −9.15 | Tyr132 | - | Thr311, Leu376, Met508, Hem601 | Hydrophobic, Aromatic |
| 23 | −3.79 | −7.50 | Tyr118 | - | Tyr118, Leu376, Hem601 | Hydrophobic |
| 24 | - | −9.73 | - | Met508 | Phe233, Thr311, Leu376, Met508, Hem601 | Hydrophobic, Aromatic |
| 25 | - | −10.20 | Tyr132 | - | Thr311, Leu376, Phe380, Met508, Hem601 | Hydrophobic |
| 26 | −4.53 | −7.72 | - | - | Phe233, Leu376, Phe380, Met508, Hem601 | Hydrophobic |
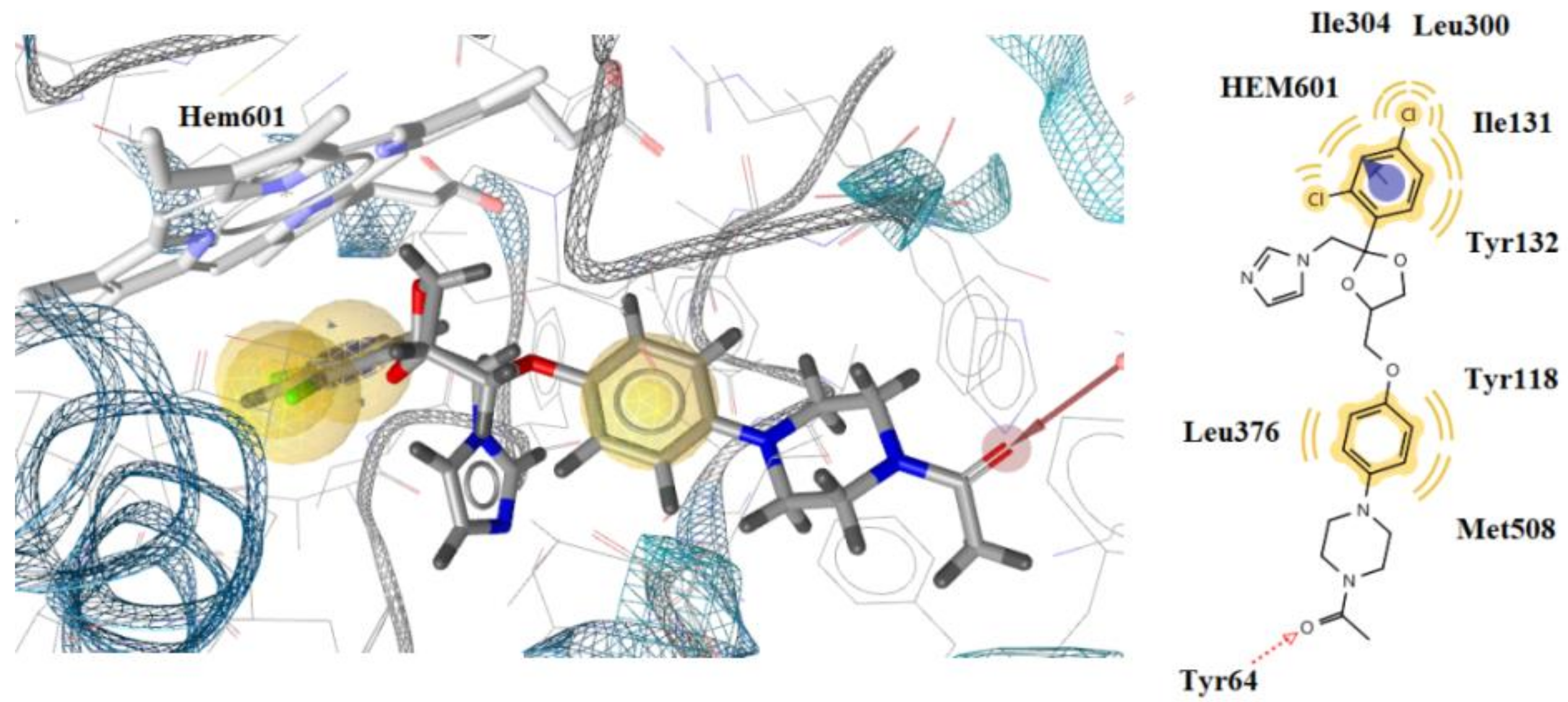
| Ket. | - | −8.23 | Tyr64 | - | Tyr118, Ile131, Tyr132, Ile304, Leu300, Leu376, Met508, Hem601 | Hydrophobic, Aromatic |
| N/N | Est. Binding Energy(kcal/mol) CYP51 of Aspergillus fumigatus 4UYM | Residues Involved in H-Bond Interactions | Hydrophobic Interactions | Interactions with HEM601 |
|---|---|---|---|---|
| 6 | −7.15 | - | Tyr122, Thr126, Val135, Ala307, Phe229, Ala303, Leu503, Hem580 | Hydrophobic |
| 7 | −10.30 | Hem508 | Phe130, Val135, Ala307, Ala303, Leu304 | Hydrogen bond |
| 8 | −11.47 | Tyr122, Hem508 | Thr126, Phe130, Val135, Ala307, Ala303, Leu503, Hem580 | Hydrophobic, hydrogen bond |
| 9 | −9.27 | Tyr122 | Thr126, Val135, Ala307, Ala303, Leu503, Hem580 | Hydrophobic, Aromatic |
| 16 | −7.91 | - | Tyr122, Thr126, Phe130, Ala307, Phe229, Ala303, Ile373, Phe504, Hem580 | Hydrophobic |
| 17 | −10.45 | Hem508 | Tyr122, Phe130, Val135, Ala307, Leu304 | Hydrogen bond |
| 18 | −8.86 | - | Phe130, Val135, Ala307, Ala303, Leu304, Hem580 | Hydrophobic, Aromatic |
| 19 | −10.82 | Hem508 | Tyr122, Thr126, Phe130, Val135, Ile373, Phe504, Hem580 | Hydrophobic, hydrogen bond |
| 20 | −8.05 | - | Tyr122, Thr126, Phe130, Ala307, Hem580 | Hydrophobic |
| 21 | −8.30 | - | Tyr122, Thr126, Phe130, Ala303, Phe504, Hem580 | Hydrophobic, Aromatic |
| 22 | −9.02 | Tyr122 | Thr126, Val135, Ala307, Leu503, Hem580 | Hydrophobic, Aromatic |
| 23 | −7.24 | - | Tyr122, Thr126, Ala307, Phe229, Ala303, Hem580 | Hydrophobic |
| 24 | −8.41 | - | Thr126, Phe130, Ala303, Phe504, Hem580 | Hydrophobic, Aromatic |
| 25 | −8.80 | - | Phe130, Val135, Ala307, Leu304, Hem580 | Hydrophobic, Aromatic |
| 26 | −7.10 | - | Tyr122, Thr126, Val135, Hem580 | Hydrophobic |
| voriconazole | −10.23 | - | Tyr122, Thr126, Phe130, Val135, Ala307, Phe229, Ala303, Ile373, Phe504, Leu503, Hem580 | Hydrophobic, Fe-binding |
| No | MW | Number of HBA a | Number of HBD b | Log Po/w (iLOGP) c | Log S d | TPSA e | Lipinski, Ghose, Veber, Egan, and Muegge Violations | Bioavailability Score | Drug-Likeness Model Score |
|---|---|---|---|---|---|---|---|---|---|
| 6 | 285.14 | 5 | 1 | −14.56 | Soluble | 49.57 | 0 | 0.55 | −0.94 |
| 7 | 364.04 | 5 | 1 | −10.84 | Soluble | 49.57 | 0 | 0.55 | −0.81 |
| 8 | 442.93 | 5 | 1 | −9.33 | Soluble | 49.57 | 0 | 0.55 | −0.62 |
| 9 | 354.03 | 5 | 1 | −10.27 | Soluble | 49.57 | 0 | 0.55 | −0.45 |
| 16 | 418.28 | 6 | 0 | −9.26 | Soluble | 49.85 | 0 | 0.55 | −0.69 |
| 17 | 487.17 | 6 | 0 | −7.33 | Poorly Soluble | 49.85 | 0 | 0.55 | 0.05 |
| 18 | 504.16 | 6 | 1 | 0.00 | Moderately Soluble | 103.05 | 1 | 0.55 | −0.27 |
| 19 | 457.15 | 5 | 0 | −7.94 | Moderately Soluble | 40.62 | 0 | 0.55 | −0.01 |
| 20 | 526.04 | 5 | 0 | −6.96 | Poorly Soluble | 40.62 | 1 | 0.55 | −0.44 |
| 21 | 425.11 | 6 | 0 | −9.97 | Moderately Soluble | 49.85 | 0 | 0.55 | −0.32 |
| 22 | 447.14 | 5 | 0 | −7.91 | Poorly Soluble | 40.62 | 0 | 0.55 | −0.23 |
| 23 | 414.07 | 4 | 0 | −1.63 | Moderately Soluble | 23.55 | 0 | 0.55 | −0.19 |
| 24 | 287.79 | 3 | 0 | −8.77 | Moderately Soluble | 6.48 | 0 | 0.55 | −1.15 |
| 25 | 360.25 | 4 | 0 | −9.78 | Moderately Soluble | 23.55 | 0 | 0.55 | −0.70 |
| 26 | 429.14 | 4 | 0 | −8.52 | Poorly Soluble | 23.55 | 0 | 0.55 | 0.17 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pogrebnoi, S.; Radul, O.; Stingaci, E.; Lupascu, L.; Valica, V.; Uncu, L.; Smetanscaia, A.; Petrou, A.; Ćirić, A.; Glamočlija, J.; et al. The Synthesis of Triazolium Salts as Antifungal Agents: A Biological and In Silico Evaluation. Antibiotics 2022, 11, 588. https://doi.org/10.3390/antibiotics11050588
Pogrebnoi S, Radul O, Stingaci E, Lupascu L, Valica V, Uncu L, Smetanscaia A, Petrou A, Ćirić A, Glamočlija J, et al. The Synthesis of Triazolium Salts as Antifungal Agents: A Biological and In Silico Evaluation. Antibiotics. 2022; 11(5):588. https://doi.org/10.3390/antibiotics11050588
Chicago/Turabian StylePogrebnoi, Serghei, Oleg Radul, Eugenia Stingaci, Lucian Lupascu, Vladimir Valica, Livia Uncu, Anastasia Smetanscaia, Anthi Petrou, Ana Ćirić, Jasmina Glamočlija, and et al. 2022. "The Synthesis of Triazolium Salts as Antifungal Agents: A Biological and In Silico Evaluation" Antibiotics 11, no. 5: 588. https://doi.org/10.3390/antibiotics11050588
APA StylePogrebnoi, S., Radul, O., Stingaci, E., Lupascu, L., Valica, V., Uncu, L., Smetanscaia, A., Petrou, A., Ćirić, A., Glamočlija, J., Soković, M., Geronikaki, A., & Macaev, F. Z. (2022). The Synthesis of Triazolium Salts as Antifungal Agents: A Biological and In Silico Evaluation. Antibiotics, 11(5), 588. https://doi.org/10.3390/antibiotics11050588










