Arctic Psychrotolerant Pseudomonas sp. B14-6 Exhibits Temperature-Dependent Susceptibility to Aminoglycosides
Abstract
1. Introduction
2. Results
2.1. Psychrotolerant Pseudomonas sp. B14-6 Shows Resistance to Streptomycin
2.2. Identification of the Determinants of Aminoglycoside Resistance
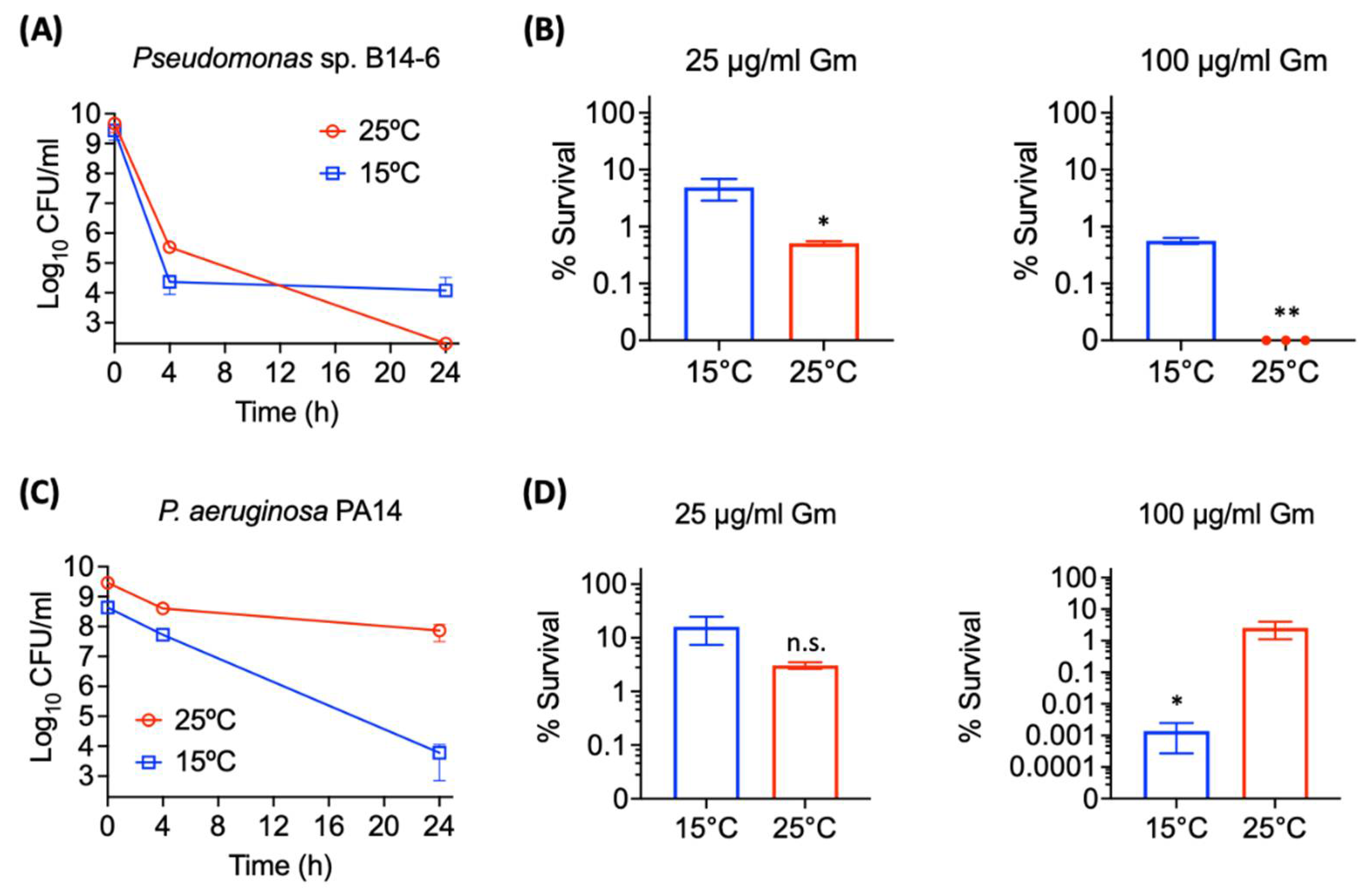
2.3. Pseudomonas sp. B14-6 Shows Reduced Susceptibility to Aminoglycosides at Low Temperature
2.4. Pseudomonas sp. B14-6 Forms a Small Portion of Persisters Only at a Low Temperature
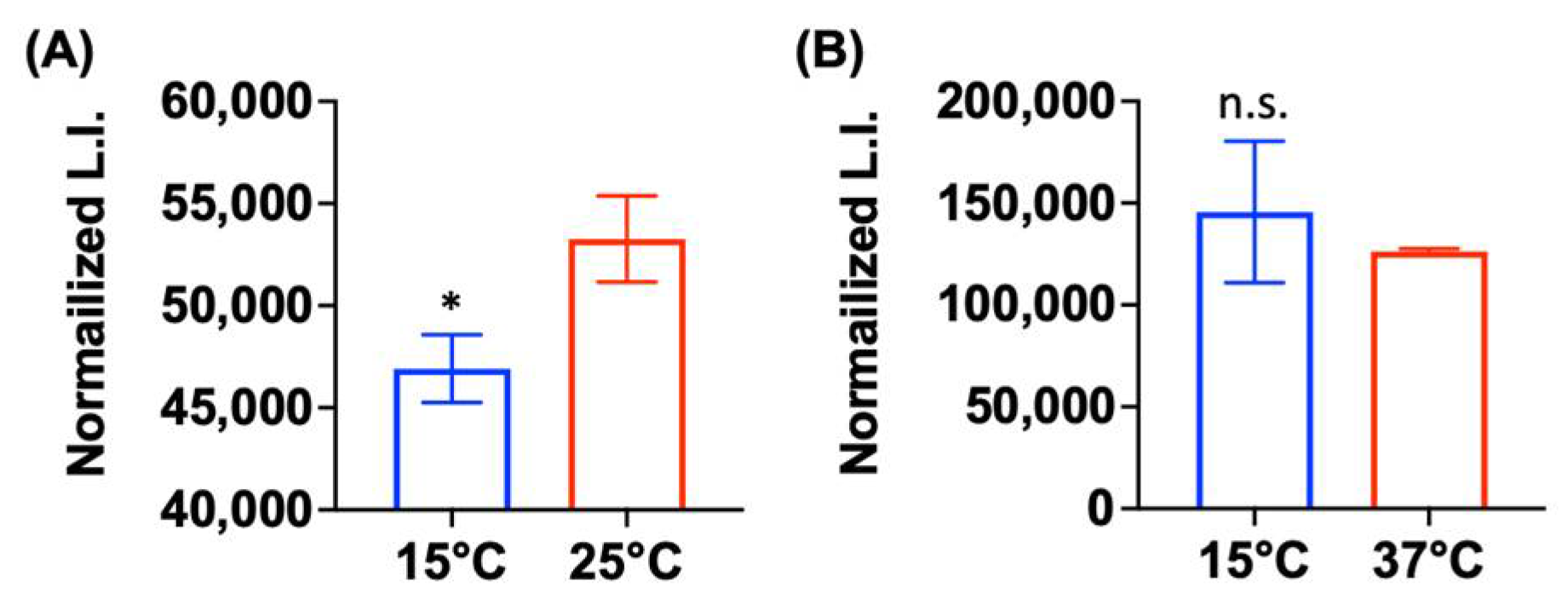
2.5. Low Metabolic Activity Is Correlated with Aminoglycoside Tolerance
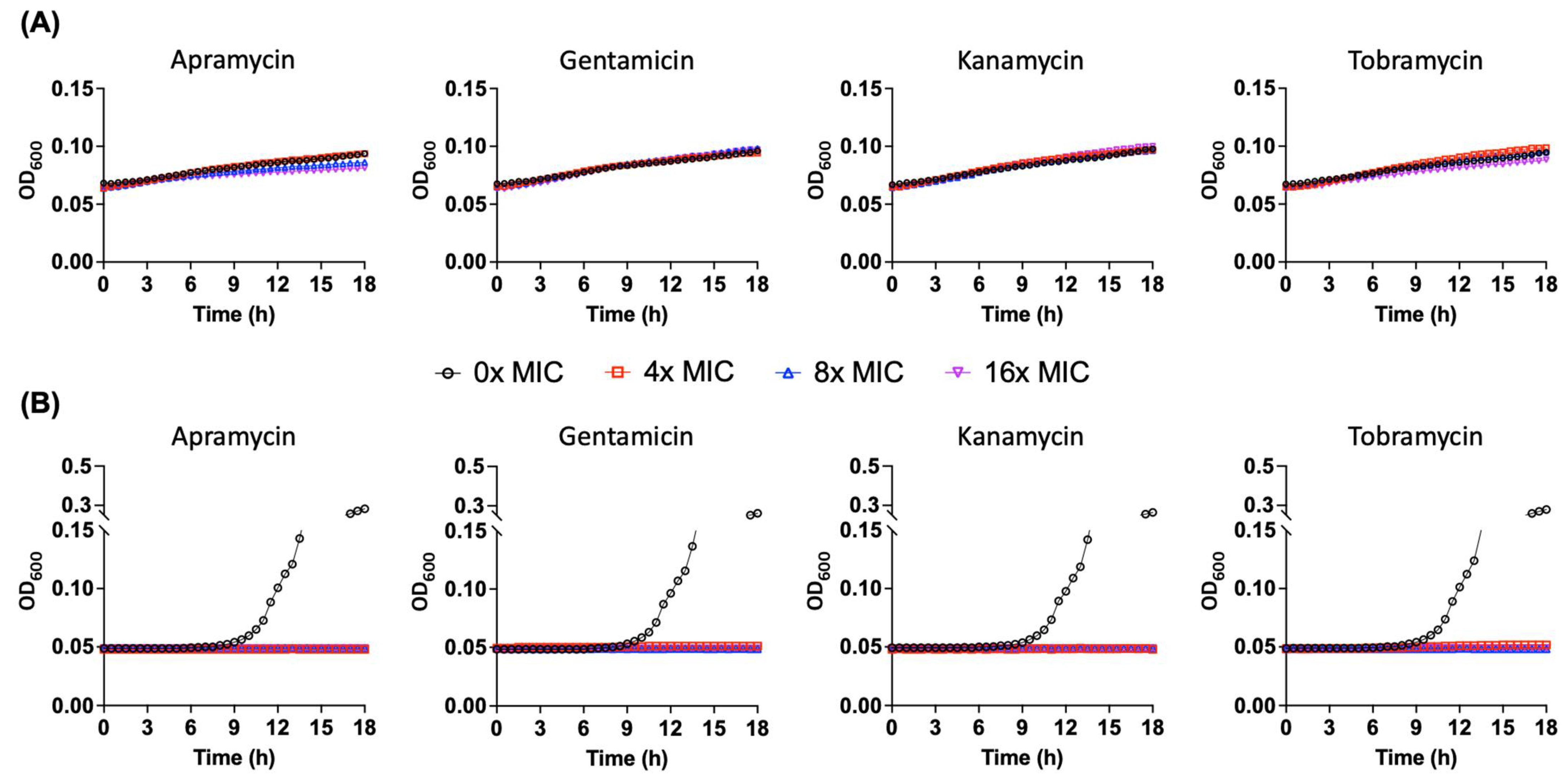
2.6. Pseudomonas sp. B14-6 Can Slowly Proliferate under Bactericidal Concentrations of Aminoglycoside
3. Discussion
4. Materials and Methods
4.1. Bacterial Strain and Growth Conditions
4.2. Chemicals
4.3. Determination of Minimum Inhibitory Concentrations (MICs)
4.4. Identification of Antibiotic Resistance Genes
4.5. Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
4.6. Determination of Aminoglycoside-Tolerant Cells
4.7. Comparison of Intracellular ATP Levels
4.8. Indentification of Partially Tolerant Slow Growing Cells
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shetty, P. The Antibiotic Paradox. Lancet Infect. Dis. 2002, 2, 704. [Google Scholar] [CrossRef]
- Lewis, K. Persister Cells. Annu. Rev. Microbiol. 2010, 64, 357–372. [Google Scholar] [CrossRef]
- Gollan, B.; Grabe, G.; Michaux, C.; Helaine, S. Bacterial Persisters and Infection: Past, Present, and Progressing. Annu. Rev. Microbiol. 2019, 73, 359–385. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Bassler, B.L. Surviving as a Community: Antibiotic Tolerance and Persistence in Bacterial Biofilms. Cell Host Microbe 2019, 26, 15–21. [Google Scholar] [CrossRef]
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.L.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic Resistance Is Ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.C.; Lee, N.; Aw, T.G. Antibiotic Resistance in Minimally Human-Impacted Environments. Int. J. Environ. Res. Public 2020, 17, 3939. [Google Scholar] [CrossRef] [PubMed]
- Goethem, M.W.V.; Pierneef, R.; Bezuidt, O.K.I.; Peer, Y.V.D.; Cowan, D.A.; Makhalanyane, T.P. A Reservoir of ‘Historical’ Antibiotic Resistance Genes in Remote Pristine Antarctic Soils. Microbiome 2018, 6, 40. [Google Scholar] [CrossRef] [PubMed]
- Makowska, N.; Zawierucha, K.; Nadobna, P.; Piątek-Bajan, K.; Krajewska, A.; Szwedyk, J.; Iwasieczko, P.; Mokracka, J.; Koczura, R. Occurrence of Integrons and Antibiotic Resistance Genes in Cryoconite and Ice of Svalbard, Greenland, and the Caucasus Glaciers. Sci. Total Environ. 2020, 716, 137022. [Google Scholar] [CrossRef]
- Hughes, V.M.; Datta, N. Conjugative Plasmids in Bacteria of the ‘Pre-Antibiotic’ Era. Nature 1983, 302, 725–726. [Google Scholar] [CrossRef]
- Perron, G.G.; Whyte, L.; Turnbaugh, P.J.; Goordial, J.; Hanage, W.P.; Dantas, G.; Desai, M.M. Functional Characterization of Bacteria Isolated from Ancient Arctic Soil Exposes Diverse Resistance Mechanisms to Modern Antibiotics. PLoS ONE 2015, 10, e0069533. [Google Scholar] [CrossRef]
- Tam, H.K.; Wong, C.M.V.L.; Yong, S.T.; Blamey, J.; González, M. Multiple-Antibiotic-Resistant Bacteria from the Maritime Antarctic. Polar Biol. 2015, 38, 1129–1141. [Google Scholar] [CrossRef]
- Shen, J.-P.; Li, Z.-M.; Hu, H.-W.; Zeng, J.; Zhang, L.-M.; Du, S.; He, J.-Z. Distribution and Succession Feature of Antibiotic Resistance Genes Along a Soil Development Chronosequence in Urumqi No.1 Glacier of China. Front. Microbiol. 2019, 10, 1569. [Google Scholar] [CrossRef]
- Gesheva, V. Distribution of Psychrophilic Microorganisms in Soils of Terra Nova Bay and Edmonson Point, Victoria Land and Their Biosynthetic Capabilities. Polar Biol. 2009, 32, 1287–1291. [Google Scholar] [CrossRef]
- Wong, C.; Tam, H.; Alias, S.; González, M.; González-Rocha, G.; Domínguez-Yévenes, M. Pseudomonas and Pedobacter Isolates from King George Island Inhibited the Growth of Foodborne Pathogens. Pol. Polar Res. 2011, 32, 3–14. [Google Scholar] [CrossRef]
- Shekh, R.M.; Singh, P.; Singh, S.M.; Roy, U. Antifungal Activity of Arctic and Antarctic Bacteria Isolates. Polar Biol. 2011, 34, 139–143. [Google Scholar] [CrossRef]
- Wright, G.D. Environmental and Clinical Antibiotic Resistomes, Same Only Different. Curr. Opin. Microbiol. 2019, 51, 57–63. [Google Scholar] [CrossRef]
- McCann, C.M.; Christgen, B.; Roberts, J.A.; Su, J.-Q.; Arnold, K.E.; Gray, N.D.; Zhu, Y.-G.; Graham, D.W. Understanding Drivers of Antibiotic Resistance Genes in High Arctic Soil Ecosystems. Sci. Total Environ. 2019, 125, 497–504. [Google Scholar] [CrossRef]
- Maisonneuve, E.; Gerdes, K. Molecular Mechanisms Underlying Bacterial Persisters. Cell 2014, 157, 539–548. [Google Scholar] [CrossRef]
- Maayer, P.D.; Anderson, D.; Cary, C.; Cowan, D.A. Some like It Cold: Understanding the Survival Strategies of Psychrophiles. EMBO Rep. 2014, 15, 508–517. [Google Scholar] [CrossRef]
- Poole, K. Aminoglycoside Resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2005, 49, 479–487. [Google Scholar] [CrossRef]
- Tseng, J.T.; Bryan, L.E.; Elzen, H.M.V.D. Mechanisms and Spectrum of Streptomycin Resistance in a Natural Population of Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 1972, 2, 136–141. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cervantes-Vega, C.; Chavez, J.; Rodriguez, M.G. Antibiotic Susceptibility of Clinical Isolates of Pseudomonas aeruginosa. Antonie Van Leeuwenhoek 1986, 52, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Choi, T.-R.; Park, Y.-L.; Song, H.-S.; Lee, S.M.; Park, S.L.; Lee, H.S.; Kim, H.-J.; Bhatia, S.K.; Gurav, R.; Lee, Y.K.; et al. Effects of a Δ-9-Fatty Acid Desaturase and a Cyclopropane-Fatty Acid Synthase from the Novel Psychrophile Pseudomonas Sp. B14-6 on Bacterial Membrane Properties. J. Ind. Microbiol. Biotechnol. 2020, 47, 1045–1057. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2019, 48, D517–D525. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; Pascale, G.D.; Ejim, L.; et al. The Comprehensive Antibiotic Resistance Database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef]
- Jackson, J.; Chen, C.; Buising, K. Aminoglycosides: How Should We Use Them in the 21st Century? Curr. Opin. Infect. Dis. 2013, 26, 516. [Google Scholar] [CrossRef]
- Ramirez, M.S.; Tolmasky, M.E. Aminoglycoside Modifying Enzymes. Drug Resist. Updates 2010, 13, 151–171. [Google Scholar] [CrossRef]
- Enne, V.I.; Delsol, A.A.; Roe, J.M.; Bennett, P.M. Evidence of Antibiotic Resistance Gene Silencing in Escherichia coli. Antimicrob. Agents Chemother. 2006, 50, 3003–3010. [Google Scholar] [CrossRef]
- Dantas, G.; Sommer, M.O. Context Matters—The Complex Interplay between Resistome Genotypes and Resistance Phenotypes. Curr. Opin. Microbiol. 2012, 15, 577–582. [Google Scholar] [CrossRef]
- Stokes, J.M.; French, S.; Ovchinnikova, O.G.; Bouwman, C.; Whitfield, C.; Brown, E.D. Cold Stress Makes Escherichia coli Susceptible to Glycopeptide Antibiotics by Altering Outer Membrane Integrity. Cell Chem. Biol. 2016, 23, 267–277. [Google Scholar] [CrossRef]
- Claudi, B.; Spröte, P.; Chirkova, A.; Personnic, N.; Zankl, J.; Schürmann, N.; Schmidt, A.; Bumann, D. Phenotypic Variation of Salmonella in Host Tissues Delays Eradication by Antimicrobial Chemotherapy. Cell 2014, 158, 722–733. [Google Scholar] [CrossRef]
- Lopatkin, A.J.; Stokes, J.M.; Zheng, E.J.; Yang, J.H.; Takahashi, M.K.; You, L.; Collins, J.J. Bacterial Metabolic State More Accurately Predicts Antibiotic Lethality than Growth Rate. Nat. Microbiol. 2019, 4, 2109–2117. [Google Scholar] [CrossRef]
- Stokes, J.M.; Lopatkin, A.J.; Lobritz, M.A.; Collins, J.J. Bacterial Metabolism and Antibiotic Efficacy. Cell Metab. 2019, 30, 251–259. [Google Scholar] [CrossRef]
- Brauner, A.; Fridman, O.; Gefen, O.; Balaban, N.Q. Distinguishing between Resistance, Tolerance and Persistence to Antibiotic Treatment. Nat. Rev. Microbiol. 2016, 14, 320–330. [Google Scholar] [CrossRef]
- Pontes, M.H.; Groisman, E.A. Slow Growth Determines Nonheritable Antibiotic Resistance in Salmonella Enterica. Sci. Signal. 2019, 12, eaax3938. [Google Scholar] [CrossRef]
- Pontes, M.H.; Groisman, E.A. A Physiological Basis for Nonheritable Antibiotic Resistance. Mbio 2020, 11, e00817-20. [Google Scholar] [CrossRef]
- Manuse, S.; Shan, Y.; Canas-Duarte, S.J.; Bakshi, S.; Sun, W.-S.; Mori, H.; Paulsson, J.; Lewis, K. Bacterial Persisters Are a Stochastically Formed Subpopulation of Low-Energy Cells. PLoS Biol. 2021, 19, e3001194. [Google Scholar] [CrossRef]
- Tuomanen, E.; Cozens, R.; Tosch, W.; Zak, O.; Tomasz, A. The Rate of Killing of Escherichia coli by Beta-Lactam Antibiotics Is Strictly Proportional to the Rate of Bacterial Growth. J. Gen. Microbiol. 1986, 132, 1297–1304. [Google Scholar] [CrossRef]
- Eng, R.H.; Padberg, F.T.; Smith, S.M.; Tan, E.N.; Cherubin, C.E. Bactericidal Effects of Antibiotics on Slowly Growing and Nongrowing Bacteria. Antimicrob. Agents Chemother. 1991, 35, 1824–1828. [Google Scholar] [CrossRef]
- Smirnova, G.V.; Oktyabrsky, O.N. Relationship between Escherichia coli Growth Rate and Bacterial Susceptibility to Ciprofloxacin. FEMS Microbiol. Lett. 2017, 365, fnx254. [Google Scholar] [CrossRef]
- Kaprelyants, A.S.; Gottschal, J.C.; Kell, D.B. Dormancy in Non-sporulating Bacteria. FEMS Microbiol. Lett. 1993, 104, 271–286. [Google Scholar] [CrossRef] [PubMed]
- Lennon, J.T.; Jones, S.E. Microbial Seed Banks: The Ecological and Evolutionary Implications of Dormancy. Nat. Rev. Microbiol. 2011, 9, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Rittershaus, E.S.C.; Baek, S.-H.; Sassetti, C.M. The Normalcy of Dormancy: Common Themes in Microbial Quiescence. Cell Host Microbe 2013, 13, 643–651. [Google Scholar] [CrossRef]
- Kaiser, P.; Regoes, R.R.; Dolowschiak, T.; Wotzka, S.Y.; Lengefeld, J.; Slack, E.; Grant, A.J.; Ackermann, M.; Hardt, W.-D. Cecum Lymph Node Dendritic Cells Harbor Slow-Growing Bacteria Phenotypically Tolerant to Antibiotic Treatment. PLoS Biol. 2014, 12, e1001793. [Google Scholar] [CrossRef] [PubMed]
- Adams, K.N.; Takaki, K.; Connolly, L.E.; Wiedenhoft, H.; Winglee, K.; Humbert, O.; Edelstein, P.H.; Cosma, C.L.; Ramakrishnan, L. Drug Tolerance in Replicating Mycobacteria Mediated by a Macrophage-Induced Efflux Mechanism. Cell 2011, 145, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Gefen, O.; Gabay, C.; Mumcuoglu, M.; Engel, G.; Balaban, N.Q. Single-Cell Protein Induction Dynamics Reveals a Period of Vulnerability to Antibiotics in Persister Bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 6145–6149. [Google Scholar] [CrossRef]
- Allison, K.R.; Brynildsen, M.P.; Collins, J.J. Metabolite-Enabled Eradication of Bacterial Persisters by Aminoglycosides. Nature 2011, 473, 216–220. [Google Scholar] [CrossRef]
- Schmidt, N.W.; Deshayes, S.; Hawker, S.; Blacker, A.; Kasko, A.M.; Wong, G.C.L. Engineering Persister-Specific Antibiotics with Synergistic Antimicrobial Functions. ACS Nano 2014, 8, 8786–8793. [Google Scholar] [CrossRef]
- Kim, W.; Zhu, W.; Hendricks, G.L.; Tyne, D.V.; Steele, A.D.; Keohane, C.E.; Fricke, N.; Conery, A.L.; Shen, S.; Pan, W.; et al. A New Class of Synthetic Retinoid Antibiotics Effective against Bacterial Persisters. Nature 2018, 556, 103–107. [Google Scholar] [CrossRef]
- Kim, W.; Zou, G.; Hari, T.P.A.; Wilt, I.K.; Zhu, W.; Galle, N.; Faizi, H.A.; Hendricks, G.L.; Tori, K.; Pan, W.; et al. A Selective Membrane-Targeting Repurposed Antibiotic with Activity against Persistent Methicillin-Resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2019, 116, 16529–16534. [Google Scholar] [CrossRef]
- Strahl, H.; Hamoen, L.W. Membrane Potential Is Important for Bacterial Cell Division. Proc. Natl. Acad. Sci. USA 2010, 107, 12281–12286. [Google Scholar] [CrossRef]
- Rahme, L.G.; Stevens, E.J.; Wolfort, S.F.; Shao, J.; Tompkins, R.G.; Ausubel, F.M. Common Virulence Factors for Bacterial Pathogenicity in Plants and Animals. Science 1995, 268, 1899–1902. [Google Scholar] [CrossRef]
- Clinical Laboratory and Standard Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, Approved Standard-Ninth Edition; CLSI Document M07-A9; CLSI: Wayne, PA, USA, 2012. [Google Scholar]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An Automatic Genome Annotation and Pathway Reconstruction Server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef]
- Kim, S.M.; Zou, G.; Kim, H.; Kang, M.; Ahn, S.; Heo, H.Y.; Kim, J.-S.; Lim, K.-M.; Ausubel, F.M.; Mylonakis, E.; et al. Antimicrobial Activity of the Membrane-Active Compound NTZDpa Is Enhanced at Low PH. Biomed. Pharmacother. 2022, 150, 112977. [Google Scholar] [CrossRef]


| Pseudomonas sp. B14-6 | P. aeruginosa PA14 | ||||
|---|---|---|---|---|---|
| Antibiotics | 15 °C | 25 °C | 15 °C | 25 °C | 37 °C |
| Gentamicin | 1 | 0.5 | 0.25 | 2 | 2 |
| Tobramycin | 0.5 | 0.5 | 0.125 | 0.5 | 0.5 |
| Apramycin | 8 | 4 | 2 | 8 | 8 |
| Kanamycin | 2 | 2 | 64 | >64 | >64 |
| Streptomycin | 64 | 32 | 2 | 16 | 32 |
| Sequence Name | Sequence Description | Sequence Length | Gene Ontologies |
|---|---|---|---|
| orf04666 | aminoglycoside phosphotransferase (aph1) | 1557 | P:metabolic process; F:transferase activity, transferring phosphorus-containing groups |
| orf05258 | aminoglycoside phosphotransferase (aph2) | 843 | P:metabolic process; F:transferase activity, transferring phosphorus-containing groups |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, M.; Choi, T.-R.; Ahn, S.; Heo, H.Y.; Kim, H.; Lee, H.S.; Lee, Y.K.; Joo, H.-S.; Yune, P.S.; Kim, W.; et al. Arctic Psychrotolerant Pseudomonas sp. B14-6 Exhibits Temperature-Dependent Susceptibility to Aminoglycosides. Antibiotics 2022, 11, 1019. https://doi.org/10.3390/antibiotics11081019
Kang M, Choi T-R, Ahn S, Heo HY, Kim H, Lee HS, Lee YK, Joo H-S, Yune PS, Kim W, et al. Arctic Psychrotolerant Pseudomonas sp. B14-6 Exhibits Temperature-Dependent Susceptibility to Aminoglycosides. Antibiotics. 2022; 11(8):1019. https://doi.org/10.3390/antibiotics11081019
Chicago/Turabian StyleKang, Minjeong, Tae-Rim Choi, Soyeon Ahn, Hee Young Heo, Hyerim Kim, Hye Soo Lee, Yoo Kyung Lee, Hwang-Soo Joo, Philip S. Yune, Wooseong Kim, and et al. 2022. "Arctic Psychrotolerant Pseudomonas sp. B14-6 Exhibits Temperature-Dependent Susceptibility to Aminoglycosides" Antibiotics 11, no. 8: 1019. https://doi.org/10.3390/antibiotics11081019
APA StyleKang, M., Choi, T.-R., Ahn, S., Heo, H. Y., Kim, H., Lee, H. S., Lee, Y. K., Joo, H.-S., Yune, P. S., Kim, W., & Yang, Y.-H. (2022). Arctic Psychrotolerant Pseudomonas sp. B14-6 Exhibits Temperature-Dependent Susceptibility to Aminoglycosides. Antibiotics, 11(8), 1019. https://doi.org/10.3390/antibiotics11081019






