Abstract
Salvia africana-lutea L., S. lanceolata L., and S. chamelaeagnea L. are used in South Africa as traditional medicines to treat infections. This paper describes an in-depth investigation into their antibacterial activities to identify bioactive compounds. Methanol extracts from 81 samples were screened against seven bacterial pathogens, using the microdilution assay. Biochemometric models were constructed using data derived from minimum inhibitory concentration (MIC) and ultra-performance liquid chromatography-mass spectrometry data. Active molecules in selected extracts were tentatively identified using high-performance thin layer chromatography (HPTLC), combined with bioautography, and finally, by analysis of active zone eluates by mass spectrometry (MS) via a dedicated interface. Salvia chamelaeagnea displayed notable activity towards all seven pathogens, and the activity, reflected by MICs, was superior to that of the other two species, as confirmed through ANOVA. Biochemometric models highlighted potentially bioactive compounds, including rosmanol methyl ether, epiisorosmanol methyl ether and carnosic acid. Bioautography assays revealed inhibition zones against A. baumannii, an increasingly multidrug-resistant pathogen. Mass spectral data of the eluted zones correlated to those revealed through biochemometric analysis. The study demonstrates the application of a biochemometric approach, bioautography, and direct MS analysis as useful tools for the rapid identification of bioactive constituents in plant extracts.
1. Introduction
Individuals with weakened immune systems are particularly susceptible to opportunistic infections caused by common bacterial pathogens, including Gram-positive Staphylococcus aureus, Bacillus cereus, Enterococcus faecium, Bacillus subtilis, and Gram-negative Escherichia coli, Acinetobacter baumannii, and Pseudomonas aeruginosa. Several Salvia species that are indigenous to southern Africa are commonly used alone, or as an ingredient of traditional polyherbal remedies, as treatments for a variety of infectious ailments. The three species, S. africana-lutea L., S. lanceolata L., and S. chamelaeagnea L., which are the subject of this research, are commonly used to treat infections related to the lungs, skin, stomach, and other infectious diseases [1]. Staphylococcus aureus, P. aeruginosa, A. baumannii, E. faecium, and B. subtilis are related to respiratory [2] and skin ailments [3], and together with E. coli, are associated with urinary tract infections [4]. Stomach ailments characterised by diarrhea, are commonly caused by B. cereus [5], E. coli, or E. faecium [6]. If these conditions are not treated fast, dehydration and secondary infections such as septicemia and gangrene can set in, often resulting in death, especially in the case of immunocompromised patients.
The antibacterial activity of methanol:chloroform (1:1, v/v) extracts of indigenous Salvia species, including the three species, has been previously investigated in vitro [7,8]. Minimum inhibitory concentrations (MICs) towards S. aureus, B. cereus, E. coli, and K. pneumoniae ranged from 0.030–8.0 mg/mL, indicating excellent to weak activity [8]. The Gram-positive pathogens were more susceptible to the extracts of two of the three species, S. africana-lutea, and S. chamelaeagnea. The same research group conducted further investigations into S. chamelaeagnea, as it displayed the best antibacterial activity [8,9]. A methanol:chloroform (1:1, v/v) extract was fractionated using column chromatography, and the most active fraction towards the tested bacterium, S. aureus, was identified utilising thin layer chromatography (TLC) and a bioautographic assay. Compounds in the active fraction were identified as carnosol, oleanolic acid, ursolic acid, and 7-O-methylepirosmanol. The highly variable MICs recorded for each of the four pure compounds reflect differences in susceptibility of the four pathogens [8]. Their study demonstrated the potential antibacterial properties of the three species towards some pathogens and justified further studies of this aspect. The shortcomings of the previous studies [7,8] are the use of single samples from one locality and the testing of extracts towards only four pathogens associated with lung and skin conditions.
The bioactivity of a plant extract is related to the presence of secondary metabolites, which include phenolic compounds, and di- and triterpenoids [10], and is dependent on the relative ratios and levels at which they occur. Chemical differences can influence the bioactivity of any given plant extract dramatically. Substantial inter- and intraspecies chemical variation within both volatile [11] and non-volatile [12] secondary metabolites of the three species was scientifically confirmed through chromatographic analyses and chemometric modelling techniques. Such variation may influence the antibacterial activity of a single specimen, which may not be representative of the species in general. This was demonstrated in another study [13] that investigated the antibacterial activity of plants from two of the three species, S. africana-lutea and S. lanceolata, towards four pathogens (E. coli, K. pneumoniae, B. cereus and S. aureus). Extracts were prepared from wild-harvested material collected from the same plant specimen during each of the four seasons of the same year. A large degree of variation in the antibacterial activities of the extracts from different samples was reported for each of the species, and the results confirmed that plants from the same species that are exposed to different environmental factors comprise different chemical constituents that can influence the bioactivity of the extracts.
Biochemometric analysis correlates chemical spectral data with biological data [14] and is a useful tool that can be applied to pre-screen plant extracts for identifying bioactive compounds with potential commercial value, without incurring unnecessary laboratory work. By combining antibacterial data (MIC values) with chromatographic data (in this case ultra-performance liquid chromatography-mass spectrometry (UPLC-MS) data), orthogonal projections to latent structures-discriminant analysis (OPLS-DA) models can be constructed and used to predict bioactive compounds [14,15]. Thin layer chromatography-direct bioautography (TLC-DB) is a relatively inexpensive and simple technique that has been utilised for the screening of plant extracts to identify potentially bioactive compounds [16,17]. This technique utilises high performance thin layer chromatography (HPTLC) to separate molecules in plant extracts, followed by a biological detection procedure [18]. Constituents that are active toward the selected pathogens are revealed, after the application of p-iodonitrotetrazolium chloride (INT), by the formation of clear circular inhibition zones around the bands. Analysis of the corresponding active bands, eluted from an identical reference HPTLC plate using a TLC-interface, by direct infusion of the sample into a mass spectrometer (MS), reveals the molecular/fragment ions of the active compounds, which enables their tentative identification. The ions of the potentially bioactive compounds highlighted by biochemometric analysis can be correlated with those observed from MS analysis of the bioactive bands, in turn confirming or discounting compounds revealed by biochemometrics. This technique has been utilised for the rapid identification of bioactive compounds in a study on propolis [19].
Apart from the few articles referenced, literature on the antimicrobial activity of the solvent extracts of the three Salvia species was found to be scant. The work reported in the available literature raised sufficient interest to pursue further investigations. The aim of this study was therefore to conduct an in-depth investigation of the antibacterial activity of the three species, using a large sample size from various localities in South Africa, to provide a more accurate overall indication of the bioactivity of the three species, identify variation in activity between the species and within the populations, as well as chemotypes that would be desirable in terms of their bioactivity for cultivation and commercialisation purposes. The MICs determined for each sample towards selected pathogens (S. aureus, B. cereus, E. faecium, B. subtilis, E. coli, P. aeruginosa and A. baumannii), were combined with UPLC-MS profiling data, using chemometric methods of analysis (OPLS-DA). This biochemometric approach was followed to identify chemical compounds that may be associated with the antibacterial activity, if any, of each species against the selected pathogens. Sample extracts from each species that displayed the best antibacterial activity (lowest MICs) towards specific pathogens were further investigated by means of TLC-DB, to identify bioactive constituents from active zones. This approach was investigated as a less complicated alternative to conventional bioassay-guided fractionation that most often utilises column chromatography.
2. Results
2.1. Antibacterial Activity
The average MICs, together with the range, determined using the microdilution assay, for the methanol extracts of the three species towards seven selected pathogens, are presented in Table 1. Also listed for each species/pathogen combination, are the overall range and average of the MICs with standard deviation (SD), positive control ciprofloxacin result, the number of active/more active samples, number of inactive/less active samples, and the statistical difference testing results.

Table 1.
Range and average (in brackets) minimum inhibitory concentrations (MICs) in mg/mL (n = 2) for crude Salvia extracts of each population (n = 5 for all populations except for Paarl where n = 6) tested against seven selected pathogens.
The antibacterial activities of the three species towards each pathogen were statistically compared to determine the significance of the variability between the three species using ANOVA single factor analysis. There was a significant difference (p < 0.05) between the activities of the three species for all seven pathogens. Comparing the three species to each other, two at a time, also indicated a significant difference in the activities of S. africana-lutea and S. chamelaeagnea for five of the pathogens (S. aureus, B. cereus, E. faecium, A. baumannii and E. coli). The activities of S. africana-lutea and S. lanceolata were significantly different for four of the pathogens (B. cereus, B. subtilis, P. aeruginosa and E. coli). Activities of S. chamelaeagnea and S. lanceolata were significantly different for all seven pathogens. Overall, S. chamelaeagnea displayed the best antibacterial activity towards the seven pathogens with average MICs 0.23–1.3 mg/mL (variance 0.007–0.64), followed by S. africana-lutea, with average MICs 0.52–3.0 mg/mL (variance 0.006–4.1), and lastly, S. lanceolata with MICs 0.46–4.2 mg/mL (variance 0.010–4.1).
Minimum inhibitory concentrations were determined for medicinally important compounds, carnosol, carnosic acid and ursolic acid, previously reported to be present in many of the extracts [12]. Activity was determined towards two selected pathogens that displayed high susceptibility across the three species toward a number of individual plant extracts, namely B. cereus and A. baumannii. Carnosol and carnosic acid displayed antimicrobial activity toward B. cereus and A. baumannii, with MICs of 0.25 mg/mL towards B. cereus, and 0.13 mg/mL and 0.062 mg/mL, respectively, towards A. baumannii. Ursolic acid displayed excellent activity against A. baumannii with an MIC of 0.031 mg/mL, although it was only weakly active towards B. cereus (MIC = 0.50 mg/mL).
2.2. Bioactive Compound Identification Using a Biochemometric Approach
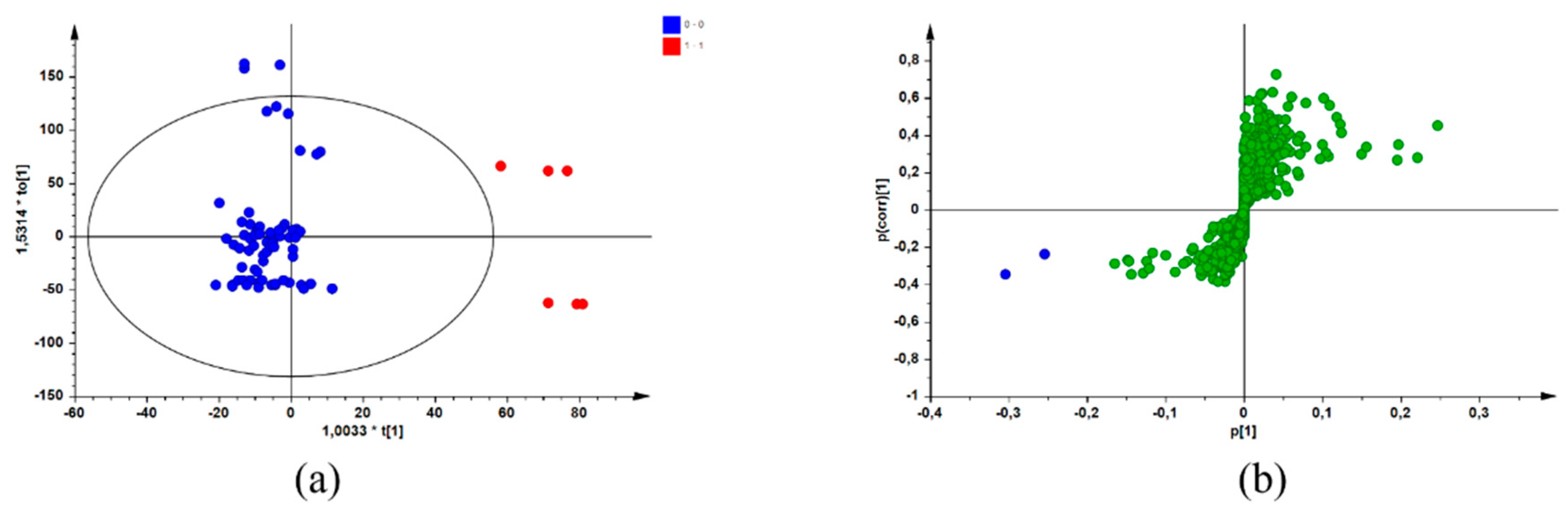
After combining the biological data (MICs) with data from a chromatographic reference technique (UPLC-MS), screening for potential bioactive compounds within the sample extracts was achieved using a biochemometric approach. By creating a data matrix comprising both datasets and using SIMCA® P + 14 software to apply multivariate analysis, an individual OPLS-DA model was constructed for each pathogen. The active/more active (Class 1) and inactive/less active (Class 2) MIC values (defined in Section 4.4) represented the Y-variables in the data matrix for each sample of the three species. These values were combined with the UPLC-MS dataset, which were selected as the X-variables [12]. Application of multivariate tools, such as OPLS-DA modelling, resulted in the identification of potential bioactive compounds for each species/pathogen combination by group-to-average comparison and through S-plots. Figure 1 is a representation of the OPLS-DA scores scatter (a) and corresponding S-plot (b) resulting from the constructed models for the pathogen/species combination of S. chamelaeagnea/A. baumannii, randomly selected for illustrative purposes. Similar plots were obtained for the other pathogen/species combinations (plots not shown). In Figure 1a, the blue dots represent the active/more active samples (Class 1), and the red dots represent the inactive/less active samples (Class 2). The blue dots on the S-plot (Figure 1a) represent the compounds associated with bioactivity. These compounds were identified from the retention time (Rt)/mass-to-charge ratio (m/z) pairs by comparison with those reported for a non-volatile metabolite variation study [12] of the same samples. The potentially bioactive compounds that were revealed for each species through the S-plots are listed in Table 2, Table 3 and Table 4, together with OPLS-DA model statistics for each species/pathogen combination.
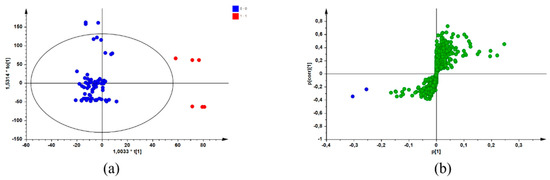
Figure 1.
OPLS-DA scores scatter plot (a) with the blue dots representing active/more active samples and the red dots representing inactive/less active samples, and the corresponding S-plot (b) with the blue dots representing potential compounds associated with bioactivity (Model constructed from S. chamelaeagnea/A. baumannii UPLC-MS/MIC combined datasets) and the green dots representing other compounds not associated with bioactivity.

Table 2.
Potentially bioactive compounds revealed through biochemometric analysis for active samples of Salvia africana-lutea towards seven selected pathogens.

Table 3.
Potentially bioactive compounds revealed through biochemometric analysis for active samples of Salvia lanceolata towards seven selected pathogens.

Table 4.
Potentially bioactive compounds revealed through biochemometric analysis for active samples of Salvia chamelaeagnea towards seven selected pathogens.
Tentative identities of the compounds were obtained from the variation study of the non-volatile compounds of the same set of samples [12]. The compounds identified for S. africana-lutea extracts (Table 2) with potential bioactivity included salvianolic acid E (Rt 3.09 min), salvianolic acid B (Rt 3.88 min), dihydroxy-dimethoxyflavone derivative (Rt 8.97 min), methyl carnosate (Rt 10.15 min), epiisorosmanol methyl ether (Rt 10.50 min) and unknown compounds at Rt 9.02 and 10.71 min. Bioactive compounds linked to the activity of the S. lanceolata extracts (Table 3) included salvianolic acid E (Rt 3.10), rosmarinic acid (Rt 3.66 min), epiisorosmanol (Rt 7.80 min), together with several unknown compounds at Rt 6.79, 8.72, 10.36, and 10.43 min. For S. chamelaeagnea extracts (Table 4), epirosmanol (Rt 7.36 min), rosmanol methyl ether (Rt 9.82 min), carnosol (Rt 9.99 min), methyl carnosate (Rt 10.15 min), carnosic acid (Rt 11.39 min) and two unknown compounds at Rt 10.74 and 12.19 min were revealed as contributors to antibacterial activity.
2.3. Thin Layer Chromatography-Direct Bioautography
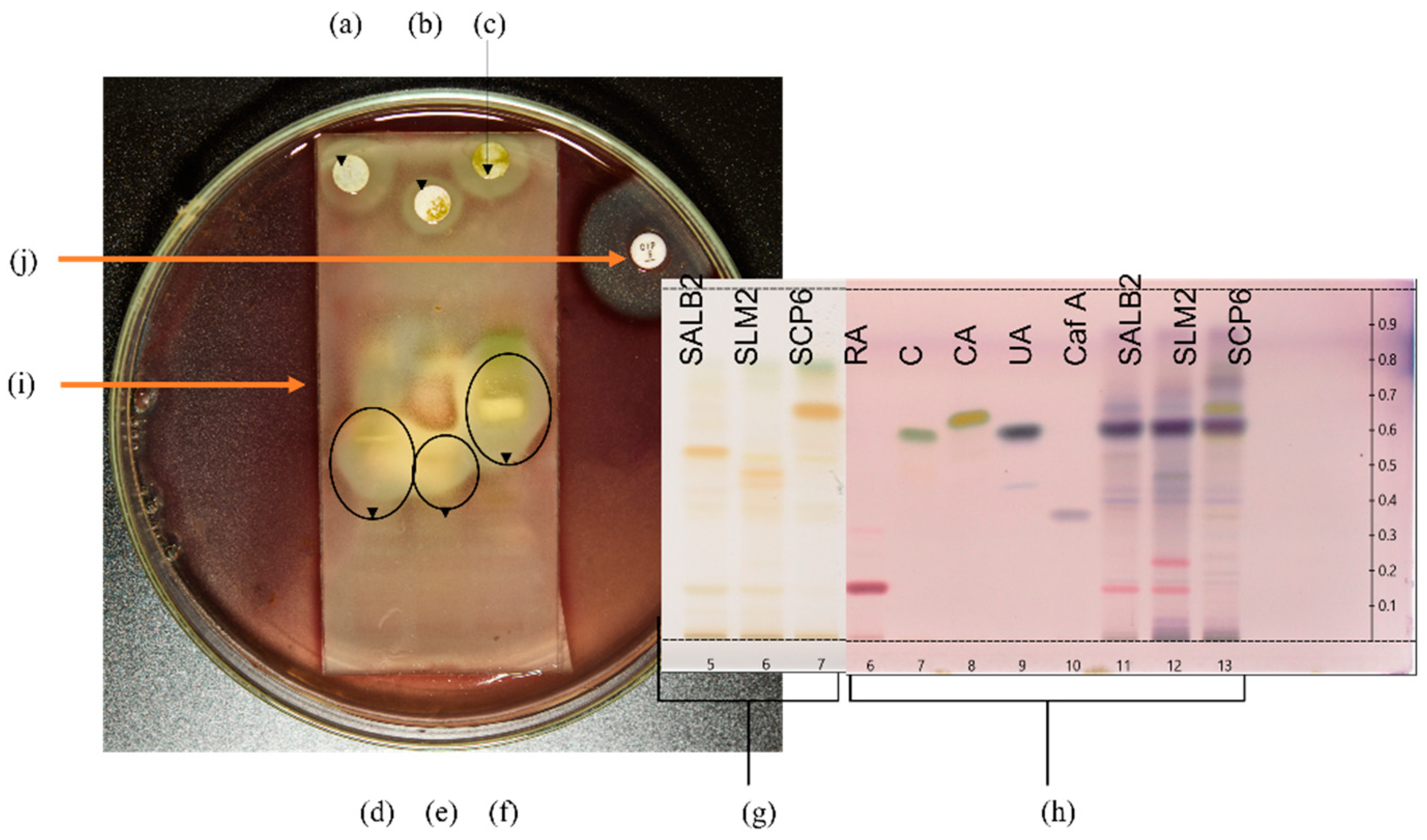
Selected plant extracts with the most promising antibacterial activity were further investigated for potentially active compounds by TLC-DB to tentatively identify active constituents and correlate to those revealed through the biochemometric approach. Therefore, the extract for each species that displayed the most promising antibacterial activity was utilised for further investigation by TLC-DB. The three extracts selected were Langebaan Sample 2 for S. africana-lutea, Mamre Sample 2 for S. lanceolata and Paarl Sample 6 for S. chamelaeagnea. The three pathogens most susceptible to the plant extracts, namely B. cereus, A. baumannii, and P. aeruginosa, were screened for inhibition by the crude extracts of the three selected samples. Only one, A. baumannii, was inhibited by the separated crude extracts, and subsequent TLC-DB analysis was conducted using this pathogen. Figure 2 represents the experimental set-up for TLC-DB.
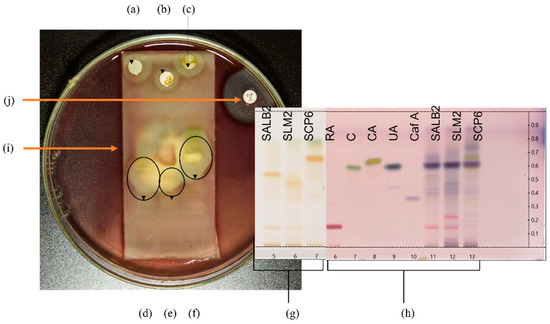
Figure 2.
Set-up for TLC-DB with HPTLC fingerprints and bioautograms. (a–c) represent sterile discs infused with crude sample extracts of S. africana-lutea, S. lanceolata and S. chamelaeagnea, respectively, with inhibition zones observed around the discs indicating activity of the whole extract. HPTLC fingerprints, (d–f) of the sample extracts, S. africana-lutea, S. lanceolata and S. chamelaeagnea, respectively, with inhibition zones around active constituents indicated by the black ovals. (g) represents HPTLC fingerprints in Tracks 5, 6 and 7 for each sample extract, SALB2, SLM2 and SCP6, on the TLC plate, as viewed under white light at 366 nm, before TLC-DB. TLC plate (h) is the reference plate and represents authentic standards and sample extracts visualised using p-anisaldehyde. Tracks 6, 7, 8, 9 and 10 represent rosmarinic acid (Rf = 0.16), carnosol (Rf = 0.59), carnosic acid (Rf = 0.66), ursolic acid (Rf = 0.64) and caffeic acid (Rf = 0.35), respectively, Tracks 11, 12 and 13 represent each sample extract for identification of constituents. (i) represents the TLC plate overlaid with agar inoculated with the selected pathogen, in this case A. baumannii. (j) is the positive control (ciprofloxacin) disc with inhibition zone observed around the disc.
The crude extracts of the three samples inhibited the growth of A. baumannii, as can be seen from the resulting inhibition zones around the discs infused with the extracts (Figure 2a–c, respectively). This provides evidence for the antibacterial activity and correlates with the low MICs obtained for the three extracts towards this pathogen. Distinct inhibition zones were observed at different sites of each fingerprint in the bioautograms of the three sample extracts (Figure 2d–f), revealing bands with activity towards the pathogen. The corresponding TLC plate depicted in Figure 2g represents the HPTLC fingerprints of the three extracts before exposure to the pathogen, and indicates bands present in each fingerprint that are not clearly visible after overlay with the inoculated agar. Selected standards (Figure 2h, Track 6—rosmarinic acid, 7–carnosol, 8—carnosic acid, 9—ursolic acid, and 10—caffeic acid) were applied to the reference plate to assist in the identification of compounds at the corresponding Rf values that may be responsible for the activity. These were analysed together with fingerprints of the extracts (Figure 2h, Track 11–13) and visualised with p-anisaldehyde-sulfuric acid reagent. Figure 2i indicates the TLC plate overlaid with agar inoculated with the selected pathogen, in this case, A. baumannii.
Comparing the bioautograms to the unexposed standards and sample fingerprints on the reference plate (Figure 2h), gave an indication of possible known compounds contributing to the antibacterial activity observed. Three compounds, carnosol, carnosic acid, and ursolic acid were identified in the fingerprint of the S. chamelaeagnea extract SCP6 (Figure 2h, Track 13) by band comparison, and in turn, could be correlated to the area where the inhibition zone was observed for this extract (Figure 2f). The three compounds therefore must contribute to the activity towards the pathogen. The reference standards could not be correlated to the active areas for the other two sample extracts (Figure 2d,e), indicating that none of these known compounds are responsible for the activity. The inhibition zone observed around the positive control, ciprofloxacin disc (Figure 2j), confirms that the A. baumannii strain tested is susceptible to conventional antibiotics and had not developed any resistance to the drug.
2.4. Hyphenated Thin Layer Chromatography-Mass Spectrometry Analysis of Bioactive Constituents
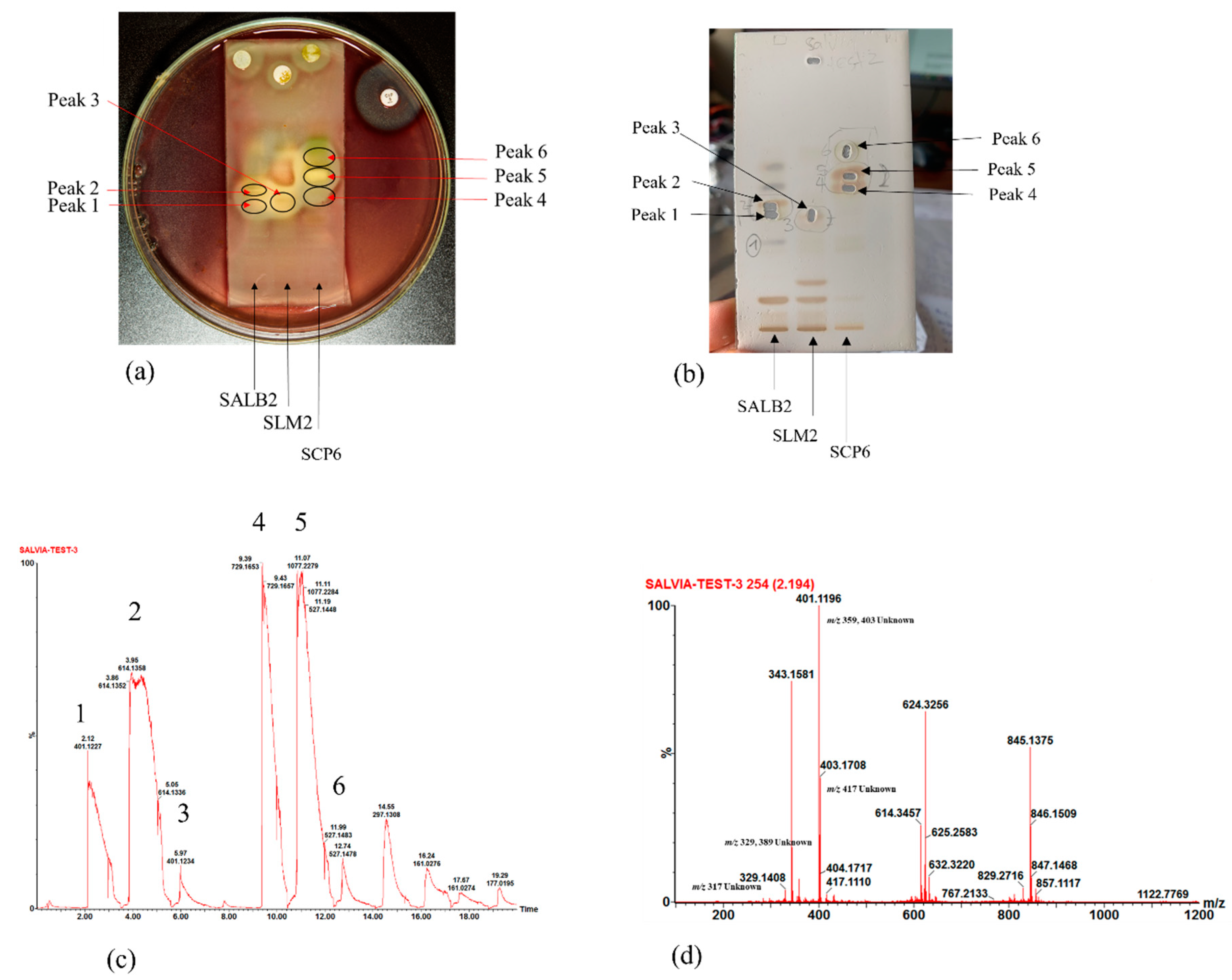
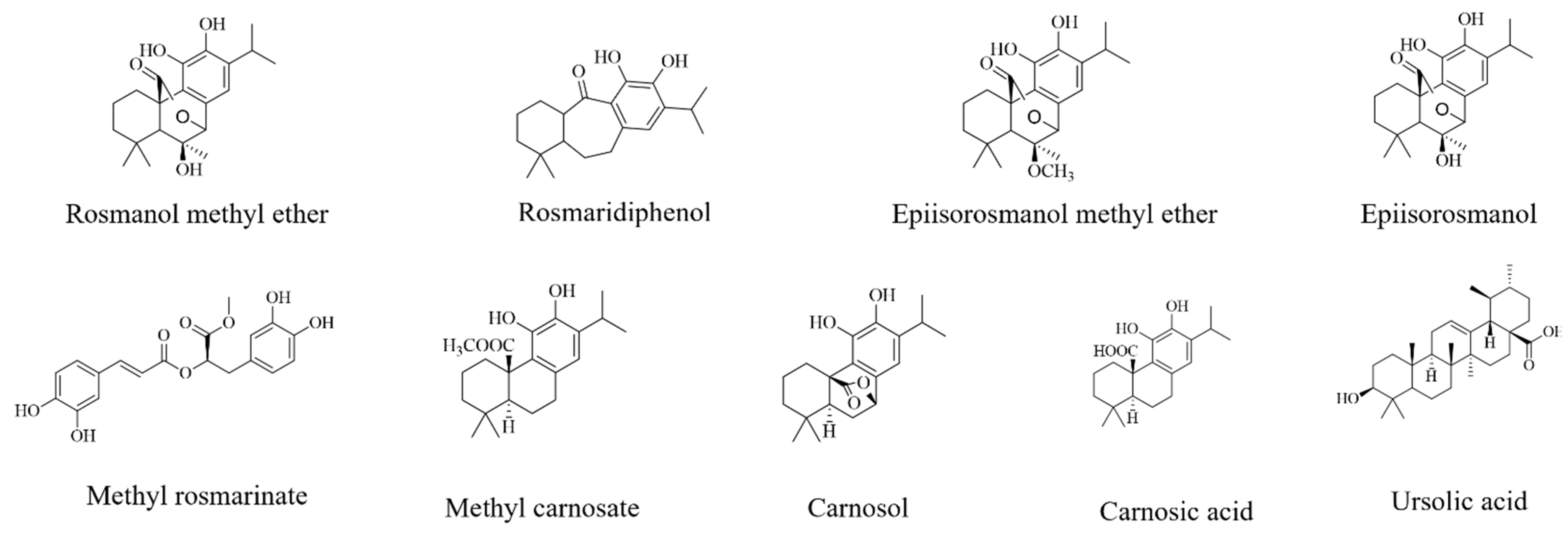
With the active bands revealed on the bioautograms, identification of the compounds was achieved using hyphenated MS analysis. The sites corresponding to zones of inhibition, where active constituents were extracted by use of the TLC-interface, are indicated on the bioautograms presented in Figure 3a. The corresponding TLC plate (Figure 3b) shows the actual sampling spots where extraction of Peaks 1–6 was performed using the TLC-sampler. Peaks 1–6, indicated in Figure 3a, correspond to Peaks 1–6 on the resulting chromatogram (generated by the software after infusing each sampled site) presented in Figure 3c. The active sites for S. africana-lutea are represented by Peaks 1 and 2, S. lanceolata by Peak 3 and S. chamelaeagnea by Peaks 4, 5 and 6. It must be noted that the observed ions are likely to be a combination of molecular ([M-H]− and fragment ions (m/z) of the compounds present within the sampled active sites. The mass spectrum obtained from the active site sampled at Peak 1 is given as an example in Figure 3d, and molecular/fragment ions can be observed. Similar mass spectra were obtained for each active site sampled at Peaks 2, 3, 4, 5, and 6 (not shown). The molecular/fragment ions of components with potential activity present in the extracted bands were detected and reported in the form of a mass spectrum displaying the m/z ratios. The m/z of the ions for each peak are listed in Table 5. Molecular structures for possible compounds identified through TLC-MS are given in Figure 4.
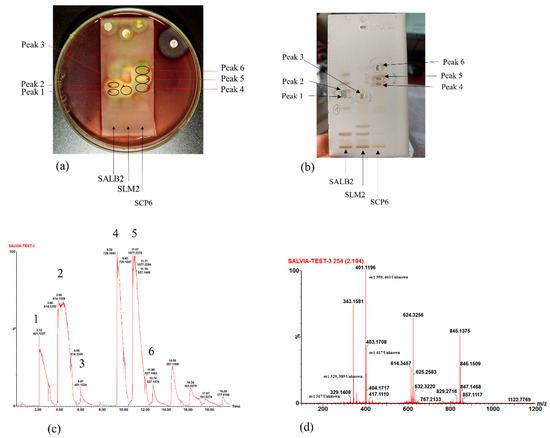
Figure 3.
(a) Bioautograms of S. africana-lutea (SALB2), S. lanceolata (SLM2) and S chamelaeagnea (SCP6) extracts indicating the active sites indicated as Peak 1–6, corresponding to the peaks indicated in (c), with (b) the corresponding enlarged TLC plate showing areas where active constituents, Peak 1–6, were extracted using the TLC-sampler. (c) Chromatogram with Peaks 1–6 resulting from direct MS analysis of active bands extracted from the TLC plate and infused directly into the MS detector for ionisation. (d) Mass spectrum obtained, in this case for Peak 1, revealing molecular/fragment ions of compounds extracted from the active site on the bioautogram.

Table 5.
Molecular/fragment ions ([M-H]− m/z) identified from active sites on bioautograms for each sample extract corresponding to peaks in Figure 3c.
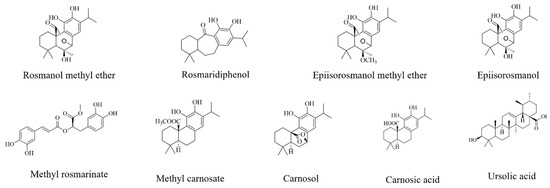
Figure 4.
Chemical structures of compounds tentatively identified by TLC-MS analysis.
3. Discussion
The microdilution assay enabled rapid, large-scale screening of extracts. The results indicated active extracts that could be earmarked for further investigation following a more focused approach. Suggested criteria for MIC values of plant extracts available from recently published literature [20] were considered as a guideline for activity. According to the author, for the purposes of ethnopharmacological research studies, MICs higher than 0.32 mg/mL reflect extracts that may still be effective, although 1.0 mg/mL was accepted as the upper limit of activity for this study. Activities were further described [20] as weak (>0.32 mg/mL), average (0.16–0.32 mg/mL), good (0.081–0.16 mg/mL), very good (0.041–0.080 mg/mL), excellent (0.021–0.040 mg/mL) or outstanding (<0.02 mg/mL), and these criteria were applied to this study.
For S. africana-lutea and S. lanceolata, most of the samples inhibited the growth of B. cereus, A. baumannii, P. aeruginosa and B. subtilis, with weak, average, or good activities (MIC ≤ 1.0 mg/mL). Salvia chamelaeagnea extracts displayed the highest degree of activity against the seven selected pathogens. Most extracts displayed weak to excellent activity, with MICs ranging from 0.038 to 1.0 mg/mL. All samples were active against B. cereus, A. baumannii and P. aeruginosa and most extracts were active towards the remaining four pathogens, S. aureus, E. faecium, B. subtilis, and E. coli. One sample from the Paarl population (Sample 6) displayed excellent activity towards B. cereus (MIC = 0.038 mg/mL), with good activity observed for 12 other samples with an MIC of 0.13 mg/mL. The same Paarl sample displayed good activity (MIC = 0.13 mg/mL) towards A. baumannii, which is notable, as this pathogen is known to be highly resistant towards conventional antibacterial agents [21]. Promising activity was exhibited by extracts for all three species towards A. baumannii. Variation in the MICs for samples from the same species and within populations was evident (Table 1) and confirms that differences in the chemical profiles of the plant extracts affect their bioactivity.
The extracts of S. chamelaeagnea contain high concentrations of carnosol (Not detected (ND)–15.1 mg/g dry weight (DW)) and carnosic acid (0.94–14.0 mg/g), as previously determined using ultra-performance liquid chromatography-photodiode array (UPLC-PDA) detection analysis [12]. The two compounds were found to be absent or not detected in the samples of S. africana-lutea and S. lanceolata. It is possible that carnosol and carnosic acid contribute towards the highly promising antimicrobial activity of several S. chamelaeagnea extracts. These compounds are present at substantial levels in sample extracts of Rosmarinus officinalis, which has widely reported antibacterial activity [22,23]. The activities of the carnosol and carnosic acid reference standards, as determined in this study, were lower than expected towards B. cereus and A. baumannii, with MICs of 0.25 mg/mL for both towards B. cereus and 0.13 mg/mL and 0.062 mg/mL, respectively, towards A. baumannii. The activity of carnosol towards B. cereus (MIC = 0.25 mg/mL) does not correlate with the higher activity (MIC = 0.02 mg/mL) previously reported [8] towards the same strain pathogen (ATCC 11778). No other reports on MICs for the two compounds towards the two pathogens could be found, apart from one study that reported a MIC of 0.078 mg/mL for carnosic acid towards B. cereus [24]. This value indicates a higher activity than that determined in this study (MIC = 0.25 mg/mL). It is therefore highly plausible that the two compounds, carnosol and carnosic acid, together with other compounds in the extracts, may contribute to the good antimicrobial activity of the extracts in which they are present towards B. cereus and A. baumannii, by acting in an additive or synergistic manner.
Ursolic acid may also contribute to a large extent to the antibacterial activity of the S. chamelaeagnea extracts, as well as that of the S. africana-lutea and S. lanceolata extracts, against the two pathogens. In this study, ursolic acid was found to be present at high concentrations in all samples representing the three species, with concentrations ranging from 7.40–38.2 mg/g [12]. The compound has been indicated in several studies to be responsible for antibacterial activity, on its own, in combination with other compounds, or as a main constituent of plant extracts. A study investigating the activity of constituents of R. officinalis extracts towards methicillin-resistant S. aureus (MRSA) reported that fractions containing ursolic acid resulted in total inhibition [25]. For ursolic acid on its own, MICs of 0.032 mg/mL, 0.064 mg/mL and 0.51 mg/mL, respectively, were reported towards S. aureus, E. coli, and P. aeruginosa [26]. In the present study, the compound displayed excellent activity against A. baumannii with a MIC of 0.031 mg/mL, although it was only weakly active towards B. cereus (MIC = 0.50 mg/mL). In a separate study [27], aimed at investigating the biofilm inhibitory activity of triterpenoids including ursolic acid, towards A. baumannii, an additive interaction of ursolic acid and ciprofloxacin in combination was reported, with a resulting MIC of 0.075 µg/mL for the combined compounds [27], which reflected better activity than the MIC of 0.16 µg/mL recorded for ciprofloxacin on its own. Other diterpene derivatives, rosmanol and 12-O-methyl carnosic acid, reported to have antimicrobial activity in Salvia officinalis extracts [28], are present in the extracts of the three species [12] tested in this study, and may contribute to the activity. It is highly likely that these compounds may act additively, or display synergistic interactions [29] with other constituents in the extracts.
In a separate study investigating the antimicrobial activity, the three species alongside 13 other South African Salvia species, were tested towards three of the seven pathogens used in the current study, namely B. cereus, E. coli and S. aureus [8]. Salvia africana-lutea was reported to be only weakly active towards B. cereus (MIC = 0.75 mg/mL) [8], which agrees with the results from the current study (Table 1). All three species were inactive toward E. coli, which is consistent with our findings that extracts were mainly inactive toward E. coli. They also reported that S. africana-lutea displayed weak activity towards S. aureus (MIC = 0.75 mg/mL). However, in the current study, most of the sample extracts were inactive toward this pathogen.
Kamatou and co-workers [8] reported S. lanceolata to be inactive towards the three pathogens, B. cereus, E. coli and S. aureus. In the current study, this was also the case for most extracts, with poor activity observed towards S. aureus and E. coli. However, the majority of samples were active towards B. cereus and for only a few samples originating mostly from the same populations, inactivity towards B. cereus was observed, affirming the insufficiency of a single sample result. In the same study [8], S. chamelaeagnea was reported to display very good activity towards S. aureus (MIC = 0.06 mg/mL) and B. cereus (MIC = 0.03 mg/mL), with weak activity (MIC = 1.0 mg/mL) towards E. coli. The results from the current study compare favourably with those reported by Kamatou and co-workers [8], since S. aureus was inhibited by a number of samples (weak and average activity), all samples were active toward B. cereus (weak to very good activity), and most samples tested displayed weak or average activity towards E. coli (Table 1).
Salvia officinalis, well known for its medicinal properties and culinary use, was reported to be inactive towards S. aureus (MIC = 5.0 mg/mL) and E. coli (MIC = 2.5 mg/mL), with weak activity against B. cereus (MIC = 0.63 mg/mL) [30]. This correlated to an extent with the activities observed for the three species towards the same three pathogens in the current study, with most extracts inactive towards S. aureus and E. coli, and mostly weak activity of S. africana-lutea and S. lanceolata. Another study reported S. officinalis to be only weakly active toward S. aureus (MIC = 0.94 mg/mL), but inactive towards E. coli and P. aeruginosa [31], corresponding to the inactivity observed in our study for S. africana-lutea and S. lanceolata towards S. aureus and E. coli. Again, there are differences in the reported results and those of the current study in relation to P. aeruginosa, with all samples of the three species screened displaying some level of activity.
In summary, the methanol plant extracts of S. africana-lutea, S. lanceolata, and S. chamelaeagnea exhibited potential as antibacterial agents towards B. cereus, A. baumannii, P. aeruginosa, and B. subtilis. Salvia chamelaeagnea was active against all seven pathogens, with very good and excellent activity reported for a few sample extracts against B. cereus and A. baumannii. The activity of the three species, and especially the activity of S. chamelaeagnea, against A. baumannii, is noteworthy. The pathogen has been reported to be persistent due to its resistance to most conventional antibiotic agents [32,33,34]. Statistically, there was a significant difference between the activities of the three species for the seven pathogens. This information supported the need for further investigation to reveal the active constituents in the extracts with the best antibacterial activities, initially using biochemometric analysis as a screening technique, followed by the more focused TLC-DB technique.
Biochemometric modelling was applied in this study to correlate the chemical components present in the highly complex extracts to the biological activity data, to assist in the identification of compounds possibly responsible for the bioactivity. Several of the compounds identified using this technique (Table 2, Table 3 and Table 4) have been reported to display antibacterial activity and are present in plant extracts with reported activity towards bacteria, therefore possibly contributing to the activity observed in this study. Salvianolic acid B, present in Salvia miltiorrhizae, tested using the microtiter plate assay, was reported to display potent antibacterial activity towards Neisseria meningitidis, a life-threatening human pathogen in some African countries [35]. Carnosol and carnosic acid, together with ursolic acid, which was not highlighted as a bioactive compound, have been reported to be responsible for the antibacterial activity of extracts of sage or S. officinalis toward MRSA [36] when tested using the microdilution method [37,38]. Salvia officinalis leaves are rich in methyl carnosate, a diterpene compound with good antibacterial activity towards B. cereus [39].
Methanolic extracts of Rosmarinus officinalis leaves, rich in rosmarinic acid, carnosol, and carnosic acid, were tested for antibacterial activity using the disc diffusion technique [23], and found to inhibit S. aureus and Listeria monocytogenes. Samples containing a higher concentration of carnosol relative to that of carnosic acid were reported to be more active. In the current study, this was the case with two S. chamelaeagnea samples, one from Paarl (MIC = 0.38 mg/mL) and one from Du Toitskloof (MIC = 0.75 mg/mL), towards S. aureus. Both samples displayed better antibacterial activity compared to the other samples for the respective populations. Overall, S. chamelaeagnea displayed the best activity towards the seven pathogens, in particular B. cereus, A. baumannii, and P. aeruginosa, possibly due to the presence of carnosol and carnosic acid. The two compounds were present at the required concentration ratios (carnosol > carnosic acid) in many of the active samples (Concentrations for individual samples given in the Supplementary material of [12]. For S. chamelaeagnea samples that were inactive towards S. aureus, carnosol was either present at a lower concentration than carnosic acid (samples from Paarl, Du Toitskloof, and Elandsberg) or the concentrations of the two compounds were almost equal (Elandsberg samples).
Many of the compounds identified for the three species have been reported in the literature to display antibacterial activity, or to be present in plant extracts with reported activity [36,39,40], supporting the strength of the biochemometric technique in highlighting compounds with antibacterial potential. Although biochemometric analysis can be used to identify compounds for preliminary screening, confirmation of their activity and identification is needed using a biological assay and a positive identification technique, such as TLC-DB and MS analysis, respectively. The application of biochemometric analysis was regarded in this study as a reliable indicator of bioactive compounds that could guide the targeted isolation of compounds with potential activity. It must be noted that ursolic acid was not highlighted as a bioactive compound from biochemometric analysis. Nevertheless, it has been reported to display antibacterial activity on its own [41] and to contribute to the activity of S. officinalis extracts [36]. The compound was present in all samples at relatively high concentrations and its contribution to the antibacterial activity of extracts for the three species should not be disregarded.
The TLC-DB technique has been successfully applied in various studies to indicate active constituents in bioautograms of other plant extracts that include S. officinalis, Thymus vulgaris, Mentha piperita extracts [28,42] and Helichrysum pedunculatum extracts [43]. Leaves from various South African tree species [44] and bark extracts from the Morus alba tree, a popular Traditional Chinese Medicine (TCM) [45] have also been assayed to reveal bioactive constituents. Antimicrobial compounds present in propolis, a resinous substance produced by honey bees, were revealed using TLC-DB, indicating the functionality of the technique with a variety of sample matrices [19]. In this study, no zones of inhibition were formed around the disks saturated with the crude sample extracts of the three species towards B. cereus and P. aeruginosa, and therefore no further investigation was conducted for these two pathogens. It is possible that the physico-chemical properties of the active constituents within the crude extracts are not optimal to promote sufficient diffusion into the water-based agar layer, resulting in no inhibition of the pathogen.
From the mass spectra resulting from TLC-MS for S. africana-lutea, the observed ions at m/z 283, 315, 329, and 359 could be correlated to the molecular/fragment ions of rosmaridiphenol (Rt 9.76 min), rosmanol methyl ether (Rt 9.81 min), epiisorosmanol methyl ether (Rt 10.50 min), and seven unknown compounds at Rt 8.71, 9.07, 10.29, 10.71, 11.50, 12.19, 12.71 min (Table 5). Compounds at Rt 9.07 and 10.71 min corresponded to the active compounds identified via biochemometric analysis. A prominent ion at m/z 401 could not be explained. The m/z of the ions observed at 345, 315, and 359 for S. lanceolata correlated to epiisorosmanol (Rt 7.83 min), rosmaridiphenol (Rt 9.76 min), epiisorosmanol methyl ether (Rt 10.50 min), and an unknown compound at Rt 6.52 min (Table 5). Epiisorosmanol was indicated as an active compound by biochemometric analysis. The presence of prominent ions at m/z 347 and 401 could not be associated with specific compounds. However, m/z 401 was also observed in the spectra of S. africana-lutea. A few other less prominent ions were observed that could not be correlated to any compounds. Mass spectra of S. chamelaeagnea highlighted ions at m/z 373, 315, 285, 329, 331, 345, 287, and 455 that correlated to six compounds, namely methyl rosmarinate (Rt 4.57 min), rosmaridephenol (Rt 9.76 min), carnosol (Rt 9.98 min), methyl carnosate (Rt 10.11 min), carnosic acid (Rt 11.39 min) and ursolic acid (Rt 13.80 min) (Table 5). Carnosol and carnosic acid corresponded to active compounds indicated by biochemometric analysis and both compounds, together with ursolic acid were confirmed to be present in the extract by means of reference standards, as illustrated in Figure 2h. A prominent ion was observed at m/z 343 that could not be explained. This ion was absent from the mass spectra of the other two species. It must be noted that the compounds tentatively identified from biochemometric analysis and TLC-DB that displayed antibacterial activity may not be active on their own as such, but the activity may be due to their combination with other compounds in the plant extract. Therefore, these compounds need to be studied on their own and in combination with each other to determine their true activity or the contribution they make.
Screening and identification of plant extracts for potential antibacterial constituents using biochemometric analysis combined with a TLC-DB approach in tandem with MS identification, is a promising alternative with several advantages that make it worth considering. It is generally faster, simpler, less expensive, specific, sensitive, and more environmentally friendly [28,46,47] than routinely used bioassay-guided fractionation, which is labour intensive, uses large quantities of plant material and solvents, and primarily focuses on major compounds that may not present bioactivity on their own [14,46]. Many samples can be screened simultaneously, and infusion of eluates into the MS is a direct approach that delivers identifications almost instantaneously, by providing information on compound ions [19]. Nevertheless, there are some limitations to this approach. With biochemometric analysis, the large number of variables from the reference dataset may complicate data visualisation and interpretation from the S-plot, resulting in false-positive identifications [14]. It is likely that some bioactive compounds are not identified, and were marked a false negative [48]. It may be due to specific peaks affected by poor separation affecting identification, as is thought to have been the case with ursolic acid, a highly bioactive and main compound in all extracts used in this study. Thin layer chromatography-bioautography may be less sensitive to the more lipophilic compounds that are not compatible with the water-based agar and thereby fail to produce inhibition zones. In addition, bands that display inhibition zones may represent a mixture of active and inactive constituents, since the resolution achieved on an HPTLC plate is not at the same level as that of more sophisticated techniques such as UPLC [47].
4. Materials and Methods
4.1. Plant Material Collection, Preparation, and Extraction
The collection of the aerial parts from various localities in South Africa, species verification, and preparation of the samples of S. africana-lutea (n = 30), S. lanceolata (n = 25), and S. chamelaeagnea (n = 26), as well as voucher specimen details, are described in two previously published articles [11,12]. Extraction of the dried aerial plant parts was performed using methanol [12]. However, after the combined supernatants of each plant extract were dried and weighed, 32 mg/mL solutions were prepared in acetone for MIC determinations. Three selected plant extracts were also prepared in acetone at 10 mg/mL for TLC-DB.
4.2. Reagents, Chemicals, and Pathogens
Tryptone Soya broth (TSB) (Oxoid; product code CM0129) and Tryptone Soya agar (TSA) (Oxoid; product code CM0131) were obtained from Quantum Biotechnologies (Pty) Ltd. (Johannesburg, South Africa) and prepared according to the manufacturer’s instructions. p-Iodonitrotetrazolium chloride (INT, Sigma-Aldrich, St Louis, MI, USA) was purchased from Merck (Germiston, South Africa). Ciprofloxacin (Sigma-Aldrich product number 17850, ≥98.0%, HPLC) was obtained from Merck (Pty) Ltd. (Modderfontein, South Africa). Seven bacterial strains, namely S. aureus (ATCC 25923), A. baumannii (ATCC 19606), B. cereus (ATCC 11778), E. coli (ATCC 8739), E. faecium (ATCC 27270), P. aeruginosa (ATCC 27853) and B. subtilis (ATCC 6051), were purchased from Davies Diagnostics (Pty) Ltd. (Randburg, South Africa). All solvents (AR grade), sulfuric and formic acids (AAR grade) were obtained from Merck (Pty) Ltd. (Modderfontein, South Africa). Ciprofloxacin discs (5 µg, Oxoid; product code CT0425B), were purchased from Thermo Fisher Scientific, Randburg, South Africa. Reference standards of individual compounds rosmarinic acid (≥98%), caffeic acid (≥98%), carnosol (100%), carnosic acid (97.6%) and ursolic acid (≥90%) were purchased from Merck (Pty) Ltd. (Modderfontein, South Africa). High-performance thin layer chromatography was performed using 20 × 10 cm HPTLC silica gel 60 F254 chromatographic plates (Merck KGaA, Darmstadt, Germany).
4.3. Antibacterial Activity Determination Using the Microdilution Assay
The MIC for each sample extract was determined towards each of the seven pathogens using the in vitro microdilution assay [49] with modifications [50]. The selected pathogens were inoculated in TSB at 37 °C for 24 h in an incubator (EcoTherm, Hartkirchen, Austria). The resulting cultures were diluted with fresh TSB to yield a concentration of approximately 1 × 106 colony forming units (CFU)/mL inoculum, which was estimated by visual comparison of turbidity to a McFarland standard (0.5) solution. Into each well of a sterile 96-well microtiter plate, 100 µL of sterile TSB broth was plated. This was followed by adding 100 µL of 32 mg/mL plant extract solution in acetone and serial dilution followed. All determinations were conducted in duplicate. The final concentrations of the plant extract in the wells from rows A to H ranged from 8.0 to 0.075 mg/mL, halving with each dilution. After sealing the plate with clear tape, it was placed overnight in an incubator at 37 °C. A 40 µL volume of INT in water solution (0.04% w/v) was subsequently plated into each well.
The plates were observed for colour change after 2–6 h, the time depending on the pathogen as indicated by the growth controls, due to the interaction of the INT solution with the bacterial organisms. The growth controls were prepared for each pathogen by mixing 100 µL TSB with the pathogen inoculum. Sterile water was used as the negative control by adding 100 µL to a well before inoculation. Positive controls were prepared by adding 100 µL of a ciprofloxacin solution (0.01 mg/mL in water) to a well and a culture control sample to confirm bacterial growth. The MIC value was determined as the lowest concentrations in each column of the microplate where no pink/purple colour was visible. The MICs for the pure compounds, carnosol, carnosic acid and ursolic acid, dissolved in methanol and further diluted in acetone, were determined using the same method, towards B. cereus and A. baumannii, two of the pathogens that were inhibited at promising MIC levels by the sample extracts. The antibacterial activities of each of the three species towards the seven pathogens were statistically compared by ANOVA single factor analysis. Significant differences between the three species were confirmed where p < 0.05.
4.4. Identification of Bioactive Compounds Using Biochemometric Analysis
Potentially bioactive compounds were identified using an untargeted OPLS-DA approach [14]. The averages of the duplicate MICs determined for the samples from each species for a specific pathogen, were combined with the corresponding UPLC-MS dataset that was utilised for a previous variation study [12]. Using SIMCA® P+ 14 software (Umetrics, Umea, Sweden), an individual OPLS-DA model was created for each pathogen. Class allocation was conducted according to activity. Samples with corresponding MIC values ≤ 1.0 mg/mL were categorised as active (Class 1), while those with MIC values > 1.0 mg/mL, as inactive (Class 2).
For a few pathogens, all sample extracts were active (MIC values ≤ 1.0 mg/mL), and the resulting MIC values were therefore classed as more active and less active, this resulted in two groups to enable the construction of a discriminant analysis (OPLS-DA) model. Bioactive compounds were thus identified for the more active samples only, and these varied from pathogen to pathogen. Pseudomonas aeruginosa was inhibited by all the samples representing the three species and therefore, for S. africana-lutea and S. chamelaeagnea, samples with MIC values ≤ 0.50 mg/mL were classed as more active (Class 1) and MIC values > 0.50 mg/mL as less active (Class 2). However, for S. lanceolata, most of the MICs for the extracts were below 0.38 mg/mL and therefore MIC values ≤ 0.38 mg/mL were classified as more active (Class 1), and the remaining MIC values > 0.38 mg/mL as less active (Class 2). The same approach was followed for the other pathogens that were susceptible to all the samples. The specific values selected are indicated in Table 6.

Table 6.
Criteria used to separate the MIC values of pathogens that displayed moderate susceptibility to all extracts of the three species into classes for biochemometric analysis and identification of marker compounds.
Loadings S-plots revealed unique variables for each species/pathogen combination that were separated from the rest of the data matrix. These variables were confirmed by group-to-average comparison and by the contribution plot. Retention time/mass-to-charge ratio pairs (Rt/m/z) were revealed and compared to those previously identified in a separate study from mass spectral data [12].
4.5. Thin Layer Chromatography-Direct Bioautography
The chromatographic fingerprints of selected extracts were prepared by HPTLC to separate the crude extract into its components and apply biological detection of compounds with antimicrobial activity using TLC-DB [19]. The sample extracts (20 µL) were applied (10 mg/mL) to the HPTLC plates as 6 mm bands and developed to an 80 mm endpoint using a semi-automated HPTLC system (CAMAG, Muttenz, Switzerland). The plates loaded with extract were developed with the optimised mobile phase (ethyl acetate:toluene:formic acid, 30:60:10). One of the smaller developed plates was kept aside to mark the identified active sites or zones for further HPTLC-MS analysis, and another was sprayed with p-anisaldehyde for visualising compounds, and this served as a reference plate for identification of compounds.
A petri dish filled with 15 mL of TSA to form a thin layer, was left to solidify. After the developed plate was sterilised under UV light for an hour in a sterile laminar flow cabinet, it was placed face-up onto the surface of the solid TSA layer. The plate was then covered with a layer of pathogen-inoculated TSA. Three pathogens, that were inhibited at the most promising MIC levels by the extracts, were used for TLC-DB, namely B. cereus, A. baumannii and P. aeruginosa. Sterile paper discs (prepared in-house by punching filter paper and subsequent autoclaving), each infused with 40 µL of the 10 mg/mL solution of a crude sample extract corresponding to the extract applied to the plate, were placed on the surface of the solidified inoculated TSA layer to evaluate inhibition by the whole extract. A ciprofloxacin disc (5 µg) was also placed on the surface of the inoculated TSA and used as a positive control. This set-up was placed in the incubator overnight at 37 °C, where after INT solution was gently poured onto the surface of the inoculated TSA. The pink/purple colour formation was observed where bacterial growth took place, while the cream white areas indicated zones of inhibition around active compounds on the bioautograms [28], crude extract discs, and positive control.
4.6. Identification of Bioactive Compounds Using Hyphenated Thin Layer Chromatography-Mass Spectrometry
The HPTLC fingerprints of the sample extracts were prepared as described in Section 4.5. The resulting plate is a separate plate, but identical to the plate used for TLC-DB, as described in Section 4.5. The target zones to be extracted were marked according to the corresponding retardation factor (Rf) of the active zones observed after TLC-DB. The zones with possible bioactivity were extracted one-by-one from the prepared plate using the semi-automated CAMAG TLC Interface (CAMAG Laboratory, Muttenz, Switzerland). Acetonitrile was used as eluent at a flow rate of 0.1 mL/min (run time: 1 min). The eluent was then analysed in negative (ESI) mode following direct inlet into the quadrupole-Time-of-Flight-mass spectrometer (QToF- MS) (Waters Xevo® G2 QToF, Waters, Milford, MA, USA) using MS settings as previously published [12]. The mass spectrum obtained revealed the m/z of ions that were compared and correlated to those reported in the previous study, enabling tentative identification of the compounds. The identified compounds were also correlated with those revealed through biochemometric analysis.
5. Conclusions
Antimicrobial screening indicated promising bioactivity for the majority of the extracts towards B. cereus, A. baumannii, P. aeruginosa, and B. subtilis, confirming the potential of these species as antibacterial agents, and substantiating their use as traditional remedies for infections associated with the pathogens. The large sample size indicated different antibacterial activities for samples from the same population, confirming the importance of considering chemical variation by incorporating many samples into the research design. Salvia chamelaeagnea was active towards all seven of the selected pathogens, displaying encouraging activity towards B. cereus and A. baumannii. Biochemometric analysis revealed compounds with potential bioactivity. Sample extracts with the best antibacterial activity identified through MIC screening were used for TLC-DB. Bioactive zones were highlighted by TLC-DB for extracts of the three species towards A. baumannii, a refractory pathogen known to be resistant to many antibacterial drugs. The technique demonstrated in this study is relatively simple for revealing potential bioactive constituents in plant extracts. Phytoconstituents present in the active zones were extracted and analysed by direct MS, which enabled the identification of potential bioactive compounds, by using data generated during the previous chemical variation study as a guide. Some of the identified compounds correlated to those predicted through biochemometric analysis. This serves as verification that a biochemometric approach can be used for predicting compounds with a high probability of good antibacterial activity. The use, in sequence, of these assays and tools, namely chromatographic analysis, the microdilution assay, biochemometric data analysis, bioautography, and HPTLC-MS, is a relatively simple, rapid, functional, and environmentally friendly approach for identifying bioactive constituents present in medicinal plants. Valuable insights into the antibacterial activity of S. africana-lutea, S. lanceolata, and S. chamelaeagnea were gained in this study that can be implemented during the selection of chemotypes for commercial development.
Author Contributions
Conceptualization, A.V. and S.V.V.; methodology, A.V., S.V.V., M.L.A.T. and W.C.; software, M.L.A.T. and W.C.; validation, M.L.A.T. and W.C.; formal analysis, M.L.A.T. and W.C.; investigation, M.L.A.T.; resources, A.V. and S.V.V.; data curation, M.L.A.T.; writing—original draft preparation, M.L.A.T.; writing—review and editing, A.V., S.C., G.K., S.V.V. and W.C.; visualization, M.L.A.T.; supervision, A.V., S.C. and G.K.; project administration, M.L.A.T.; funding acquisition, A.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Research Foundation (NRF Research Chairs program, NRF Grant No. 86923) of South Africa, the South African Medical Research Council (SAMRC, EMU Grant No. 23015) and Tshwane University of Technology (TUT).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors appreciate financial support from the National Research Foundation (NRF Research Chairs program) of South Africa, the South African Medical Research Council (SAMRC), and Tshwane University of Technology (TUT). We thank John Manning for sample collection and species identification.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Watt, J.M.; Breyer-Brandwijk, M.G. The Medicinal and Poisonous Plants of Southern and Eastern Africa, 2nd ed.; Livingstone Ltd.: Edinburgh, UK, 1962. [Google Scholar]
- Bocanegra-García, V.; Del Rayo Camacho-Corona, M.; Ramírez-Cabrera, M.; Rivera, G.; Garza-Gonzlez, E. The bioactivity of plant extracts against representative bacterial pathogens of the lower respiratory tract. BMC Res. Notes 2009, 2, 95. [Google Scholar] [CrossRef] [PubMed]
- Hanphanphoom, S.; Krajangsang, S.; Thophon, S.; Waranusantigul, P.; Kangwanrangsan, N.; Sukhumaporn, K. Antimicrobial activity of Chromolaena odorata extracts against bacterial human skin infections. Mod. Appl. Sci. 2016, 10, 159–171. [Google Scholar] [CrossRef]
- Mishra, M.P.; Padhy, R.N. In vitro antibacterial efficacy of 21 Indian timber-yielding plants against multidrug-resistant bacteria causing urinary tract infection. Osong Public Heal. Res. Perspect. 2013, 4, 347–357. [Google Scholar] [CrossRef]
- Jensen, G.B.; Hansen, B.M.; Eilenberg, J.; Mahillon, J. The hidden lifestyles of Bacillus cereus and relatives. Environ. Microbiol. 2003, 5, 631–640. [Google Scholar] [CrossRef]
- Li, L.; Yan, Q.; Ma, N.; Chen, X.; Li, G.; Liu, M. Analysis of intestinal flora and inflammatory cytokine levels in children with non-infectious diarrhea. Transl. Pediatr. 2021, 10, 1340–1345. [Google Scholar] [CrossRef] [PubMed]
- Kamatou, G.P.P.; Viljoen, A.M.; Gono-Bwalya, A.B.; Van Zyl, R.L.; Van Vuuren, S.F.; Lourens, A.C.U.; Başer, K.H.C.; Demirci, B.; Lindsey, K.L.; Van Staden, J.; et al. The in vitro pharmacological activities and a chemical investigation of three South African Salvia species. J. Ethnopharmacol. 2005, 102, 382–390. [Google Scholar] [CrossRef]
- Kamatou, G.P.P.; Van Vuuren, S.F.; Van Heerden, F.R.; Seaman, T.; Viljoen, A.M. Antibacterial and antimycobacterial activities of South African Salvia species and isolated compounds from S. chamelaeagnea. S. Afr. J. Bot. 2007, 73, 552–557. [Google Scholar] [CrossRef]
- Kamatou, G.P.P. Indigenous Salvia Species—An Investigation of Their Pharmacological Activities and Phytochemistry; University of Witwatersrand: Johannesburg, South Africa, 2006. [Google Scholar]
- Afonso, A.F.; Pereira, O.R.; Fernandes, Â.S.F.; Calhelha, R.C.; Silva, A.M.S.; Ferreira, I.C.F.R.; Cardoso, S.M. The health-benefits and phytochemical profile of Salvia apiana and Salvia farinacea Var. Victoria Blue decoctions. Antioxidants 2019, 8, 241. [Google Scholar] [CrossRef]
- Lim Ah Tock, M.J.; Kamatou, G.P.P.; Combrinck, S.; Sandasi, M.; Viljoen, A.M. A chemometric assessment of essential oil variation of three Salvia species indigenous to South Africa. Phytochemistry 2020, 172, 112249. [Google Scholar] [CrossRef]
- Lim Ah Tock, M.J.; Chen, W.; Combrinck, S.; Sandasi, M.; Kamatou, G.P.P.; Viljoen, A.M. Exploring the phytochemical variation of non-volatile metabolites within three South African Salvia species using UPLC-MS fingerprinting and chemometric analysis. Fitoterapia 2021, 152, 104940. [Google Scholar] [CrossRef]
- Kamatou, G.P.P.; Van Zyl, R.L.; Van Vuuren, S.F.; Figueiredo, A.C.; Barroso, J.G.; Pedro, L.G.; Viljoen, A.M. Seasonal variation in essential oil composition, oil toxicity and the biological activity of solvent extracts of three South African Salvia species. S. Afr. J. Bot. 2008, 74, 230–237. [Google Scholar] [CrossRef]
- Kellogg, J.J.; Todd, D.A.; Egan, J.M.; Raja, H.A.; Oberlies, N.H.; Kvalheim, O.M.; Cech, N.B. Biochemometrics for Natural Products Research: Comparison of data analysis approaches and application to identification of bioactive compounds. J. Nat. Prod. 2016, 79, 376–386. [Google Scholar] [CrossRef]
- Suleman, T.; Van Vuuren, S.; Sandasi, M.; Viljoen, A.M. Antimicrobial activity and chemometric modelling of South African Propolis. J. Appl. Microbiol. 2015, 119, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Choma, I.; Jesionek, W. TLC-Direct Bioautography as a high throughput method for detection of antimicrobials in plants. Chromatography 2015, 2, 225–238. [Google Scholar] [CrossRef]
- Dewanjee, S.; Gangopadhyay, M.; Bhattacharya, N.; Khanra, R.; Dua, T.K. Bioautography and its scope in the field of natural product chemistry. J. Pharm. Anal. 2015, 5, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Marston, A. Thin-layer chromatography with biological detection in phytochemistry. J. Chromatogr. A 2011, 1218, 2676–2683. [Google Scholar] [CrossRef]
- Kasote, D.; Ahmad, A.; Chen, W.; Combrinck, S.; Viljoen, A. HPTLC-MS as an efficient hyphenated technique for the rapid identification of antimicrobial compounds from Propolis. Phytochem. Lett. 2015, 11, 326–331. [Google Scholar] [CrossRef]
- Eloff, J.N. A proposal towards a rational classification of the antimicrobial activity of acetone tree leaf extracts in a search for new antimicrobials. Planta Med. 2021, 87, 836–840. [Google Scholar] [CrossRef]
- Sato, Y.; Unno, Y.; Miyazaki, C.; Ubagai, T.; Ono, Y. Multidrug-resistant Acinetobacter Baumannii resists reactive oxygen species and survives in macrophages. Sci. Rep. 2019, 9, 17462. [Google Scholar] [CrossRef]
- Andrade, J.M.; Faustino, C.; Garcia, C.; Ladeiras, D.; Reis, C.P.; Rijo, P. Rosmarinus officinalis L.: An update review of its phytochemistry and biological activity. Futur. Sci. OA 2018, 4, FSO283. [Google Scholar] [CrossRef]
- Jordán, M.J.; Lax, V.; Rota, M.C.; Lorán, S.; Sotomayor, J.A. Relevance of carnosic acid, carnosol, and rosmarinic acid concentrations in the in vitro antioxidant and antimicrobial activities of Rosmarinus officinalis (L.) Methanolic Extracts. J. Agric. Food Chem. 2012, 60, 9603–9608. [Google Scholar] [CrossRef] [PubMed]
- Klancnik, A.; Kolar, M.H.; Abramovi, H.; Mozina, S.S. In vitro antimicrobial and antioxidant activity of commercial of commercial rosemary extract formulations. J. Food Prot. 2009, 72, 1744–1752. [Google Scholar] [CrossRef] [PubMed]
- Khin, M.; Knowles, S.L.; Crandall, W.J.; Jones, D.D.; Oberlies, N.H.; Cech, N.B.; Houriet, J. Capturing the antimicrobial profile of Rosmarinus officinalis against methicillin-resistant Staphylococcus aureus (MRSA) with bioassay-guided fractionation and bioinformatics. J. Pharm. Biomed. Anal. 2021, 197, 113965. [Google Scholar] [CrossRef]
- Do Nascimento, P.G.G.; Lemos, T.L.G.; Bizerra, A.M.C.; Arriaga, Â.M.C.; Ferreira, D.A.; Santiago, G.M.P.; Braz-Filho, R.; Galberto, J.; Costa, M.; Br, A.M.C.A. Antibacterial and antioxidant activities of ursolic acid and derivatives. Molecules 2014, 19, 1317–1327. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.P.; Mitra, A.; Bhattacharya, A.; Sen, A. Quorum quenching activity of pentacyclic triterpenoids leads to inhibition of biofilm formation by Acinetobacter baumannii. Biofouling 2020, 36, 922–937. [Google Scholar] [CrossRef] [PubMed]
- Jesionek, W.; Majer-Dziedzic, B.; Horváth, G.; Móricz, Á.M.; Choma, I.M. Screening of antibacterial compounds in Salvia officinalis L. Tincture Using Thin-Layer Chromatography-Direct Bioautography and Liquid Chromatography-Tandem mass spectrometry techniques. J. Planar Chromatogr. Mod. TLC 2017, 30, 357–362. [Google Scholar] [CrossRef]
- Caesar, L.K.; Cech, N.B.; Kubanek, J.; Linington, R.; Luesch, H. Synergy and antagonism in natural product extracts: When 1 + 1 does not equal 2. Nat. Prod. Rep. 2019, 36, 845–936. [Google Scholar] [CrossRef]
- Hemeg, H.A.; Moussa, I.M.; Ibrahim, S.; Dawoud, T.M.; Alhaji, J.H.; Mubarak, A.S.; Kabli, S.A.; Alsubki, R.A.; Tawfik, A.M.; Marouf, S.A. Antimicrobial effect of different herbal plant extracts against different microbial population. Saudi J. Biol. Sci. 2020, 27, 3221–3227. [Google Scholar] [CrossRef]
- Afonso, A.F.; Pereira, O.R.; Fernandes, Â.; Calhelha, R.C.; Silva, A.M.S.; Ferreira, R.C.F.; Cardoso, S.M. Phytochemical composition and bioactive effects of Salvia africana, Salvia officinalis “Icterina” and Salvia mexicana aqueous extracts. Molecules 2019, 24, 4327. [Google Scholar] [CrossRef]
- Yang, C.H.; Su, P.W.; Moi, S.H.; Chuang, L.Y. Biofilm formation in Acinetobacter baumannii: Genotype-phenotype correlation. Molecules 2019, 24, 1849. [Google Scholar] [CrossRef]
- Almosa, A.A.O.; Al-Salamah, A.A.; Maany, D.A.; El-Abd, M.A.E.; Ibrahim, A.S.S. Biofilm formation by clinical Acinetobacter Baumannii strains and its effect on antibiotic resistance. Egypt. J. Chem. 2021, 64, 1615–1625. [Google Scholar] [CrossRef]
- Sharifipour, E.; Shams, S.; Esmkhani, M.; Khodadadi, J.; Fotouhi-Ardakani, R.; Koohpaei, A.; Doosti, Z.; Ej Golzari, S. Evaluation of bacterial co-infections of the respiratory tract in COVID-19 patients admitted to ICU. BMC Infect. Dis. 2020, 20, 646. [Google Scholar] [CrossRef]
- Huttunen, S.; Toivanen, M.; Liu, C.; Tikkanen-Kaukanen, C. Novel anti-infective potential of salvianolic acid B against human serious pathogen Neisseria meningitidis. BMC Res. Notes 2016, 9, 25. [Google Scholar] [CrossRef][Green Version]
- Ghorbani, A.; Esmaeilizadeh, M. Pharmacological properties of Salvia officinalis and its components. J. Tradit. Complement. Med. 2017, 7, 433–440. [Google Scholar] [CrossRef]
- Horiuchi, K.; Shiota, S.; Hatano, T.; Yoshida, T.; Kuroda, T.; Tsuchiya, T. Antimicrobial activity of oleanolic acid from Salvia officinalis and related compounds on vancomycin-resistant Enterococci (VRE). Biol. Pharm. Bull. 2007, 30, 1147–1149. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, K.; Shiota, S.; Kuroda, T.; Hatano, T.; Yoshida, T.; Tsuchiya, T. Potentiation of antimicrobial activity of aminoglycosides by carnosol from Salvia officinalis. Biol. Pharm. Bull. 2007, 30, 287–290. [Google Scholar] [CrossRef] [PubMed]
- Climati, E.; Mastrogiovanni, F.; Valeri, M.; Salvini, L.; Bonechi, C.; Mamadalieva, N.Z.; Egamberdieva, D.; Taddei, A.R.; Tiezzi, A. Methyl carnosate, an antibacterial diterpene isolated from Salvia officinalis leaves. Nat. Prod. Commun. 2013, 8, 429–430. [Google Scholar] [CrossRef] [PubMed]
- Sharma, Y.; Velamuri, R.; Fagan, J.; Schaefer, J. Full-spectrum analysis of bioactive compounds in rosemary (Rosmarinus officinalis L.) as influenced by different extraction methods. Molecules 2020, 25, 4599. [Google Scholar] [CrossRef]
- Jesus, J.A.; Lago, J.H.G.; Laurenti, M.D.; Yamamoto, E.S.; Passero, L.F.D. Antimicrobial activity of oleanolic and ursolic acids: An update. Evidence-based Complement. Altern. Med. 2015, 2015, 620472. [Google Scholar] [CrossRef]
- Jesionek, W.; Grzelak, E.M.; Majer-Dziedzi, B.; Chom, I.M. Thin-layer chromatography - direct bioautography for the screening of antimicrobial properties of plant extracts. J. Planar Chromatogr. Mod. TLC 2013, 26, 109–113. [Google Scholar] [CrossRef]
- Dilika, F.; Meyer, J.J.M. Antibacterial Activity of Helichrysum pedunculatum callus cultures. S. Afr. J. Bot. 1998, 64, 312–313. [Google Scholar] [CrossRef]
- Suleiman, M.M.; McGaw, L.J.; Naidoo, V.; Eloff, J.N. Detection of antimicrobial compounds by bioautography of different extracts of leaves of South African tree species. African J. Tradit., Compliment. Altern. Med. 2010, 7, 64–78. [Google Scholar] [CrossRef] [PubMed]
- Ristivojević, P.M.; Tahir, A.; Malfent, F.; Opsenica, D.M.; Rollinger, J.M. High-performance thin-layer chromatography/bioautography and liquid chromatography-mass spectrometry hyphenated with chemometrics for the quality assessment of Morus alba samples. J. Chromatogr. A 2019, 1594, 190–198. [Google Scholar] [CrossRef]
- Cornejo-Báez, A.A.; Peña-Rodríguez, L.M.; Álvarez-Zapata, R.; Vázquez-Hernández, M.; Sánchez-Medina, A. Chemometrics: A complementary tool to guide the isolation of pharmacologically active natural products. Drug Discov. Today 2020, 25, 27–37. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, Y.; Wang, R.; Wang, Z.; Yang, B.; Kuang, H. An evolving technology that integrates classical methods with continuous technological developments: Thin-layer chromatography bioautography. Molecules 2021, 26, 4647. [Google Scholar] [CrossRef] [PubMed]
- Caesar, L.K.; Kellogg, J.J.; Kvalheim, O.M.; Cech, R.A.; Cech, N.B. Integration of biochemometrics and molecular networking to identify bioactive constituents of ashitaba (Angelica Keiskei Koidzumi). Planta Med. 2018, 84, 721. [Google Scholar] [CrossRef] [PubMed]
- Eloff, J.N. A sensitive and quick microplate method to determine the minimal inhibitory concentration of plant extracts for bacteria. Planta Med. 1998, 64, 711–713. [Google Scholar] [CrossRef] [PubMed]
- Van Vuuren, S.F.; Nkwanyana, M.N.; de Wet, H. Antimicrobial evaluation of plants used for the treatment of diarrhoea in a rural community in Northern Maputaland, KwaZulu-Natal, South Africa. BMC Complement. Altern. Med. 2015, 15, 53. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).