Prevalence, Antibiotic-Resistance, and Replicon-Typing of Salmonella Strains among Serovars Mainly Isolated from Food Chain in Marche Region, Italy
Abstract
:1. Introduction
2. Results
2.1. Salmonella Strains
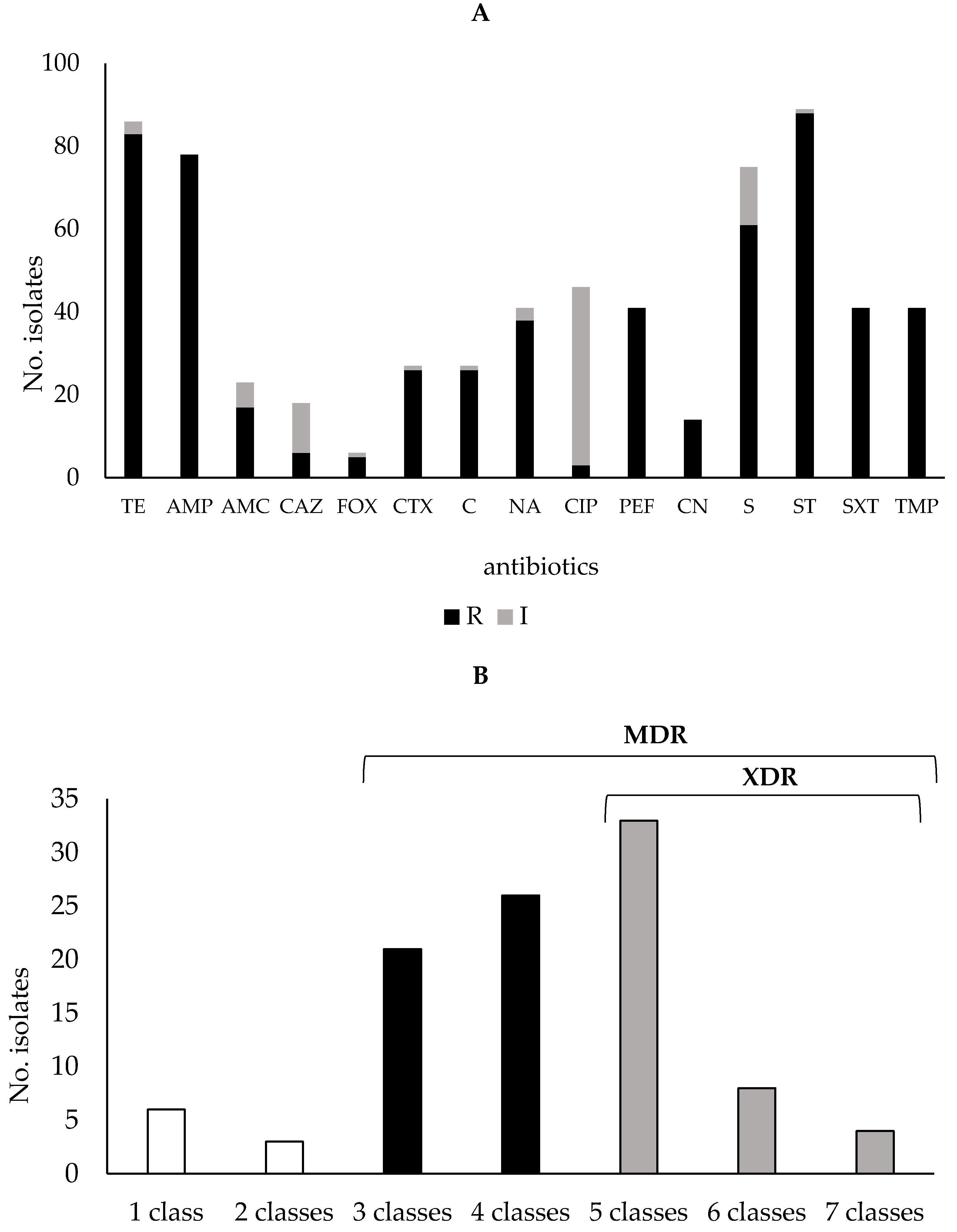
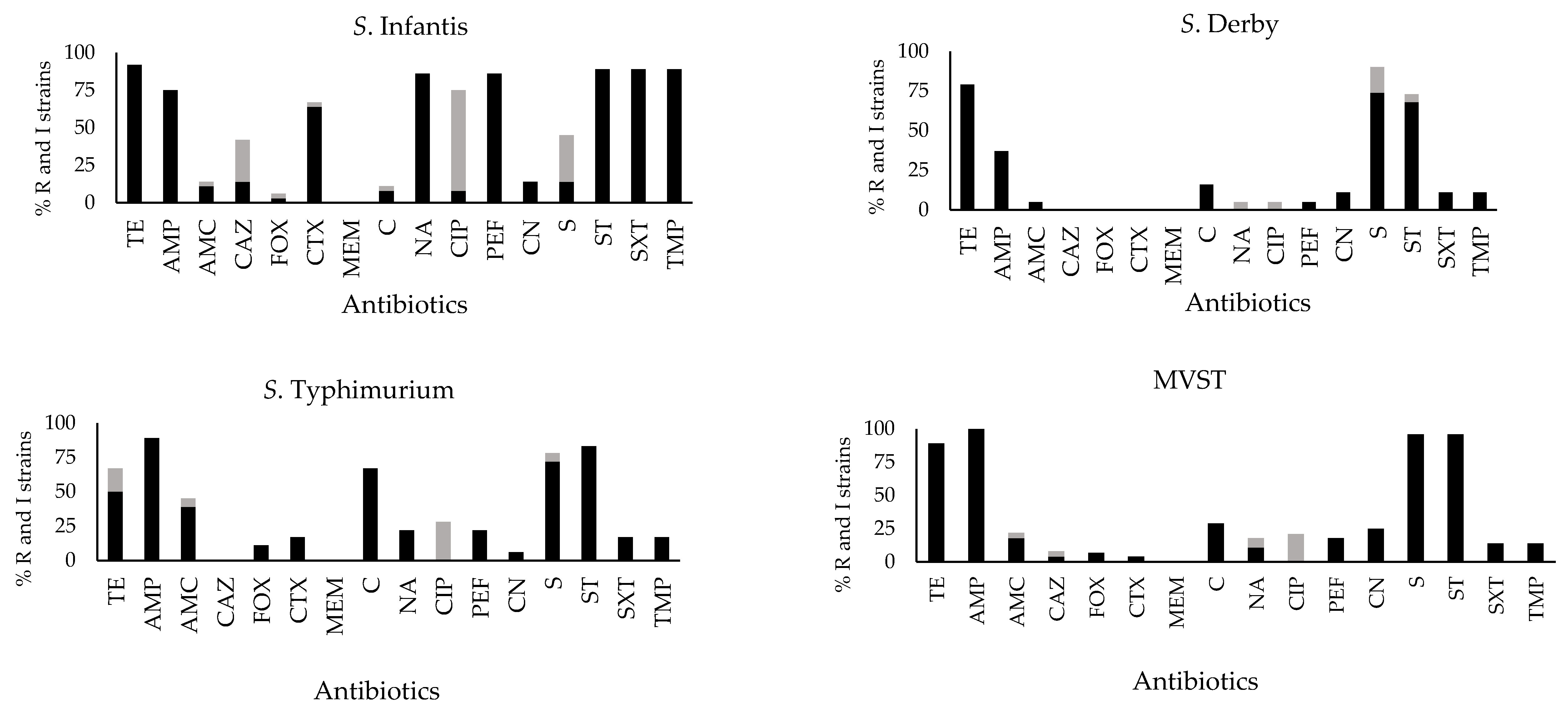
2.2. Antibiotic Susceptibility Profiles
2.3. Tetracycline Resistance Genes
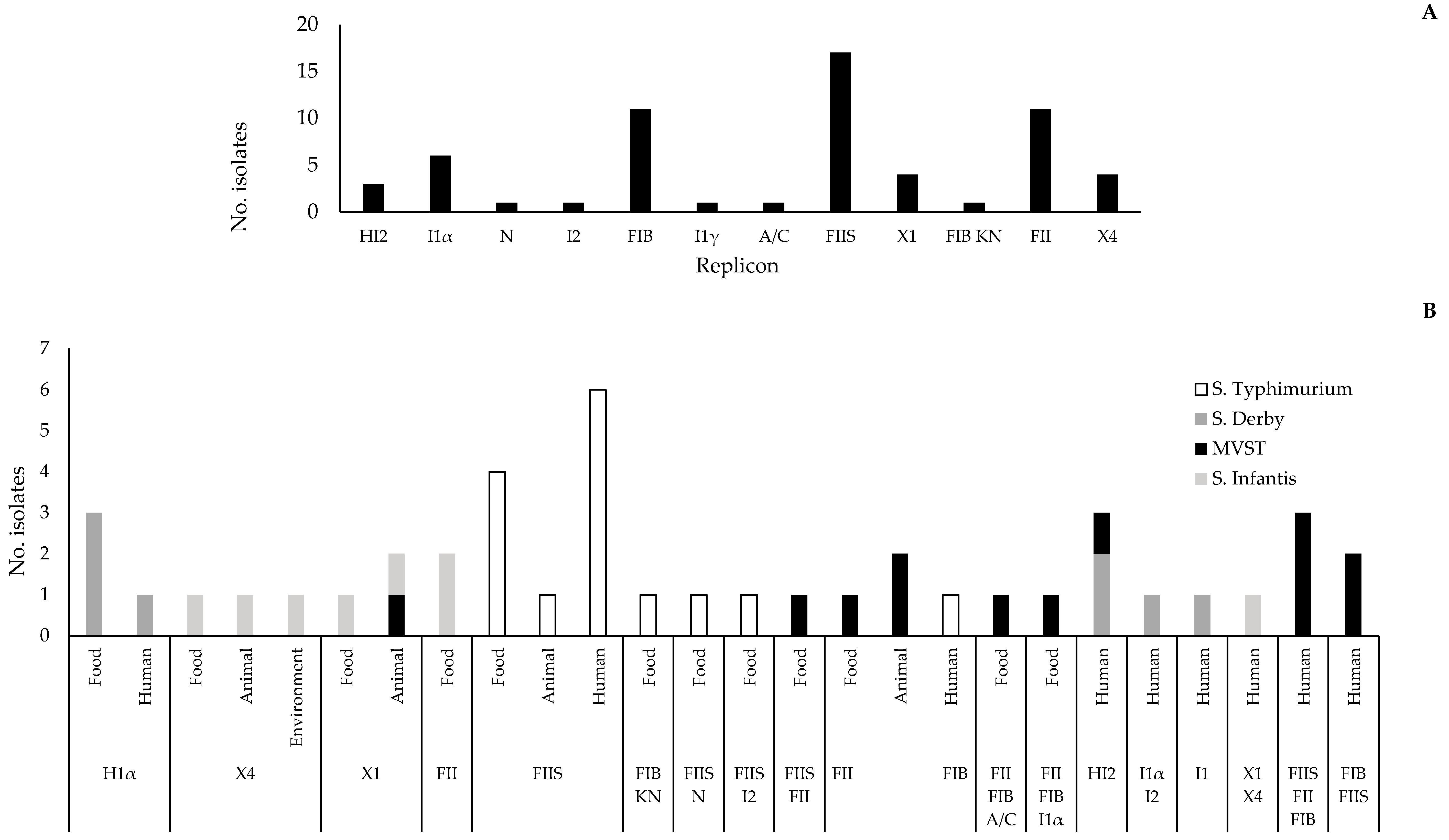
2.4. Replicon Typing
3. Discussion
4. Materials and Methods
4.1. Study Design and Selection of Strains
4.2. Serotyping Analysis and Antibiotic Susceptibility Testing
4.3. Bacterial DNA Extraction and Plasmid Typing
4.4. PCR screening of Tetracycline Resistance Genes
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gal-Mor, O. Persistent Infection and Long-Term Carriage of Typhoidal and Nontyphoidal Salmonellae. Clin. Microbiol. Rev. 2018, 32, e00088-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haselbeck, A.H.; Panzner, U.; Im, J.; Baker, S.; Meyer, C.G.; Marks, F. Current perspectives on invasive nontyphoidal Salmonella disease. Curr. Opin. Infect. Dis. 2017, 30, 498–503. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union One Health 2019 Zoonoses Report. EFSA J. 2021, 19, e06406. [Google Scholar]
- Ehuwa, O.; Jaiswal, A.K.; Jaiswal, S. Salmonella, Food Safety and Food Handling Practices. Foods 2021, 10, 907. [Google Scholar] [CrossRef] [PubMed]
- Keithlin, J.; Sargeant, J.M.; Thomas, M.K.; Fazil, A. Systematic review and meta-analysis of the proportion of non-typhoidal Salmonella cases that develop chronic sequelae. Epidemiol. Infect. 2015, 143, 1333–1351. [Google Scholar] [CrossRef] [Green Version]
- Ao, T.T.; Feasey, N.A.; Gordon, M.A.; Keddy, K.H.; Angulo, F.J.; Crump, J.A. Global burden of invasive nontyphoidal Salmonella disease, 2010. Emerg. Infect. Dis. 2015, 21, 941–949. [Google Scholar] [CrossRef] [Green Version]
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O’Brien, S.J.; International Collaboration on Enteric Disease “Burden of Illness” Studies. The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef] [Green Version]
- V.T. Nair, D.; Venkitanarayanan, K.; Kollanoor Johny, A. Antibiotic-Resistant Salmonella in the Food Supply and the Potential Role of Antibiotic Alternatives for Control. Foods 2018, 7, 167. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Biswas, S.; Paudyal, N.; Pan, H.; Li, X.; Fang, W.; Yue, M. Antibiotic Resistance in Salmonella Typhimurium Isolates Recovered from the Food Chain Through National Antimicrobial Resistance Monitoring System Between 1996 and 2016. Front. Microbiol. 2019, 10, 985. [Google Scholar] [CrossRef] [Green Version]
- Founou, L.L.; Founou, R.C.; Essack, S.Y. Antibiotic Resistance in the Food Chain: A Developing Country-Perspective. Front. Microbiol. 2016, 7, 1881. [Google Scholar] [CrossRef]
- Verraes, C.; Van Boxstael, S.; Van Meervenne, E.; Van Coillie, E.; Butaye, P.; Catry, B.; de Schaetzen, M.-A.; Van Huffel, X.; Imberechts, H.; Dierick, K.; et al. Antimicrobial resistance in the food chain: A review. Int. J. Environ. Res. Public Health 2013, 10, 2643–2669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennani, H.; Mateus, A.; Mays, N.; Eastmure, E.; Stärk, K.D.C.; Häsler, B. Overview of Evidence of Antimicrobial Use and Antimicrobial Resistance in the Food Chain. Antibiotics 2020, 9, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pezzella, C.; Ricci, A.; DiGiannatale, E.; Luzzi, I.; Carattoli, A. Tetracycline and streptomycin resistance genes, transposons, and plasmids in Salmonella enterica isolates from animals in Italy. Antimicrob. Agents Chemother. 2004, 48, 903–908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- European Food Safety Authority (EFSA); Aerts, M.; Battisti, A.; Hendriksen, R.; Kempf, I.; Teale, C.; Tenhagen, B.; Veldman, K.; Wasyl, D.; Guerra, B.; et al. A Technical specifications on harmonised monitoring of antimicrobial resistance in zoonotic and indicator bacteria from food-producing animals and food. EFSA J. 2019, 17, e05709. [Google Scholar]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Monnet, D.L. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Staffolani, M.; Medici, L.; Napoleoni, M.; Fisichella, S. Data on isolates of enteric bacteria from human clinical cases, animals, food and environment samples, in the year 2015 in Marche Region (Italy). Sanità Pubblica Vet. 2016, 95. Available online: http://www.spvet.it/archivio/numero-95/626.html (accessed on 9 May 2022).
- Staffolani, M.; Medici, L.; Napoleoni, M.; Fisichella, S. Data on isolates of enteric bacteria from human clinical cases, animals, food and environment samples, in the year 2016 in Marche Region (Italy). Sanità Pubblica Vet. 2017, 101. Available online: http://www.spvet.it/archivio/numero-101/660.html (accessed on 9 May 2022).
- Napoleoni, M.; Medici, L.; Staffolani, M.; Fisichella, S. Data on isolates of enteric bacteria from human clinical cases, animals, food and environment samples, in the year 2017 in Marche Region (Italy). Sanità Pubblica Vet. 2018, 110. Available online: http://www.spvet.it/archivio/numero-110/688.html (accessed on 9 May 2022).
- Napoleoni, M.; Medici, L.; Staffolani, M.; Fisichella, S. Data on isolates of enteric bacteria from human clinical cases, animals, food and environment samples in the year 2018 in Marche Region (Italy). Sanità Pubblica Vet. 2019, 114. Available online: http://spvet.it/archivio/numero-114/697.html (accessed on 9 May 2022).
- Napoleoni, M.; Staffolani, M.; Pazzaglia, I.; Fisichella, S. Data on isolates of enteric bacteria from human clinical cases, animals, food and environment samples in the year 2019 in Marche Region (Italy). Sanità Pubblica Vet. 2020, 118. Available online: http://spvet.it/archivio/numero-118/705.html (accessed on 9 May 2022).
- Napoleoni, M.; Silenzi, V.; Staffolani, M.; Guidi, F.; Blasi, G.; Fisichella, S.; Rocchegiani, E. Data on isolates of enteric bacteria from human clinical cases, animals, food and environment samples in the year 2020 in Marche Region (Italy). Sanità Pubblica Vet. 2021, 125. Available online: http://www.spvet.it/archivio/numero-125/718.html (accessed on 9 May 2022).
- Leati, M.; Cibin, V.; Mancin, M.; Pestelli, P.; Barco, L. Enter-Vet Report Dati 2019; Centro di Referenza Nazionale per le Salmonellosi; Istituto Zooprofilattico Sperimentale delle Venezie: Legnaro (PD), Italy, 2022; pp. 1–38. [Google Scholar]
- Ferrari, R.G.; Rosario, D.K.A.; Cunha-Neto, A.; Mano, S.B.; Figueiredo, E.E.S.; Conte-Junior, C.A. Worldwide Epidemiology of Salmonella Serovars in Animal-Based Foods: A Meta-analysis. Appl. Environ. Microbiol. 2019, 85, e00591-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crim, S.M.; Iwamoto, M.; Huang, J.Y.; Griffin, P.M.; Gilliss, D.; Cronquist, A.B.; Cartter, M.; Tobin-D’Angelo, M.; Blythe, D.; Smith, K.; et al. Incidence and Trends of Infection with Pathogens Transmitted Commonly Through Food—Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2006–2013. MMWR Morb. Mortal. Wkly. Rep. 2020, 63, 328–332. [Google Scholar]
- Gomes-Neves, E.; Antunes, P.; Tavares, A.; Themudo, P.; Cardoso, M.F.; Gärtner, F.; Costa, J.M.; Peixe, L. Salmonella cross-contamination in swine abattoirs in Portugal: Carcasses, meat and meat handlers. Int. J. Food Microbiol. 2012, 157, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, R.G.; Panzenhagen, P.H.N.; Conte-Junior, C.A. Phenotypic and Genotypic Eligible Methods for Salmonella Typhimurium Source Tracking. Front. Microbiol. 2017, 8, 2587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.; Yan, M.; Liu, L.; Lai, J.; Chan, E.W.; Chen, S. Comparative characterization of nontyphoidal Salmonella isolated from humans and food animals in China, 2003–2011. Heliyon 2018, 4, e00613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerouanton, A.; Rose, V.; Weill, F.X.; Granier, S.A.; Denis, M. Genetic diversity and antimicrobial resistance profiles of Salmonella enterica serotype derby isolated from pigs, pork, and humans in France. Foodborne Pathog. Dis. 2013, 10, 977–984. [Google Scholar] [CrossRef] [Green Version]
- Alba, P.; Leekitcharoenphon, P.; Carfora, V.; Amoruso, R.; Cordaro, G.; Di Matteo, P.; Engage-Eurl-Ar Network Study Group. Molecular epidemiology of Salmonella Infantis in Europe: Insights into the success of the bacterial host and its parasitic pESI-like megaplasmid. Microb. Genom. 2020, 6, e000365. [Google Scholar] [CrossRef]
- Mughini-Gras, L.; van Hoek, A.H.; Cuperus, T.; Dam-Deisz, C.; van Overbeek, W.; van den Beld, M.; Wit, B.; Rapallini, M.; Wullings, B.; Franz, E.; et al. Prevalence, risk factors and genetic traits of Salmonella Infantis in Dutch broiler flocks. Vet Microbiol. 2021, 258, 109120. [Google Scholar] [CrossRef]
- Franco, A.; Leekitcharoenphon, P.; Feltrin, F.; Alba, P.; Cordaro, G.; Iurescia, M.; Battisti, A. Emergence of a Clonal Lineage of Multidrug-Resistant ESBL-Producing Salmonella Infantis Transmitted from Broilers and Broiler Meat to Humans in Italy between 2011 and 2014. PLoS ONE 2015, 10, e0144802. [Google Scholar] [CrossRef] [Green Version]
- Dionisi, A.M.; Lucarelli, C.; Benedetti, I.; Owczarek, S.; Luzzi, I. Molecular characterisation of multidrug-resistant Salmonella enterica serotype Infantis from humans, animals and the environment in Italy. Int. J. Antimicrob. Agents 2011, 38, 384–389. [Google Scholar] [CrossRef] [PubMed]
- Elnekave, E.; Hong, S.L.; Lim, S.; Johnson, T.J.; Perez, A.; Alvarez, J. Comparing serotyping with whole-genome sequencing for subtyping of non-typhoidal Salmonella enterica: A large-scale analysis of 37 serotypes with a public health impact in the USA. Microb. Genom. 2020, 6, mgen000425. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union Summary Report on Antimicrobial Resistance in Zoonotic and Indicator Bacteria from Humans, Animals and Food in 2018/2019. EFSA J. 2021, 19, e06490. [Google Scholar]
- Frech, G.; Schwarz, S. Molecular analysis of tetracycline resistance in Salmonella enterica subsp. enterica serovars Typhimurium, enteritidis, Dublin, Choleraesuis, Hadar and Saintpaul: Construction and application of specific gene probes. J. Appl. Microbiol. 2000, 89, 633–641. [Google Scholar] [CrossRef]
- European Medicines Agency; European Surveillance of Veterinary Antimicrobial Consumption. Sales of Veterinary Antimicrobial Agents in 31 European Countries in 2019 and 2020; Eleventh ESVAC Report EMA/58183/2021; 2021; Available online: https://www.ema.europa.eu/en/documents/report/sales-veterinary-antimicrobial-agents-31-european-countries-2019-2020-trends-2010-2020-eleventh_en.pdf (accessed on 9 May 2022).
- WHO. Critically Important Antimicrobials for Human Medicine, 6th Revision. 2019. Available online: https://www.who.int/foodsafety/publications/antimicrobials-sixth/en/https://www.who.int/publications/i/item/9789241515528 (accessed on 9 May 2022).
- European Union (EU). Commission Notice—Guidelines for the prudent use of antimicrobials in veterinary medicine (2015/C 299/04). Off. J. Eur. Union 2015, 58, 7–26. Available online: https://ec.europa.eu/health/system/files/2016-11/2015_prudent_use_guidelines_en_0.pdf (accessed on 9 May 2022).
- Gargano, V.; Sciortino, S.; Gambino, D.; Costa, A.; Agozzino, V.; Reale, S.; Vicari, D. Antibiotic Susceptibility Profile and Tetracycline Resistance Genes Detection in Salmonella spp. Strains Isolated from Animals and Food. Antibiotics 2021, 10, 809. [Google Scholar] [CrossRef]
- Bacci, C.; Lanzoni, E.; Vismarra, A.; Alpigiani, I.; Nuvoloni, R.; Bonardi, S.; Brindani, F. Antibiotic resistance and resistance genes in Salmonella enterica isolated from pork meat and pig carcasses in Northern Italy. Large Animal. Rev. 2014, 20, 201–207. [Google Scholar]
- García-Soto, S.; Abdel-Glil, M.Y.; Tomaso, H.; Linde, J.; Methner, U. Emergence of Multidrug-Resistant Salmonella enterica Subspecies enterica Serovar Infantis of Multilocus Sequence Type 2283 in German Broiler Farms. Front. Microbiol. 2020, 11, 1741. [Google Scholar] [CrossRef]
- Grossman, T.H. Tetracycline Antibiotics and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025387. [Google Scholar] [CrossRef] [Green Version]
- Lauteri, C.; Maggio, F.; Serio, A.; Festino, A.R.; Paparella, A.; Vergara, A. Overcoming Multidrug Resistance in Salmonella spp. Isolates Obtained From the Swine Food Chain by Using Essential Oils: An in vitro Study. Front. Microbiol. 2022, 12, 808286. [Google Scholar] [CrossRef]
- Peruzy, M.F.; Capuano, F.; Proroga, Y.T.R.; Cristiano, D.; Carullo, M.R.; Murru, N. Antimicrobial Susceptibility Testing for Salmonella Serovars Isolated from Food Samples: Five-Year Monitoring (2015–2019). Antibiotics 2020, 9, 365. [Google Scholar] [CrossRef]
- Pavelquesi, S.L.S.; de Oliveira Ferreira, A.C.A.; Rodrigues, A.R.; de Souza Silva, C.M.; Orsi, D.C.; da Silva, I.C.R. Presence of Tetracycline and Sulfonamide Resistance Genes in Salmonella spp.: Literature Review. Antibiotics 2021, 10, 1314. [Google Scholar] [CrossRef] [PubMed]
- Camarda, A.; Pugliese, N.; Pupillo, A.; Oliva, M.; Circella, E.; Dionisi, A.M.; Ricci, A.; Legretto, M.; Caroli, A.; Pazzani, C. Resistance genes, phage types and pulsed field gel electrophoresis pulsotypes in Salmonella enterica strains from laying hen farms in southern Italy. Int. J. Environ. Res. Public Health 2013, 10, 3347–3362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tawyabur, M.; Islam, M.; Sobur, M.; Hossain, M.; Mahmud, M.; Paul, S.; Rahman, M. Isolation and Characterization of Multidrug-Resistant Escherichia coli and Salmonella spp. from Healthy and Diseased Turkeys. Antibiotics 2020, 9, 770. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A. Plasmids in Gram negatives: Molecular typing of resistance plasmids. Int. J. Med. Microbiol. 2011, 301, 654–658. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 2009, 53, 2227–2238. [Google Scholar] [CrossRef] [Green Version]
- Oluwadare, M.; Lee, M.D.; Grim, C.J.; Lipp, E.K.; Cheng, Y.; Maurer, J.J. The Role of the Salmonella spvB IncF Plasmid and Its Resident Entry Exclusion Gene traS on Plasmid Exclusion. Front. Microbiol. 2020, 11, 949. [Google Scholar] [CrossRef]
- Di Cesare, A.; Losasso, C.; Barco, L.; Eckert, E.M.; Conficoni, D.; Sarasini, G.; Corno, G.; Ricci, A. Diverse distribution of Toxin-Antitoxin II systems in Salmonella enterica serovars. Sci. Rep. 2016, 6, 28759. [Google Scholar] [CrossRef] [Green Version]
- European Union. Commission Regulation (EC) No 2073/2005 of 15 November 2005 on Microbiological Criteria for Foodstuffs. Off. J. Eur. Union 2005, 50, 1–26. Available online: https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX:32005R2073 (accessed on 9 May 2022).
- ISO 6579-1:2017/A1:2020; Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 1: Detection of Salmonella spp.—Amendment 1 Broader Range of Incubation Temperatures, Amendment to the Status of Annex D, and Correction of the Composition. International Organization for Standardization: Geneva, Switzerland, 2020.
- Italian Ministry of Health. National Control Plan for Salmonellosis in Poultry 2019–2021 (Piano Nazionale di Controllo delle Salmonellosi negli Avicoli 2019–2021). Directorate General for Animal Health and Veterinary Medicines of the Italian Ministry of Health. 2018. Available online: https://www.salute.gov.it/imgs/C_17_pubblicazioni_2849_allegato.pdf (accessed on 9 May 2022).
- ISO/TR 6579-3:2014; Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 3: Guidelines for Serotyping of Salmonella spp. International Organization for Standardization: Geneva, Switzerland, 2014.
- Jarlier, V.; Nicolas, M.H.; Fournier, G.; Philippon, A. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in enterobacteriaceae: Hospital prevalence and susceptibility patterns. Clin. Infect. Dis. 1988, 10, 867–878. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 32nd ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022. [Google Scholar]
- Barbadoro, P.; Bencardino, D.; Carloni, E.; Omiccioli, E.; Ponzio, E.; Micheletti, R.; Andreoni, F. Carriage of carbapenem-resistant enterobacterales in adult patients admitted to a university hospital in italy. Antibiotics 2021, 10, 61. [Google Scholar] [CrossRef]
- European Reference Laboratory for Antimicrobial Resistance (EURL-AR, Technical University of Denmark, National Food Institute). List of Primers for Detection of Antimicrobial Resistance Genes. Available online: https://www.eurl-ar.eu/CustomerData/Files/Folders/25-resourcer/459_primerliste-til-web-07-11-2018.pdf (accessed on 9 May 2022).
- Sengeløv, G.; Agersø, Y.; Halling-Sørensen, B.; Baloda, S.B.; Andersen, J.S.; Jensen, L.B. Bacterial antibiotic resistance levels in Danish farmland as a result of treatment with pig manure slurry. Environ. Int. 2003, 28, 587–595. [Google Scholar] [CrossRef]
- Miranda, C.D.; Kehrenberg, C.; Ulep, C.; Schwarz, S.; Roberts, M.C. Diversity of tetracycline resistance genes in bacteria from Chilean salmon farms. Antimicrob. Agents Chemother. 2003, 47, 883–888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agersø, Y.; Sandvang, D. Class 1 integrons and tetracycline resistance genes in Alcaligenes, Arthrobacter, and Pseudomonas spp. isolated from pigsties and manured soil. Appl. Environ. Microbiol. 2005, 71, 7941–7947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Source | Serovars | Strain n. |
|---|---|---|
| Animals | S. Infantis | 8 |
| S. Derby | 1 | |
| S. Typhimurium | 1 | |
| MVST | 6 | |
| Environment | S. Infantis | 5 |
| S. Derby | 1 | |
| S. Typhimurium | 0 | |
| MVST | 3 | |
| Foods | S. Infantis | 11 |
| S. Derby | 8 | |
| S. Typhimurium | 7 | |
| MVST | 9 | |
| Humans | S. Infantis | 12 |
| S. Derby | 10 | |
| S. Typhimurium | 10 | |
| MVST | 10 | |
| Total | 102 |
| tet Genes | Resistant Strains | Intermediate Strains |
|---|---|---|
| tetA | 10 2 S. Derby, 7 S. Infantis, 1 S. Typhimurium | 0 |
| tetB | 27 1 S. Derby, 24 MVST, 2 S. Typhimurium | 0 |
| tetC | 0 | 0 |
| tetD | 0 | 0 |
| tetG | 3 S. Typhimurium | 2 S. Typhimurium |
| tetM | 1 MVST | 0 |
| Target Gene | Primer Sequence | Amplicon Size (bp) | Tm (°C) | Ref. |
|---|---|---|---|---|
| tetA | 5′-GTAATTCTGAGCACTGTCGC-3′ | 956 | 57 | [60] |
| 5′-CTGCCTGGACAACATTGCTT-3′ | ||||
| tetB | 5′-CTCAGTATTCCAAGCCTTTG-3′ | 414 | 52 | [60] |
| 5′-ACTCCCCTGAGCTTGAGGGG-3′ | ||||
| tetC | 5′-GGTTGAAGGCTCTCAAGGGC-3′ | 505 | 65 | [60] |
| 5′-CCTCTTGCGGGATATCGTCC-3′ | ||||
| tetD | 5′-CATCCATCCGGAAGTGATAGC-3′ | 436 | 57 | [61] |
| 5′-GGATATCTCACCGCATCTGC-3′ | ||||
| tetG | 5′-GCAGCGAAAGCGTATTTGCG-3′ | 662 | 62 | [62] |
| 5′-TCCGAAAGCTGTCCAAGCAT-3′ | ||||
| tetM | 5′-GTTAAATAGTGTTCTTGGAG-3′ | 657 | 45 | [62] |
| 5′-CTAAGATATGGCTCTAACAA-3′ |
| Species | Strain Name | tet Gene |
|---|---|---|
| E. coli | tetA, NCTC 50078 | tetA |
| E. coli | tetB, CSH50::Tn10 | tetB |
| E. coli | tetC DO7 pBR 322, Tet | tetC |
| E. coli | tetD C600 psl 106, Tet tetDx2 | tetD |
| S. Typhimurium | P502212 DT104 sul1 and tetG | tetG |
| Staph. intermedius | 2567, chromosomal tetM | tetM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Russo, I.; Bencardino, D.; Napoleoni, M.; Andreoni, F.; Schiavano, G.F.; Baldelli, G.; Brandi, G.; Amagliani, G. Prevalence, Antibiotic-Resistance, and Replicon-Typing of Salmonella Strains among Serovars Mainly Isolated from Food Chain in Marche Region, Italy. Antibiotics 2022, 11, 725. https://doi.org/10.3390/antibiotics11060725
Russo I, Bencardino D, Napoleoni M, Andreoni F, Schiavano GF, Baldelli G, Brandi G, Amagliani G. Prevalence, Antibiotic-Resistance, and Replicon-Typing of Salmonella Strains among Serovars Mainly Isolated from Food Chain in Marche Region, Italy. Antibiotics. 2022; 11(6):725. https://doi.org/10.3390/antibiotics11060725
Chicago/Turabian StyleRusso, Ilaria, Daniela Bencardino, Maira Napoleoni, Francesca Andreoni, Giuditta Fiorella Schiavano, Giulia Baldelli, Giorgio Brandi, and Giulia Amagliani. 2022. "Prevalence, Antibiotic-Resistance, and Replicon-Typing of Salmonella Strains among Serovars Mainly Isolated from Food Chain in Marche Region, Italy" Antibiotics 11, no. 6: 725. https://doi.org/10.3390/antibiotics11060725
APA StyleRusso, I., Bencardino, D., Napoleoni, M., Andreoni, F., Schiavano, G. F., Baldelli, G., Brandi, G., & Amagliani, G. (2022). Prevalence, Antibiotic-Resistance, and Replicon-Typing of Salmonella Strains among Serovars Mainly Isolated from Food Chain in Marche Region, Italy. Antibiotics, 11(6), 725. https://doi.org/10.3390/antibiotics11060725







