Nisin Mutant Prevention Concentration and the Role of Subinhibitory Concentrations on Resistance Development by Diabetic Foot Staphylococci
Abstract
:1. Introduction
2. Results and Discussion
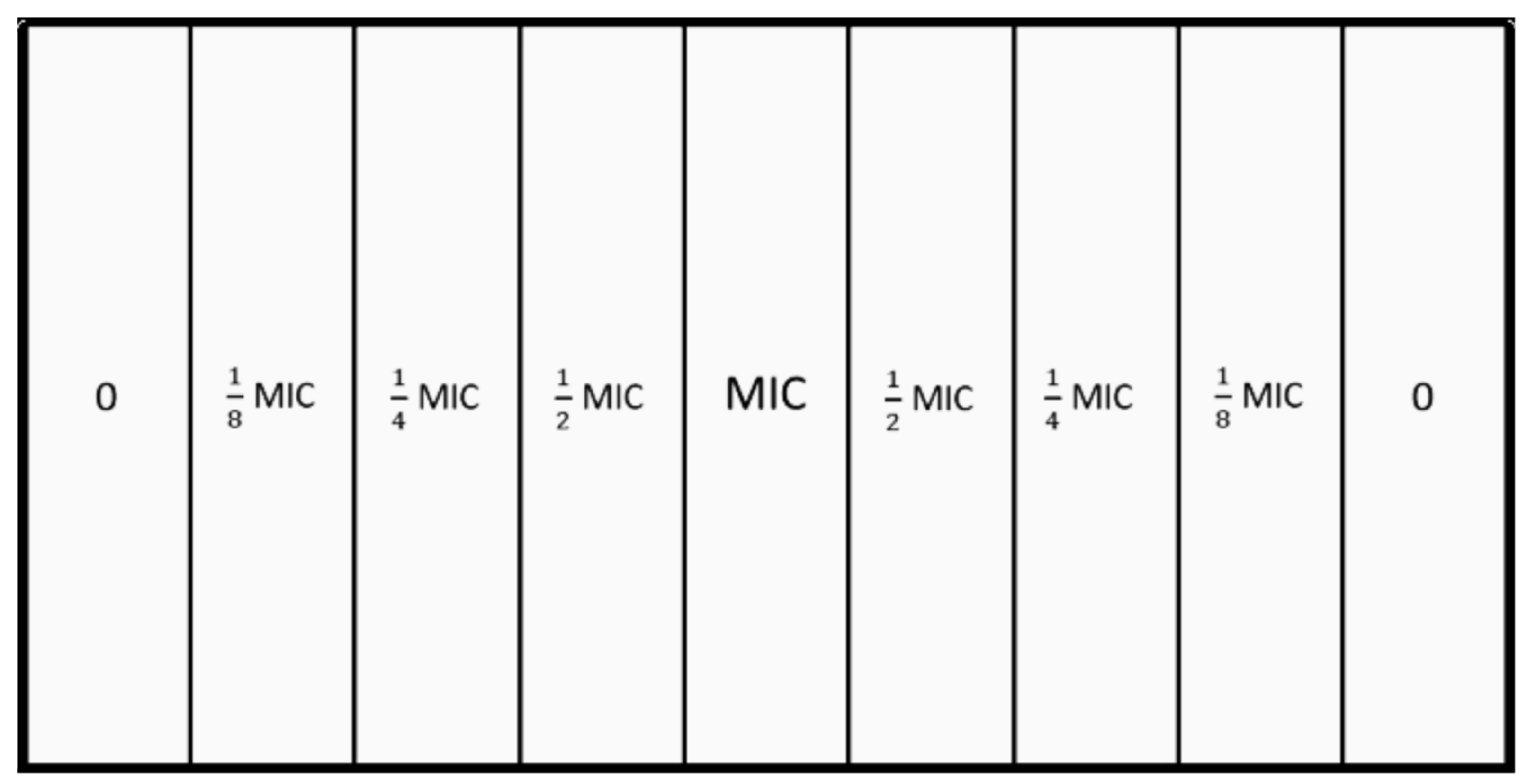
2.1. Mutant Selection Window
2.2. Horizontal Gene Transfer
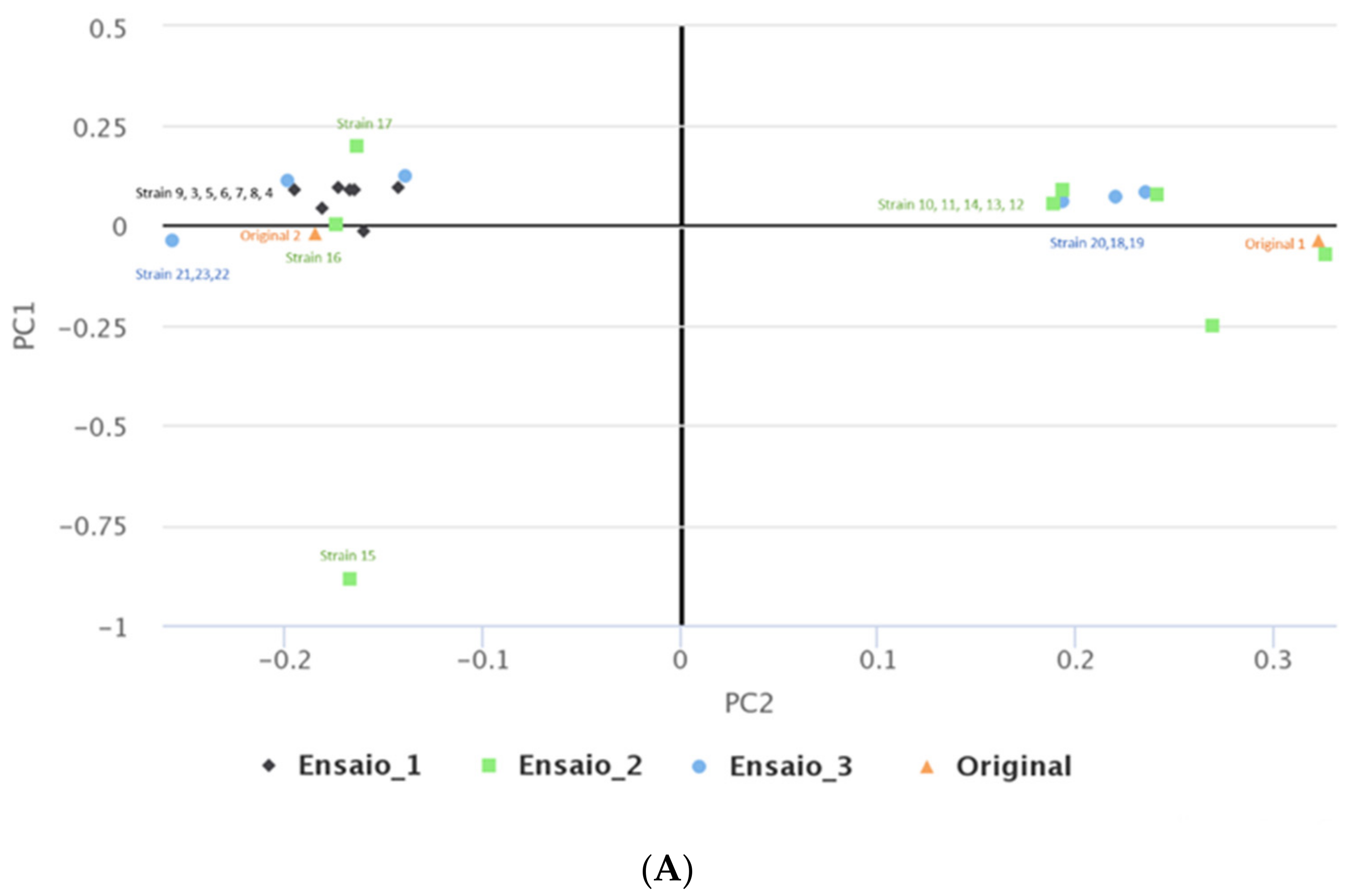
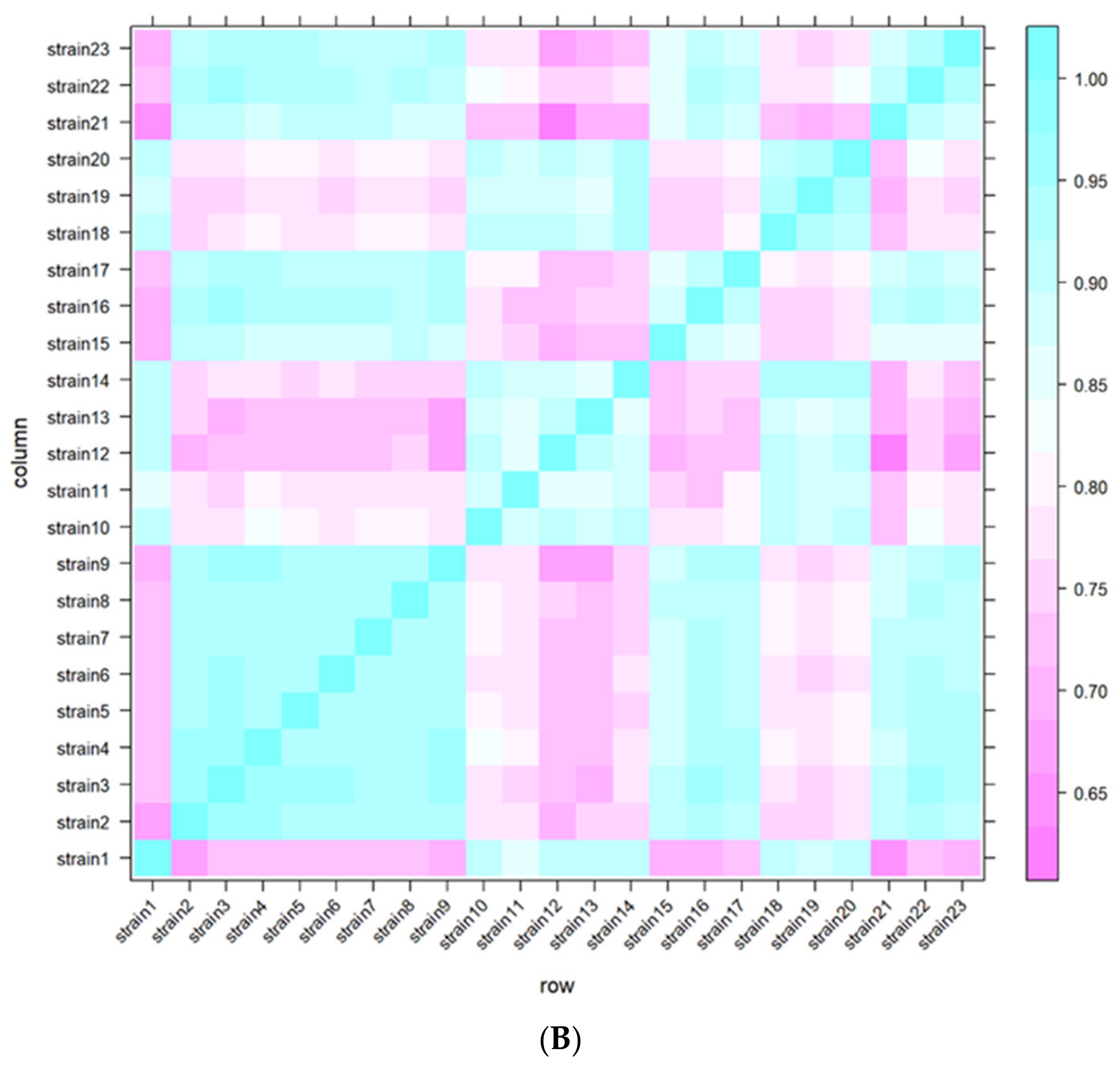
2.3. MEGA-Plate Assays and Variant Call Analysis
3. Materials and Methods
3.1. Bacterial Isolates
3.2. Nisin Solution
3.3. Determination of the Mutant Prevention Concentration
3.4. Horizontal Gene Transfer
3.4.1. DNA Extraction
3.4.2. Multiplex PCR for vanA Detection
3.4.3. Horizontal Gene Transfer protocol
3.4.4. PCR for pSK41-like Plasmid Detection
3.5. MEGA-Plate Assays and Variant Call Analysis
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, N. Preventing Foot Ulcers in Patients with Diabetes. JAMA 2005, 293, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Skrepnek, G.H.; Mills, J.L.; Lavery, L.A.; Armstrong, D.G. Health Care Service and Outcomes Among an Estimated 6.7 Million Ambulatory Care Diabetic Foot Cases in the U.S. Diabetes Care 2017, 40, 936–942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Citron, D.M.; Goldstein, E.J.C.; Merriam, C.V.; Lipsky, B.A.; Abramson, M.A. Bacteriology of Moderate-to-Severe Diabetic Foot Infections and In Vitro Activity of Antimicrobial Agents. J. Clin. Microbiol. 2007, 45, 2819–2828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hobizal, K.B.; Wukich, D.K. Diabetic Foot Infections: Current Concept Review. Diabet. Foot Ankle 2012, 3, 18409. [Google Scholar] [CrossRef] [PubMed]
- Weigel, L.M.; Clewell, D.B.; Gill, S.R.; Clark, N.C.; McDougal, L.K.; Flannagan, S.E.; Kolonay, J.F.; Shetty, J.; Killgore, G.E.; Tenover, F.C. Genetic Analysis of a High-Level Vancomycin-Resistant Isolate of Staphylococcus aureus. Science 2003, 302, 1569–1571. [Google Scholar] [CrossRef]
- Sujatha, S.; Praharaj, I. Glycopeptide Resistance in Gram-Positive Cocci: A Review. Interdiscip. Perspect. Infect. Dis. 2012, 2012, 781679. [Google Scholar] [CrossRef] [Green Version]
- Mendes, J.J.; Marques-Costa, A.; Vilela, C.; Neves, J.; Candeias, N.; Cavaco-Silva, P.; Melo-Cristino, J. Clinical and Bacteriological Survey of Diabetic Foot Infections in Lisbon. Diabetes Res. Clin. Pract. 2012, 95, 153–161. [Google Scholar] [CrossRef]
- Kos, V.N.; Desjardins, C.A.; Griggs, A.; Cerqueira, G.; Van Tonder, A.; Holden, M.T.G.; Godfrey, P.; Palmer, K.L.; Bodi, K.; Mongodin, E.F.; et al. Comparative Genomics of Vancomycin-Resistant Staphylococcus aureus Strains and Their Positions within the Clade Most Commonly Associated with Methicillin-Resistant S. Aureus Hospital-Acquired Infection in the United States. mBio 2012, 3, e00112-12. [Google Scholar] [CrossRef] [Green Version]
- Gardete, S.; Tomasz, A. Mechanisms of Vancomycin Resistance in Staphylococcus aureus. J. Clin. Investig. 2014, 124, 2836–2840. [Google Scholar] [CrossRef]
- McDougal, L.K.; Fosheim, G.E.; Nicholson, A.; Bulens, S.N.; Limbago, B.M.; Shearer, J.E.S.; Summers, A.O.; Patel, J.B. Emergence of Resistance among USA300 Methicillin-Resistant Staphylococcus aureus Isolates Causing Invasive Disease in the United States. Antimicrob. Agents Chemother. 2010, 54, 3804–3811. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.; Clark, N.; Patel, J.B. PSK41-like Plasmid Is Necessary for Inc18-like VanA Plasmid Transfer from Enterococcus faecalis to Staphylococcus aureus in Vitro. Antimicrob. Agents Chemother. 2013, 57, 212–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gottrup, F.; Apelqvist, J. Present and New Techniques and Devices in the Treatment of DFU: A Critical Review of Evidence. Diabetes Metab. Res. Rev. 2012, 28, 64–71. [Google Scholar] [CrossRef]
- Bahar, A.A.; Ren, D. Antimicrobial Peptides. Pharm. Basel Switz. 2013, 6, 1543–1575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Gallo, R.L. Antimicrobial Peptides. Curr. Biol. 2016, 26, R14–R19. [Google Scholar] [CrossRef] [PubMed]
- Abee, T.; Delves-Broughton, J. Bacteriocins—Nisin. In Food Preservatives; Russell, N.J., Gould, G.W., Eds.; Springer US: Boston, MA, USA, 2003; pp. 146–178. [Google Scholar] [CrossRef]
- Hassan, M.; Kjos, M.; Nes, I.F.; Diep, D.B.; Lotfipour, F. Natural Antimicrobial Peptides from Bacteria: Characteristics and Potential Applications to Fight against Antibiotic Resistance. J. Appl. Microbiol. 2012, 113, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, S.A.; Truscott, F.; Dickman, R.; Ward, J.; Tabor, A.B. Simplified Lipid II-Binding Antimicrobial Peptides: Design, Synthesis and Antimicrobial Activity of Bioconjugates of Nisin Rings A and B with Pore-Forming Peptides. Bioorg. Med. Chem. 2018, 26, 5691–5700. [Google Scholar] [CrossRef]
- Field, D.; Cotter, P.D.; Hill, C.; Ross, R.P. Bioengineering Lantibiotics for Therapeutic Success. Front. Microbiol. 2015, 6, 1363. [Google Scholar] [CrossRef] [Green Version]
- Lagedroste, M.; Reiners, J.; Smits, S.H.J.; Schmitt, L. Systematic Characterization of Position One Variants within the Lantibiotic Nisin. Sci. Rep. 2019, 9, 935. [Google Scholar] [CrossRef]
- Zhou, H.; Fang, J.; Tian, Y.; Lu, X.Y. Mechanisms of Nisin Resistance in Gram-Positive Bacteria. Ann. Microbiol. 2014, 64, 413–420. [Google Scholar] [CrossRef]
- Shin, J.M.; Gwak, J.W.; Kamarajan, P.; Fenno, J.C.; Rickard, A.H.; Kapila, Y.L. Biomedical Applications of Nisin. J. Appl. Microbiol. 2016, 120, 1449–1465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, R.; Veiga, A.S.; Tavares, L.; Castanho, M.; Oliveira, M. Bacterial Biofilms in Diabetic Foot Ulcers: Potential Alternative Therapeutics. In Microbial Biofilms—Importance and Applications; IntechOpen: London, UK, 2016. [Google Scholar] [CrossRef] [Green Version]
- Cunha, E.; Trovão, T.; Pinheiro, A.; Nunes, T.; Santos, R.; Moreira da Silva, J.; São Braz, B.; Tavares, L.; Veiga, A.S.; Oliveira, M. Potential of Two Delivery Systems for Nisin Topical Application to Dental Plaque Biofilms in Dogs. BMC Vet. Res. 2018, 14, 375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xilin, Z.; Drlica, K. Restricting the Selection of Antibiotic-Resistant Mutant Bacteria: Measurement and Potential Use of the Mutant Selection Window. J. Infect. Dis. 2002, 185, 561–565. [Google Scholar] [CrossRef]
- Cairns, B.J.; Payne, R.J.H. Bacteriophage Therapy and the Mutant Selection Window. Antimicrob. Agents Chemother. 2008, 52, 4344–4350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drlica, K. The Mutant Selection Window and Antimicrobial Resistance. J. Antimicrob. Chemother. 2003, 52, 11–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drlica, K.; Zhao, X. Mutant Selection Window Hypothesis Updated. Clin. Infect. Dis. 2007, 44, 681–688. [Google Scholar] [CrossRef] [PubMed]
- Vasilchenko, A.S.; Rogozhin, E.A. Sub-inhibitory effects of antimicrobial peptides. Front Microbiol. 2019, 10, 1160. [Google Scholar] [CrossRef] [Green Version]
- Gottschalk, S.; Gottlieb, C.T.; Vestergaard, M.; Hansen, R.; Gram, L.; Ingmer, H.; Thomsen, L.E. Amphibian antimicrobial peptide fallaxin analogue FL9 affects virulence gene expression and DNA replication in Staphylococcus aureus. J. Med. Microbiol. 2015, 64, 1504–1513. [Google Scholar] [CrossRef] [Green Version]
- Polikanov, Y.S.; Aleksashin, N.A.; Beckert, B.; Wilson, D.N. The mechanisms of action of ribosome-targeting peptide antibiotics. Front. Mol. Biosci. 2018, 5, 48. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, T.; Harada, K.; Kataoka, Y. Mutant Prevention Concentration of Orbifloxacin: Comparison between Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus pseudintermedius of Canine Origin. Acta Vet. Scand. 2013, 55, 37. [Google Scholar] [CrossRef] [Green Version]
- López, Y.; Tato, M.; Gargallo-Viola, D.; Cantón, R.; Vila, J.; Zsolt, I. Mutant Prevention Concentration of Ozenoxacin for Quinolone-Susceptible or -Resistant Staphylococcus aureus and Staphylococcus epidermidis. PLoS ONE 2019, 14, e0223326. [Google Scholar] [CrossRef]
- Raj Dwivedi, G.; Khwaja, S.; Singh Negi, A.; Panda, S.S.; Swaroop Sanket, A.; Pati, S.; Chand Gupta, A.; Bawankule, D.U.; Chanda, D.; Kant, R.; et al. Design, Synthesis and Drug Resistance Reversal Potential of Novel Curcumin Mimics Van D: Synergy Potential of Curcumin Mimics. Bioorganic Chem. 2021, 106, 104454. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.; Gomes, D.; Macedo, H.; Barros, D.; Tibério, C.; Veiga, A.S.; Tavares, L.; Castanho, M.; Oliveira, M. Guar gum as a new antimicrobial peptide delivery system against diabetic foot ulcers Staphylococcus aureus isolates. J. Med. Microbiol. 2016, 65, 1092–1099. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, S.; Nakano, Y.; Watanabe, A. A Correlation between Reduced Susceptibilities to Vancomycin and Daptomycin among the MRSA Isolates Selected in Mutant Selection Window of Both Vancomycin and Daptomycin. J. Infect. Chemother. 2014, 20, 752–756. [Google Scholar] [CrossRef] [PubMed]
- Hasper, H.E.; Kramer, N.E.; Smith, J.L.; Hillman, J.D.; Zachariah, C.; Kuipers, O.P.; de Kruijff, B.; Breukink, E. An Alternative Bactericidal Mechanism of Action for Lantibiotic Peptides That Target Lipid II. Science 2006, 313, 1636–1637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breukink, E.; de Kruijff, B. Lipid II as a Target for Antibiotics. Nat. Rev. Drug Discov. 2006, 5, 321–332. [Google Scholar] [CrossRef]
- Cunha, E.; Janela, R.; Costa, M.; Tavares, L.; Veiga, A.S.; Oliveira, M. Nisin Influence on the Antimicrobial Resistance Ability of Canine Oral Enterococci. Antibiotics 2020, 9, 890. [Google Scholar] [CrossRef]
- Fernández, L.; Delgado, S.; Herrero, H.; Maldonado, A.; Rodríguez, J.M. The Bacteriocin Nisin, an Effective Agent for the Treatment of Staphylococcal Mastitis During Lactation. J. Hum. Lact. 2008, 24, 311–316. [Google Scholar] [CrossRef]
- Younes, M.; Aggett, P.; Aguilar, F.; Crebelli, R.; Dusemund, B.; Filipič, M.; Frutos, M.J.; Galtier, P.; Gundert-Remy, U.; Kuhnle, G.G.; et al. Safety of nisin (E 234) as a food additive in the light of new toxicological data and the proposed extension of use. EFSA J. 2017, 15, e05063. [Google Scholar] [CrossRef]
- WHO. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Available online: http://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/ (accessed on 11 February 2021).
- Hsu, S.-T.D.; Breukink, E.; Tischenko, E.; Lutters, M.A.G.; de Kruijff, B.; Kaptein, R.; Bonvin, A.M.J.J.; van Nuland, N.A.J. The Nisin–Lipid II Complex Reveals a Pyrophosphate Cage That Provides a Blueprint for Novel Antibiotics. Nat. Struct. Mol. Biol. 2004, 11, 963–967. [Google Scholar] [CrossRef] [Green Version]
- Bader, M.S. Diabetic Foot Infection. Am. Fam. Physician 2008, 78, 71–79. [Google Scholar]
- Butler, M.S.; Hansford, K.A.; Blaskovich, M.A.T.; Halai, R.; Cooper, M.A. Glycopeptide Antibiotics: Back to the Future. J. Antibiot. 2014, 67, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Mottola, C.; Semedo-Lemsaddek, T.; Mendes, J.J.; Melo-Cristino, J.; Tavares, L.; Cavaco-Silva, P.; Oliveira, M. Molecular Typing, Virulence Traits and Antimicrobial Resistance of Diabetic Foot Staphylococci. J. Biomed. Sci. 2016, 23, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossi, F.; Diaz, L.; Wollam, A.; Panesso, D.; Zhou, Y.; Rincon, S.; Narechania, A.; Xing, G.; Di Gioia, T.S.R.; Doi, A.; et al. Transferable Vancomycin Resistance in a Community-Associated MRSA Lineage. N. Engl. J. Med. 2014, 370, 1524–1531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, J.M.; Kulhankova, K.; Stach, C.S.; Vu, B.G.; Salgado-Pabón, W. Phenotypes and Virulence among Staphylococcus aureus USA100, USA200, USA300, USA400, and USA600 Clonal Lineages. mSphere 2016, 1, e00071-16. [Google Scholar] [CrossRef] [Green Version]
- Friães, A.; Resina, C.; Manuel, V.; Lito, L.; Ramirez, M.; Melo-Cristino, J. Epidemiological Survey of the First Case of Vancomycin-Resistant Staphylococcus aureus Infection in Europe. Epidemiol. Infect. 2015, 143, 745–748. [Google Scholar] [CrossRef]
- Kohler, V.; Vaishampayan, A.; Grohmann, E. Broad-Host-Range Inc18 Plasmids: Occurrence, Spread and Transfer Mechanisms. Plasmid 2018, 99, 11–21. [Google Scholar] [CrossRef]
- Wistrand-Yuen, E.; Knopp, M.; Hjort, K.; Koskiniemi, S.; Berg, O.G.; Andersson, D.I. Evolution of High-Level Resistance during Low-Level Antibiotic Exposure. Nat. Commun. 2018, 9, 1599. [Google Scholar] [CrossRef] [Green Version]
- Falord, M.; Mäder, U.; Hiron, A.; Dbarbouillé, M.; Msadek, T. Investigation of the Staphylococcus aureus GraSR regulon reveals novel links to virulence, stress response and cell wall signal transduction pathways. PLoS ONE 2011, 6, e21323. [Google Scholar] [CrossRef]
- Falord, M.; Karimova, G.; Hiron, A.; Msadeka, T. GraXSR proteins interact with the VraFG ABC transporter to form a five-component system required for cationic antimicrobial peptide sensing and resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2012, 56, 1047–1058. [Google Scholar] [CrossRef] [Green Version]
- Boles, B.R.; Thoende, M.; Roth, A.J.; Horswill, A.R. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS ONE. 2010, 5, e10146. [Google Scholar] [CrossRef] [Green Version]
- El Shazely, B.; Yu, G.; Johnston, P.R.; Rolff, J. Resistance Evolution Against Antimicrobial Peptides in Staphylococcus aureus Alters Pharmacodynamics Beyond the MIC. Front. Microbiol. 2020, 11, 103. [Google Scholar] [CrossRef]
- Hiron, A.; Falord, M.; Valle, J.; Débarbouillé, M.; Msadek, T. Bacitracin and nisin resistance in Staphylococcus aureus: A novel pathway involving the BraS/BraR two-component system (SA2417/SA2418) and both the BraD/BraE and VraD/VraE ABC transporters. Mol. Microbiol. 2011, 81, 602–622. [Google Scholar] [CrossRef] [PubMed]
- Randall, C.P.; Gupta, A.; Utley-Drew, B.; Lee, S.Y.; Morrison-Williams, G.; O’Neill, A.J. Acquired nisin resistance in Staphylococcus aureus involves constitutive activation of an intrinsic peptide antibiotic detoxification module. MSphere 2018, 3, e00633-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arii, K.; Kawada-Matsuo, M.; Oogai, Y.; Noguchi, K.; Komatsuzawa, H. Single mutations in BraRS confer high resistance against nisin A in Staphylococcus aureus. Microbiol. Open 2019, 8, e791. [Google Scholar] [CrossRef] [Green Version]
- Sinel, C.; Jaussaud, C.; Auzou, M.; Giard, J.C.; Cattoir, V. Mutant prevention concentrations of daptomycin for Enterococcus faecium clinical isolates. Int. J. Antimicrob. Agents 2016, 48, 449–452. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Trujillo, E.; Pérez-Roth, E.; Méndez-Alvarez, S.; Claverie-Martín, F. Multiplex PCR for simultaneous detection of enterococcal genes VanA and VanB and staphylococcal genes MecA, IleS-2 and FemB. Int. Microbiol. Off. J. Span. Soc. Microbiol. 2003, 6, 113–115. [Google Scholar] [CrossRef]
- Niederhäusern, S.; Bondi, M.; Messi, P.; Iseppi, R.; Sabia, C.; Manicardi, G.; Anacarso, I. Vancomycin-resistance transferability from VanA enterococci to Staphylococcus aureus. Curr. Microbiol. 2011, 62, 1363–1367. [Google Scholar] [CrossRef]
- Albrecht, V.S.; Zervos, M.J.; Kaye, K.S.; Tosh, P.K.; Arshad, S.; Hayakawa, K.; Kallen, A.J.; McDougal, L.K.; Limbago, B.M.; Guh, A.Y. Prevalence of and risk factors for vancomycin-resistant Staphylococcus aureus precursor organisms in southeastern Michigan. Infect. Control Hosp. Epidemiol. 2014, 35, 1531–1534. [Google Scholar] [CrossRef]
- Caryl, J.A.; O’Neill, A.J. Complete Nucleotide Sequence of PGO1, the prototype conjugative plasmid from the staphylococci. Plasmid 2009, 62, 35–38. [Google Scholar] [CrossRef]
- Baym, M.; Lieberman, T.M.; Kelsic, E.D.; Chait, R.; Gross, R.; Yelin, I.; Kishony, R. Spatiotemporal microbial evolution on antibiotic landscapes. Science 2016, 353, 1147–1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, A.; Som, A. Decoding molecular markers and transcriptional circuitry of naive and primed states of human pluripotency. Stem Cell Res. 2021, 53, 102334. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [Green Version]
- Dimmer, E.C.; Huntley, R.P.; Barrell, D.G.; Binns, D.; Draghici, S.; Camon, E.B.; Hubank, M.; Talmud, F.J.; Apweiler, R.; Lovering, R.C. The gene ontology—Providing a functional role in proteomic studies. Proteomics 2008, 8. [Google Scholar] [CrossRef]
- Dereeper, A.; Homa, F.; Andres, G.; Sempere, G.; Sarah, G.; Hueber, Y.; Dufayard, J.F.; Ruiz, M. SNiPlay3: A web-based application for exploration and large-scale analyses of genomic variations. Nucleic Acids Res. 2015, 43, W295–W300. [Google Scholar] [CrossRef] [Green Version]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]




| Isolate Identification | mecA Gene | Clonal Complex | MPC Value | MPC/MIC Ratio |
|---|---|---|---|---|
| S. aureus A1.1 | + | 5 | >720 | - |
| S. aureus A5.2 | - | 8 | >720 | - |
| S. aureus A6.3 | - | 7 | >720 | - |
| S. aureus B3.2 | - | 5 | >720 | - |
| S. aureus B3.3 | - | 5 | >720 | - |
| S. aureus B7.3 | + | 5 | >720 | - |
| S. aureus B13.1 | + | 5 | 720 | 144 |
| S. aureus B14.2 | + | 22 | >720 | - |
| S. aureus Z1.1 | + | 22 | >720 | - |
| S. aureus Z2.2 | - | 5 | >720 | - |
| S. aureus Z3.1 | - | 5 | >720 | - |
| S. aureus Z5.2 | - | 5 | >720 | - |
| S. aureus Z12.2 | - | 5 | 540 | 43 |
| S. aureus Z14.1 | - | 5 | >720 | - |
| S. aureus Z16.1 | + | 5 | >720 | - |
| S. aureus Z17.2 | - | 30 | 360 | 72 |
| S. aureus Z21.1 | + | 5 | >720 | - |
| S. aureus Z21.3 | + | 5 | >720 | - |
| S. aureus Z23.2 | - | 45 | 540 | 108 |
| S. aureus Z25.2 | - | 182 | 540 | 43 |
| S. aureus Z27.2 | - | 5 | >720 | - |
| S. aureus Z27.3 | - | 5 | >720 | - |
| S. aureus Z32.2 | - | 5 | 360 | 29 |
| S. aureus ATCC 29213 | - | ? | >720 | - |
| Isolate Identification | Control (Not Supplemented) | 2 Round Supplemented with Nisin (Sub-MIC Value of 5.63 µg/mL) | 3 Round Supplemented with Vancomycin (Sub-MIC Value of 0.28 µg/mL) |
|---|---|---|---|
| S. aureus A1.1 | - | - | - |
| S. aureus A5.2 | - | - | - |
| S. aureus A6.3 | - | - | - |
| S. aureus B3.2 | - | - | - |
| S. aureus B3.3 | - | - | - |
| S. aureus B7.3 | - | - | - |
| S. aureus B13.1 | - | - | - |
| S. aureus B14.2 | - | - | - |
| S. aureus Z1.1 | - | - | - |
| S. aureus Z2.2 | - | - | - |
| S. aureus Z3.1 | - | - | - |
| S. aureus Z5.2 | + (vanA detected) | - | - |
| S. aureus Z12.2 | - | - | - |
| S. aureus Z14.1 | - | - | - |
| S. aureus Z16.1 | - | - | - |
| S. aureus Z17.2 | - | - | - |
| S. aureus Z21.1 | - | - | - |
| S. aureus Z21.3 | - | - | - |
| S. aureus Z23.2 | - | - | - |
| S. aureus Z25.2 | - | - | - |
| S. aureus Z27.2 | - | - | - |
| S. aureus Z27.3 | - | - | - |
| S. aureus Z32.2 | - | - | - |
| S. aureus ATCC 29213 | - | - | - |
| Isolate Code | Assay | Original S. aureus Strain | MEGA-Plate Division Concentration |
|---|---|---|---|
| E1(2b) | 1 | S. aureus Z25.2 | 1/8 MIC-2.8125 µg mL−1 |
| E1(3b) | 1 | S. aureus Z25.2 | 1/4 MIC-5.625 µg mL−1 |
| E1(4b) | 1 | S. aureus Z25.2 | 1/2 MIC-11.25 µg mL−1 |
| E1(5b) | 1 | S. aureus Z25.2 | MIC-22.5 µg mL−1 |
| E1(6a) | 1 | S. aureus Z25.2 | 1/2 MIC-11.25 µg mL−1 |
| E1(7b) | 1 | S. aureus Z25.2 | 1/4 MIC-5.625 µg mL−1 |
| E1(8b) | 1 | S. aureus Z25.2 | 1/8 MIC-2.8125 µg mL−1 |
| E2(2b) | 2 | S. aureus ATCC 29213 | 1/8 MIC-2.8125 µg mL−1 |
| E2(3a) | 2 | S. aureus ATCC 29213 | 1/4 MIC-5.625 µg mL−1 |
| E2(4a) | 2 | S. aureus ATCC 29213 | 1/2 MIC-11.25 µg mL−1 |
| E2(5a) | 2 | S. aureus ATCC 29213 | MIC-22.5 µg mL−1 |
| E2(5b) | 2 | S. aureus ATCC 29213 | MIC-22.5 µg mL−1 |
| E2(6a) | 2 | S. aureus Z25.2 | 1/2 MIC-11.25 µg mL−1 |
| E2(7b) | 2 | S. aureus Z25.2 | 1/4 MIC-5.625 µg mL−1 |
| E2(8b) | 2 | S. aureus Z25.2 | 1/8 MIC-2.8125 µg mL−1 |
| E3(2a) | 3 | S. aureus ATCC 29213 | 1/8 MIC-2.8125 µg mL−1 |
| E3(3a) | 3 | S. aureus ATCC 29213 | 1/4 MIC-5.625 µg mL−1 |
| E3(4a) | 3 | S. aureus ATCC 29213 | 1/2 MIC-11.25 µg mL−1 |
| E3(6b) | 3 | S. aureus Z25.2 | 1/2 MIC-11.25 µg mL−1 |
| E3(7b) | 3 | S. aureus Z25.2 | 1/4 MIC-5.625 µg mL−1 |
| E3(8b) | 3 | S. aureus Z25.2 | 1/8 MIC-2.8125 µg mL−1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa, M.; Meirinhos, C.; Cunha, E.; Gomes, D.; Pereira, M.; Dias, R.; Tavares, L.; Oliveira, M. Nisin Mutant Prevention Concentration and the Role of Subinhibitory Concentrations on Resistance Development by Diabetic Foot Staphylococci. Antibiotics 2022, 11, 972. https://doi.org/10.3390/antibiotics11070972
Costa M, Meirinhos C, Cunha E, Gomes D, Pereira M, Dias R, Tavares L, Oliveira M. Nisin Mutant Prevention Concentration and the Role of Subinhibitory Concentrations on Resistance Development by Diabetic Foot Staphylococci. Antibiotics. 2022; 11(7):972. https://doi.org/10.3390/antibiotics11070972
Chicago/Turabian StyleCosta, Margarida, Cláudia Meirinhos, Eva Cunha, Diana Gomes, Marcelo Pereira, Ricardo Dias, Luís Tavares, and Manuela Oliveira. 2022. "Nisin Mutant Prevention Concentration and the Role of Subinhibitory Concentrations on Resistance Development by Diabetic Foot Staphylococci" Antibiotics 11, no. 7: 972. https://doi.org/10.3390/antibiotics11070972
APA StyleCosta, M., Meirinhos, C., Cunha, E., Gomes, D., Pereira, M., Dias, R., Tavares, L., & Oliveira, M. (2022). Nisin Mutant Prevention Concentration and the Role of Subinhibitory Concentrations on Resistance Development by Diabetic Foot Staphylococci. Antibiotics, 11(7), 972. https://doi.org/10.3390/antibiotics11070972






