Proteomic Comparison of Ivermectin Sensitive and Resistant Staphylococcus aureus Clinical Isolates Reveals Key Efflux Pumps as Possible Resistance Determinants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates and Resistance Classification
2.2. Effects of IVM on Bacterial Growth
2.3. Time Kill-Kinetics
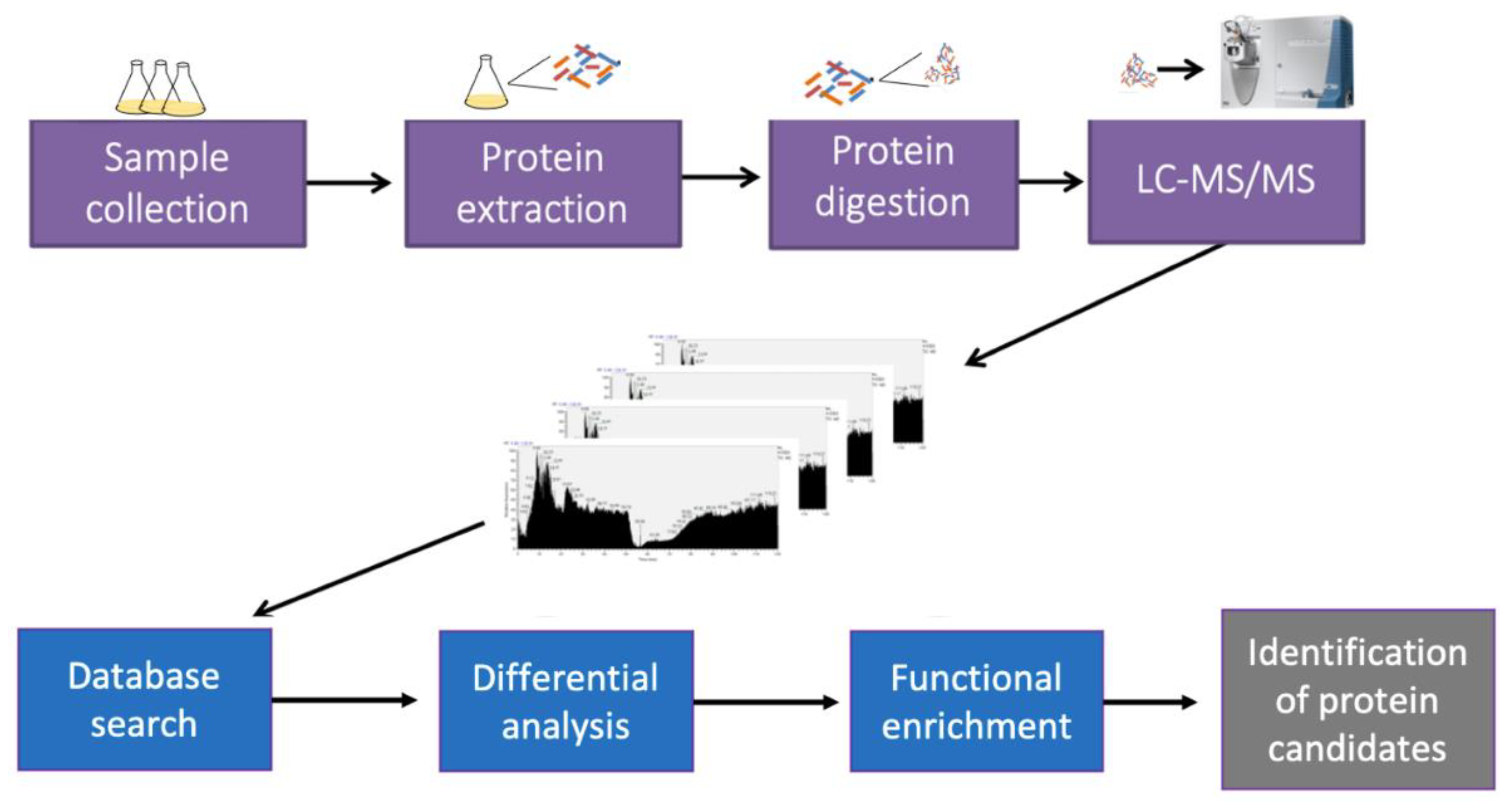
2.4. Protein Extraction and Digestion
2.5. Mass Spectrometry Analysis (2D LC−MS/MS)
2.6. Database Searching
2.7. Bioinformatics
2.8. Extraction of RNA
2.9. Reverse Transcription and Quantitative PCR
3. Results
3.1. Inhibition of S. aureus Growth by IVM as Evidenced by Growth Curves and Time-Kill Kinetics
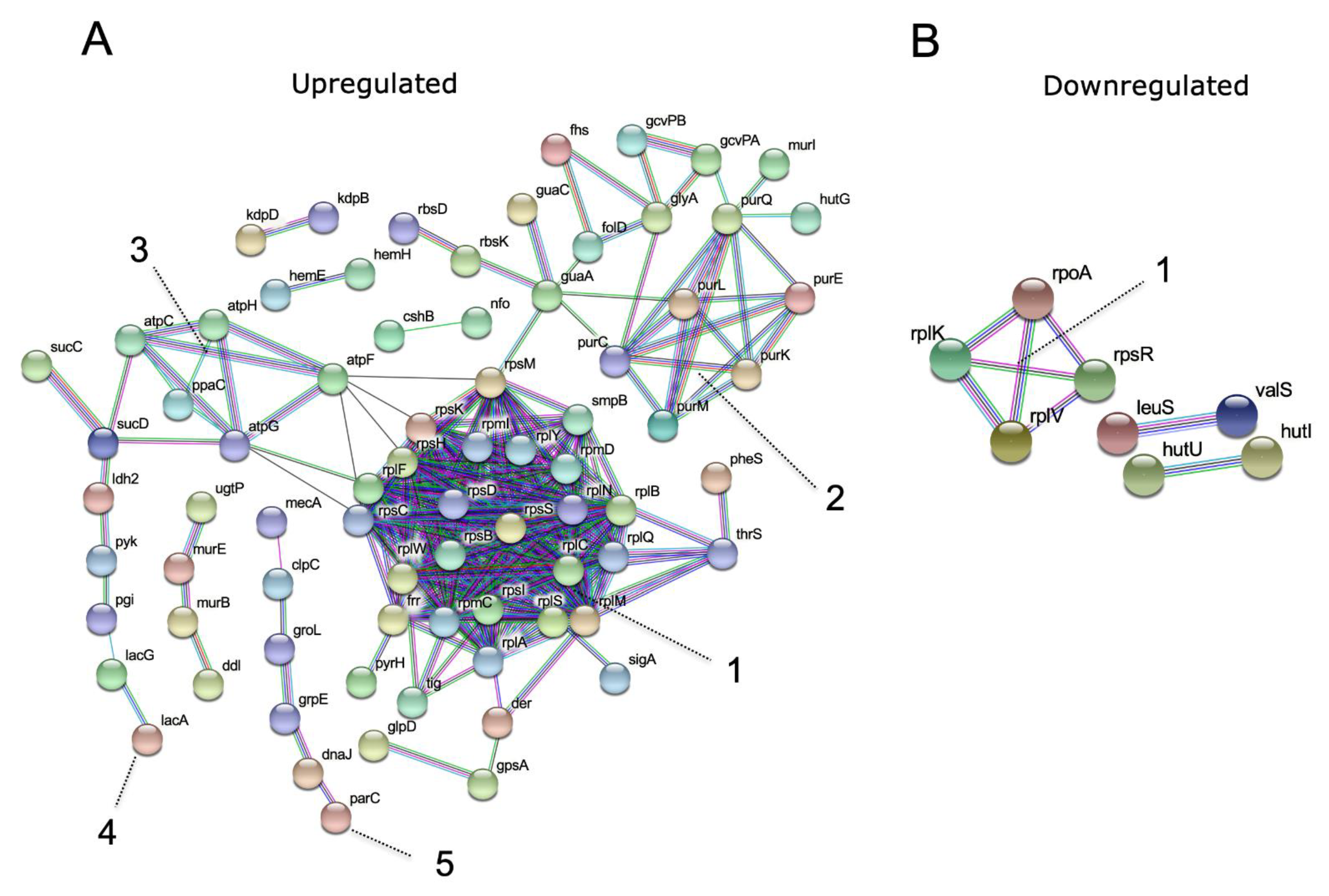
3.2. Proteomic Analysis
3.3. Validation of the Overexpressed Bacterial Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Wolstenholme, A.; Rogers, A. Glutamate-gated chloride channels and the mode of action of the avermectin/milbemycin anthelmintics. Parasitology 2005, 131, S85. [Google Scholar] [CrossRef]
- Geary, T.G. Ivermectin 20 years on: Maturation of a wonder drug. Trends Parasitol. 2005, 21, 530–532. [Google Scholar] [CrossRef]
- Kobylinski, K.C.; Foy, B.D.; Richardson, J.H. Ivermectin inhibits the sporogony of Plasmodium falciparum in Anopheles gambiae. Malar. J. 2012, 11, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashraf, S.; Chaudhry, U.; Raza, A.; Ghosh, D.; Zhao, X. In vitro activity of ivermectin against Staphylococcus aureus clinical isolates. Antimicrob. Resist. Infect. Control 2018, 7, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Mastrangelo, E.; Pezzullo, M.; De Burghgraeve, T.; Kaptein, S.; Pastorino, B.; Dallmeier, K.; de Lamballerie, X.; Neyts, J.; Hanson, A.M.; Frick, D.N. Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: New prospects for an old drug. J. Antimicrob. Chemother. 2012, 67, 1884–1894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashraf, S.; Beech, R.N.; Hancock, M.A.; Prichard, R.K. Ivermectin binds to Haemonchus contortus tubulins and promotes stability of microtubules. Int. J. Parasitol. 2015, 45, 647–654. [Google Scholar] [CrossRef]
- Ashraf, S.; Mani, T.; Beech, R.; Prichard, R. Macrocyclic lactones and their relationship to the SNPs related to benzimidazole resistance. Mol. Biochem. Parasitol. 2015, 201, 128–134. [Google Scholar] [CrossRef]
- Ashraf, S.; Prichard, R. Ivermectin exhibits potent anti-mitotic activity. Vet. Parasitol. 2016, 226, 1–4. [Google Scholar] [CrossRef]
- Sharmeen, S.; Skrtic, M.; Sukhai, M.A.; Hurren, R.; Gronda, M.; Wang, X.; Fonseca, S.B.; Sun, H.; Wood, T.E.; Ward, R. The antiparasitic agent ivermectin induces chloride-dependent membrane hyperpolarization and cell death in leukemia cells. Blood J. Am. Soc. Hematol. 2010, 116, 3593–3603. [Google Scholar] [CrossRef] [Green Version]
- Lim, L.E.; Vilchèze, C.; Ng, C.; Jacobs Jr, W.R.; Ramón-García, S.; Thompson, C.J. Anthelmintic avermectins kill Mycobacterium tuberculosis, including multidrug-resistant clinical strains. Antimicrob. Agents Chemother. 2013, 57, 1040–1046. [Google Scholar] [CrossRef] [Green Version]
- Omansen, T.F.; Porter, J.L.; Johnson, P.D.; van der Werf, T.S.; Stienstra, Y.; Stinear, T.P. In-vitro activity of avermectins against Mycobacterium ulcerans. PLoS Negl. Trop. Dis. 2015, 9, e0003549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chambers, H.F.; DeLeo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riekerink, R.O.; Barkema, H.; Kelton, D.; Scholl, D. Incidence rate of clinical mastitis on Canadian dairy farms. J. Dairy Sci. 2008, 91, 1366–1377. [Google Scholar] [CrossRef] [PubMed]
- Jevons, M.P.; Coe, A.; Parker, M. Methicillin resistance in staphylococci. Lancet 1963, 7287, 904–907. [Google Scholar] [CrossRef]
- Grundmann, H.; Aanensen, D.M.; Van Den Wijngaard, C.C.; Spratt, B.G.; Harmsen, D.; Friedrich, A.W.; Group, E.S.R.L.W. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: A molecular-epidemiological analysis. PLoS Med. 2010, 7, e1000215. [Google Scholar] [CrossRef] [Green Version]
- Dashtbani-Roozbehani, A.; Brown, M.H. Efflux Pump Mediated Antimicrobial Resistance by Staphylococci in Health-Related Environments: Challenges and the Quest for Inhibition. Antibiotics 2021, 10, 1502. [Google Scholar] [CrossRef]
- Lindqvist, R. Estimation of Staphylococcus aureus growth parameters from turbidity data: Characterization of strain variation and comparison of methods. Appl. Environ. Microbiol. 2006, 72, 4862–4870. [Google Scholar] [CrossRef] [Green Version]
- McKay, G.A.; Beaulieu, S.; Arhin, F.F.; Belley, A.; Sarmiento, I.; Parr, T., Jr.; Moeck, G. Time-kill kinetics of oritavancin and comparator agents against Staphylococcus aureus, Enterococcus faecalis and Enterococcus faecium. J. Antimicrob. Chemother. 2009, 63, 1191–1199. [Google Scholar] [CrossRef] [Green Version]
- Schallenberger, M.A.; Niessen, S.; Shao, C.; Fowler, B.J.; Romesberg, F.E. Type I signal peptidase and protein secretion in Staphylococcus aureus. J. Bacteriol. 2012, 194, 2677–2686. [Google Scholar] [CrossRef]
- Kislinger, T.; Gramolini, A.O.; MacLennan, D.H.; Emili, A. Multidimensional protein identification technology (MudPIT): Technical overview of a profiling method optimized for the comprehensive proteomic investigation of normal and diseased heart tissue. J. Am. Soc. Mass Spectrom. 2005, 16, 1207–1220. [Google Scholar] [CrossRef] [Green Version]
- Sivagnanam, K.; Raghavan, V.G.; Shah, M.; Hettich, R.L.; Verberkmoes, N.C.; Lefsrud, M.G. Comparative shotgun proteomic analysis of Clostridium acetobutylicum from butanol fermentation using glucose and xylose. Proteome Sci. 2011, 9, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shteynberg, D.; Nesvizhskii, A.I.; Moritz, R.L.; Deutsch, E.W. Combining results of multiple search engines in proteomics. Mol. Cell. Proteom. 2013, 12, 2383–2393. [Google Scholar] [CrossRef] [Green Version]
- McCormack, A.L.; Eng, J.K.; Yates III, J.R. Peptide sequence analysis on quadrupole mass spectrometers. Methods 1994, 6, 274–283. [Google Scholar] [CrossRef]
- Dorfer, V.; Pichler, P.; Stranzl, T.; Stadlmann, J.; Taus, T.; Winkler, S.; Mechtler, K. MS Amanda, a universal identification algorithm optimized for high accuracy tandem mass spectra. J. Proteome Res. 2014, 13, 3679–3684. [Google Scholar] [CrossRef] [PubMed]
- Ishihama, Y.; Oda, Y.; Tabata, T.; Sato, T.; Nagasu, T.; Rappsilber, J.; Mann, M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteom. 2005, 4, 1265–1272. [Google Scholar] [CrossRef] [Green Version]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote) omics data. Nat. Methods 2016, 13, 731. [Google Scholar] [CrossRef]
- Huntley, R.P.; Sawford, T.; Mutowo-Meullenet, P.; Shypitsyna, A.; Bonilla, C.; Martin, M.J.; O’Donovan, C. The GOA database: Gene ontology annotation updates for 2015. Nucleic Acids Res. 2015, 43, D1057–D1063. [Google Scholar] [CrossRef]
- Ogawara, H. Comparison of antibiotic resistance mechanisms in antibiotic-producing and pathogenic bacteria. Molecules 2019, 24, 3430. [Google Scholar] [CrossRef] [Green Version]
- Tsuge, K.; Ohata, Y.; Shoda, M. Gene yerP, involved in surfactin self-resistance in Bacillus subtilis. Antimicrob. Agents Chemother. 2001, 45, 3566–3573. [Google Scholar] [CrossRef] [Green Version]
- Altman, D.R.; Sullivan, M.J.; Chacko, K.I.; Balasubramanian, D.; Pak, T.R.; Sause, W.E.; Kumar, K.; Sebra, R.; Deikus, G.; Attie, O. Genome plasticity of agr-defective Staphylococcus aureus during clinical infection. Infect. Immun. 2018, 86, e00331-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomlinson, K.L.; Lung, T.W.F.; Dach, F.; Annavajhala, M.K.; Gabryszewski, S.J.; Groves, R.A.; Drikic, M.; Francoeur, N.J.; Sridhar, S.H.; Smith, M.L. Staphylococcus aureus induces an itaconate-dominated immunometabolic response that drives biofilm formation. Nat. Commun. 2021, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Reverdy, A.; Chen, Y.; Hunter, E.; Gozzi, K.; Chai, Y. Protein lysine acetylation plays a regulatory role in Bacillus subtilis multicellularity. PLoS ONE 2018, 13, e0204687. [Google Scholar] [CrossRef]
- Hinchliffe, P.; Greene, N.P.; Paterson, N.G.; Crow, A.; Hughes, C.; Koronakis, V. Structure of the periplasmic adaptor protein from a major facilitator superfamily (MFS) multidrug efflux pump. FEBS Lett. 2014, 588, 3147–3153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Ma, Y.; Liu, P.; Li, X. Multidrug resistance operon emrAB contributes for chromate and ampicillin co-resistance in a Staphylococcus strain isolated from refinery polluted river bank. SpringerPlus 2016, 5, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Omote, H.; Hiasa, M.; Matsumoto, T.; Otsuka, M.; Moriyama, Y. The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol. Sci. 2006, 27, 587–593. [Google Scholar] [CrossRef]
- Kaatz, G.W.; McAleese, F.; Seo, S.M. Multidrug resistance in Staphylococcus aureus due to overexpression of a novel multidrug and toxin extrusion (MATE) transport protein. Antimicrob. Agents Chemother. 2005, 49, 1857–1864. [Google Scholar] [CrossRef] [Green Version]
- Tan, X.; Xie, H.; Zhang, B.; Zhou, J.; Dou, Z.; Wang, X.; Wang, N. A Novel Ivermectin-Derived Compound D4 and Its Antimicrobial/Biofilm Properties against MRSA. Antibiotics 2021, 10, 208. [Google Scholar] [CrossRef]
- Raza, A.; Kopp, S.R.; Bagnall, N.H.; Jabbar, A.; Kotze, A.C. Effects of in vitro exposure to ivermectin and levamisole on the expression patterns of ABC transporters in Haemonchus contortus larvae. Int. J. Parasitol. Drugs Drug Resist. 2016, 6, 103–115. [Google Scholar] [CrossRef] [Green Version]
- Raza, A.; Qamar, A.G.; Hayat, K.; Ashraf, S.; Williams, A.R. Anthelmintic resistance and novel control options in equine gastrointestinal nematodes. Parasitology 2019, 146, 425–437. [Google Scholar] [CrossRef]
- Baraka, O.Z.; Mahmoud, B.M.; Marschke, C.K.; Geary, T.G.; Homeida, M.M.; Williams, J.F. Ivermectin distribution in the plasma and tissues of patients infected with Onchocerca volvulus. Eur. J. Clin. Pharmacol. 1996, 50, 407–410. [Google Scholar] [CrossRef] [PubMed]
- Dadarkar, S.S.; Deore, M.D.; Gatne, M.M. Comparative evaluation of acute toxicity of ivermectin by two methods after single subcutaneous administration in rats. Regul. Toxicol. Pharmacol. 2007, 47, 257–260. [Google Scholar] [CrossRef] [PubMed]





| Gene Size (bp) | Sense | (5′–3′) |
|---|---|---|
| mepA 132 | Forward: | CGCGATTGCAAGTTATGGTATC |
| Reverse: | ACGTCTTTCATACGGCCTTTA | |
| swrC 100 | Forward: | CAACGCTGGAACGAGTGTAA |
| Reverse: | ATCAGGTTCAGTAGGCGAAATG | |
| Emrb 105 | Forward | CCACCATATTGCGATGCTAATTC |
| Reverse | GACAACGCAAACTACACAACAT | |
| gyraseA 110 | Forward: | CAGAGCTCGTTCGTGACAAG |
| Reverse: | AGCATTTGCATCCTTACGCA |
| Protein | Gene Name | Fold Change/Unique | Protein Function |
|---|---|---|---|
| A0A0D3Q904 | ecsA3 | 1.65 | ABC transporter, ATP-binding protein EcsA |
| A0A0D6GCY7 | mepA | 2.21 | MATE family efflux transporter (Multi antimicrobial extrusion protein (Na(+)/drug antiporter) |
| A0A0D6GDE2 | ytrB1 | 2.29 | ABC transporter ATP-binding protein |
| A0A0D6GZT9 | msbA2 | unique | ABC transporter ATP-binding protein |
| A0A0D6H6R4 | emrB4 | unique | MFS transporter (Multidrug resistance transporter) |
| A0A0Z0Q5C1 | norB3 | unique | Permease |
| Q9RQG6 | swrC | 1.70 | RND (Resistance nodulation division multidrug transporter) |
| X5DZI9 | BTN44_13755 | unique | MFS transporter (Major facilitator family transporter) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ashraf, S.; Parrine, D.; Bilal, M.; Chaudhry, U.; Lefsrud, M.; Zhao, X. Proteomic Comparison of Ivermectin Sensitive and Resistant Staphylococcus aureus Clinical Isolates Reveals Key Efflux Pumps as Possible Resistance Determinants. Antibiotics 2022, 11, 759. https://doi.org/10.3390/antibiotics11060759
Ashraf S, Parrine D, Bilal M, Chaudhry U, Lefsrud M, Zhao X. Proteomic Comparison of Ivermectin Sensitive and Resistant Staphylococcus aureus Clinical Isolates Reveals Key Efflux Pumps as Possible Resistance Determinants. Antibiotics. 2022; 11(6):759. https://doi.org/10.3390/antibiotics11060759
Chicago/Turabian StyleAshraf, Shoaib, Débora Parrine, Muhammad Bilal, Umer Chaudhry, Mark Lefsrud, and Xin Zhao. 2022. "Proteomic Comparison of Ivermectin Sensitive and Resistant Staphylococcus aureus Clinical Isolates Reveals Key Efflux Pumps as Possible Resistance Determinants" Antibiotics 11, no. 6: 759. https://doi.org/10.3390/antibiotics11060759
APA StyleAshraf, S., Parrine, D., Bilal, M., Chaudhry, U., Lefsrud, M., & Zhao, X. (2022). Proteomic Comparison of Ivermectin Sensitive and Resistant Staphylococcus aureus Clinical Isolates Reveals Key Efflux Pumps as Possible Resistance Determinants. Antibiotics, 11(6), 759. https://doi.org/10.3390/antibiotics11060759







