Genomic and Phenotypic Characterization of Cutibacterium acnes Bacteriophages Isolated from Acne Patients
Abstract
1. Introduction
2. Results
2.1. Phage Isolation and Host Ranges
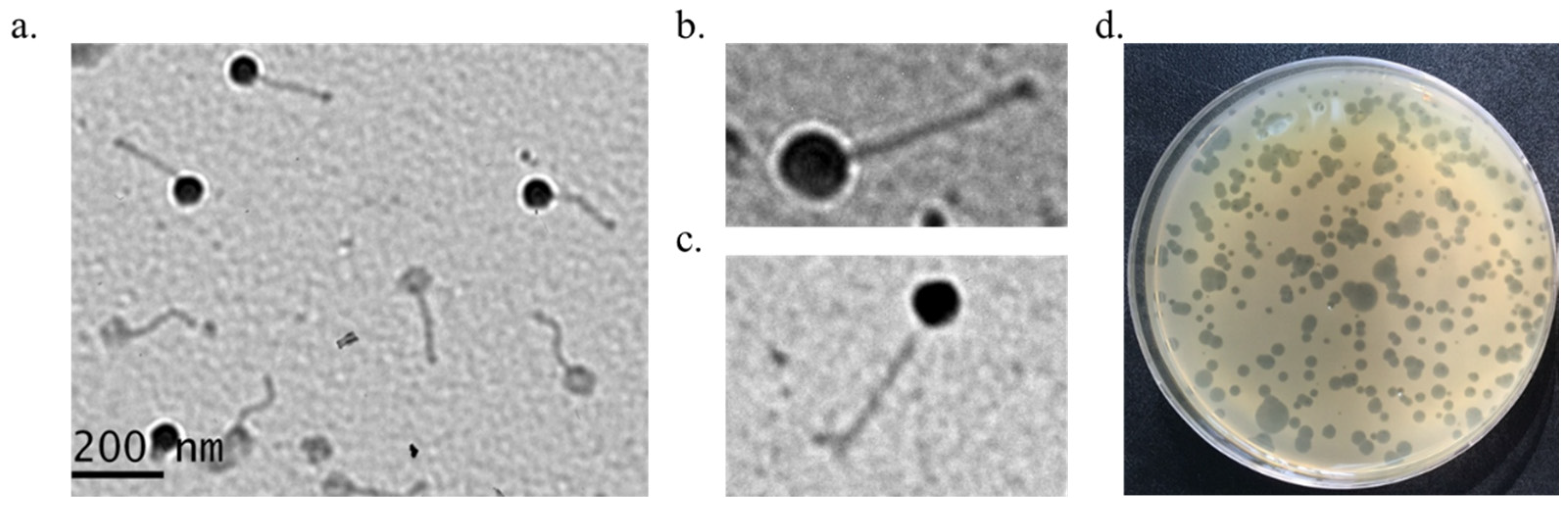
2.2. Morphological Analysis
2.3. Genomic DNA Characterization of Three Phages, 2012-15, P1, and P2
2.4. Hydrophobic Characteristics of the Capsid and Tape Measure Proteins of the 2012-15 Phage
2.5. Adsorption Rate and One-Step Growth Curve of Phage 2012-15
2.6. Phage 2012-15 Stabilities at Different Temperatures and pHs
3. Discussion
4. Materials and Methods
4.1. Phage Isolation
4.2. Bacterial Culture
4.3. Determination of Bacterial Host Range
4.4. Transmission Electron Microscope Analysis
4.5. Genome Sequencing
4.6. Bioinformatic Analysis
4.7. Adsorption Assay and One-Step Growth Curve
4.8. Temperature and pH Stability Test
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pauzenberger, L.; Heller, V.; Ostermann, R.C.; Laky, B.; Heuberer, P.R.; Anderl, W. Cutibacteriuma (formerly Propionibacterium acnes) contamination of the surgical field during shoulder arthroscopy. J. Arthroscop. Relat. Surg. 2019, 35, 1750–1757. [Google Scholar] [CrossRef]
- Ridberg, S.; Hellmark, B.; Nilsdotter, Å.; Söderquist, B. Cutibacterium acnes (formerly Propionibacterium acnes) isolated from prosthetic joint infections is less susceptible to oxacillin than to benzylpenicillin. J. Bone Jt. Infect. 2019, 4, 106–110. [Google Scholar] [CrossRef]
- Rozas, M.; de Ruijter, A.H.; Fabrega, M.J.; Zorgani, A.; Guell, M.; Paetzold, B.; Brillet, F. From dysbiosis to healthy skin: Major contributions of Cutibacterium acnes to skin homeostasis. Microorganisms 2021, 9, 628. [Google Scholar] [CrossRef] [PubMed]
- Claesen, J.; Spagnolo, J.B.; Ramos, S.F.; Kurita, K.L.; Byrd, A.L.; Aksenov, A.A.; Melnik, A.V.; Wong, W.R.; Wang, S.; Hernandez, R.D.; et al. A Cutibacterium acnes antibiotic modulates human skin microbiota composition in hair follicles. Sci. Transl. Med. 2020, 12, eaay5445. [Google Scholar] [CrossRef] [PubMed]
- Platsidaki, E.; Dessinioti, C. Recent advances in understanding Propionibacterium acnes (Cutibacterium acnes) in acne. F1000Research 2018, 7, 1953. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, A.M.; Gallo, R.L. Host-microbiome interactions and recent progress into understanding the biology of acne vulgaris. Microbiome 2018, 6, 177. [Google Scholar] [CrossRef]
- Berthelot, J.; Corvec, S.; Hayem, G. SAPHO, autophagie, IL-1, FoxO1, et Propionibacterium (Cutibacterium) acnes. Revue. Du. Rhumatisme. 2018, 85, 132–137. [Google Scholar] [CrossRef]
- Rukavina, I. SAPHO syndrome: A review. J. Child Orthop. 2015, 9, 19–27. [Google Scholar] [CrossRef]
- Fakhri, G.; Tayeh, C.; Dbaibo, G.; Sedawy, O.E.; Halim, N.A.; Bitar, F.; Arabi, M. Cardiac tamponade caused by Cutibacterium acnes: An updated and comprehensive review of the literature. Can. J. Infect. Dis. Med. Microbiol. 2020, 2020, 9598210. [Google Scholar] [CrossRef]
- Rossin, E.J.; Tieger, M.; Rao, N.A.; O’Hearn, T.M.; Eliott, D.; Wu, D.M. Two cases of Cutibacterium acnes (C acnes) endophthalmitis manifesting with unusual epiretinal deposits. Ophthalmic Surg. Lasers Imaging Retin. 2022, 53, 164–167. [Google Scholar] [CrossRef]
- Boisrenoult, P. Cutibacterium acnes prosthetic joint infection: Diagnosis and treatment. Orthop. Traumatol-Surg. Res. 2018, 104, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-J.; Kim, S.; Mun, J.-U.; Bae, K.-C.; Kim, J.; Kyung, H.-S. Diagnosis of periprosthetic joint bacterial infections by culture of sonication fluid from infected implants. J. Orthop. Surg. 2019, 27, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Brüggemann, H.; Al-Zeer, M.A. Bacterial signatures and their inflammatory potentials associated with prostate cancer. APMIS 2020, 128, 80–91. [Google Scholar] [CrossRef]
- Slayton, R.B.; Toth, D.; Lee, B.Y.; Tanner, W.; Bartsch, S.M.; Khader, K.; Wong, K.; Brown, K.; McKinnell, J.A.; Ray, W.; et al. Vital signs: Estimated effects of a coordinated approach for action to reduce antibiotic-resistant infections in health care facilities—United States. Am. J. Transplant. 2015, 64, 826–831. [Google Scholar] [CrossRef]
- Band, V.I.; Weiss, D.S. Heteroresistance: A cause of unexplained antibiotic treatment failure? PLoS Pathog. 2019, 15, e1007726. [Google Scholar] [CrossRef]
- Castillo, D.E.; Nanda, S.; Keri, J.E. Propionibacterium acnes (Cutibacterium) bacteriophage therapy in acne: Current evidence and future perspectives. Derm. Ther. 2019, 9, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Karadag, A.S.; Aslan, M.; Kayıran, M.; Wu, C.-Y.; Chen, W.; Parish, L.C. Antibiotic resistance in acne: Changes, consequences, and concerns. J. Eur. Acad. Derm. Venereol. 2021, 35, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Layton, A.M.; Thiboutot, D.; Tan, J. Reviewing the global burden of acne: How can we improve care to reduce this burden? Br. J. Derm. 2021, 184, 219–225. [Google Scholar] [CrossRef]
- Adesanya, O.; Oduselu, T.; Akin-Ajani, O.; Olubusuyi, M.; Adewumi, O.M.; Ademowo, O.G. An example of bacteriophage therapy: An emerging player in the fight against anti-microbial resistance. AIMS Microbiol. 2020, 6, 204–230. [Google Scholar] [CrossRef]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage therapy: An alternative to antibiotics in an age of multidrug resistance. World J. Gastrointest Pharm. 2017, 8, 162–173. [Google Scholar] [CrossRef]
- Wittebole, X.; De Roock, S.; Opal, S.M. A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence 2014, 5, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Rehman, S.; Ali, Z.; Khan, M.; Bostan, N.; Naseem, S. The dawn of phage therapy. Rev. Med. Virol. 2019, 29, 2041. [Google Scholar] [CrossRef]
- Mayslich, C.; Grange, P.A.; Dupin, N. Cutibacterium acnes as an opportunistic pathogen: An update of its virulence-associated factors. Microorganisms 2021, 9, 303. [Google Scholar] [CrossRef] [PubMed]
- Nazipi, S.; Stødkilde, K.; Scavenius, C.; Brüggemann, H. The skin bacterium Propionibacterium acnes employs two variants of hyaluronate lyase with distinct properties. Microorganisms 2017, 5, 57. [Google Scholar] [CrossRef] [PubMed]
- Łobocka, M.; Dąbrowska, K.; Górski, A. Engineered bacteriophage therapeutics: Rationale, challenges and future. BioDrugs 2021, 35, 255–280. [Google Scholar] [CrossRef] [PubMed]
- Guerin, E.; Hill, C. Shining light on human gut bacteriophages. Front. Cell. Infect. Microbiol. 2020, 10, 481. [Google Scholar] [CrossRef]
- Brzin, B. Studies on the Corynebacterium acnes. Acta Pathol. Microbiol. Scand. 1964, 60, 599–608. [Google Scholar] [CrossRef]
- Brüggemann, H.; Lood, R. Bacteriophages infecting Propionibacterium acnes. BioMed. Res. Int. 2013, 2013, 705741. [Google Scholar] [CrossRef]
- Kim, S.; Song, H.; Lee, W.-J.; Kim, J. Antimicrobial susceptibility and characterization of Propionibacterium acnes by multilocus sequence typing and repetitive-sequence-based PCR. J. Bacteriol. Virol. 2016, 46, 135–141. [Google Scholar] [CrossRef]
- McDowell, A.; Barnard, E.; Nagy, I.; Gao, A.; Tomida, S.; Li, H.; Eady, A.; Cove, J.; Nord, C.E.; Patrick, S. An expanded multilocus sequence typing scheme for Propionibacterium acnes: Investigation of ‘pathogenic’, ‘commensal’ and antibiotic strains. PLoS ONE 2012, 7, e41480. [Google Scholar] [CrossRef]
- Marinelli, L.J.; Fitz-Gibbon, S.; Hayes, C.; Bowman, C.; Inkeles, M.; Loncaric, A.; Russell, D.A.; Jacobs-Sera, D.; Cokus, S.; Pellegrini, M.; et al. Propionibacterium acnes Bacteriophages Display Limited Genetic Diversity and Broad Killing Activity against Bacterial Skin Isolates. mBio 2012, 3, e00279-12. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.S.; Huang, J.S.; Jayathilaka, G.D.L.P.; Lateef, S.S.; Gupta, S. Production of Antipeptide Antibodies. In Immunoelectron Microscopy. Methods in Molecular Biology; Schwartzbach, S., Osafune, T., Eds.; Humana Press: Totowa, NJ, USA, 2010; Volume 657, pp. 93–108. [Google Scholar]
- Mahony, J.; Stockdale, S.R.; Collins, B.; Spinelli, S.; Douillard, F.P.; Cambillau, C.; van Sinderen, D. Lactococcus lactis phage TP901–1 as a model for Siphoviridae virion assembly. Bacteriophage 2016, 6, e1123795. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ramasamy, S.; Barnard, E.; Dawson, T.L.; Li, H. The role of the skin microbiota in acne pathophysiology. Br. J. Dermatol. 2019, 181, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Eun, D.H.; Kim, S.M.; Kim, J.; Lee, W.J. Efficacy of bacteriophages in Propionibacterium acnes-induced inflammation in mice. Ann. Dermatol. 2019, 31, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Kim, S.; Kim, S.M.; Seol, S.Y.; Kim, J. Characterization of induced Staphylococcus aureus bacteriophage SAP-26 and its anti-biofilm activity with rifampicin. Biofouling 2011, 27, 1087–1093. [Google Scholar] [CrossRef]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef]
- Eddy, S.R. Profile hidden Markov models. Bioinformatics 1998, 14, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Kim, O.-S.; Cho, Y.-J.; Lee, K.; Yoon, S.-H.; Kim, M.; Na, H.; Park, S.-C.; Jeon, Y.S.; Lee, J.-H.; Yi, H.; et al. Introducing EzTaxon-e: A prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 2012, 62, 716–721. [Google Scholar] [CrossRef]
- Todd, M.; Lowe, D.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar]
- Overbeek, R.; Begley, T.; Butler, R.M.; Choudhuri, J.V.; Chuang, H.-Y.; Cohoon, M.; de Crécy-Lagard, V.; Diaz, N.; Disz, T.; Edwards, R.; et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005, 33, 5691–5702. [Google Scholar] [CrossRef] [PubMed]
- Pruitt, K.D.; Tatusova, T.; Klimke, W.; Maglott, D.R. NCBI Reference Sequences: Current status, policy and new initiatives. Nucleic Acids Res. 2009, 37, D32–D36. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Zavaljevski, N.; Desai, V.; Reifman, J. Genome-wide enzyme annotation with precision control: Catalytic families (CatFam) databases. Proteins Struct. Funct. Bioinform. 2009, 74, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple Alignment of Conserved Genomic Sequence with Rearrangements. Genome. Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, J.S.; Agarwala, R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics 2007, 23, 1073–1079. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Rahman, M.; Seol, S.Y.; Yoon, S.S.; Kim, J. Pseudomonas aeruginosa Bacteriophage PA1Ø Requires Type IV Pili for Infection and Shows Broad Bactericidal and Biofilm Removal Activities. Appl. Environ. Microbiol. 2012, 78, 6380–6385. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, S.H.; Rahman, M.; Kim, J. Characterization of a Salmonella enteritidis bacteriophage showing broad lytic activity against gram-negative enteric bacteria. J. Clin. Microbiol. 2018, 56, 917–925. [Google Scholar] [CrossRef]



| Phage | Multi-Locus Sequence Type of C. acnes Clinical Isolates | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ST1 | ST2 | ST6 | ST22 | ST25 | ST53 | ST69 | ST115 | |||||||||
| 109 | 893 | 2874 | 105 | 2875 | 1490 | 1912 | 114 | 106 | 2877 | 113 | 2878 | 391 | 2876 | 1475 | 112 | |
| 2012-15 | + | + | + | + | + | + | + | + | + | + | − | − | + | + | + | + |
| P1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| P2 | + | + | + | + | + | + | + | + | + | + | − | − | + | + | + | + |
| P3 | + | + | + | + | + | + | + | + | + | + | − | − | + | + | + | + |
| P4 | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + |
| P5 | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + |
| P6 | + | + | + | + | + | + | + | + | + | + | − | − | + | + | + | + |
| P7 | + | + | + | + | + | + | + | + | + | + | − | − | + | + | + | + |
| P8 | + | + | + | + | + | + | + | + | + | + | − | − | + | + | − | + |
| P9 | + | + | + | + | + | + | + | + | + | + | − | − | + | + | + | + |
| P10 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| P11 | + | + | + | + | − | + | + | + | + | + | − | − | + | + | + | + |
| P12 | + | + | + | + | − | + | + | + | + | + | − | − | + | + | + | + |
| P13 | + | + | + | + | − | + | + | + | + | + | − | − | + | + | + | + |
| P14 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Phage | Structural Protein (Number of Amino Acids) | Acidic Amino Acids (%) | Basic Amino Acids (%) | Neutral Amino Acids (%) | Hydrophobic Amino Acids (%) |
|---|---|---|---|---|---|
| 2012-15 phage | Tape measure protein (921) | 3.58 | 8.9 | 31.81 | 55.7 |
| Capsid protein (323) | 10.84 | 9.91 | 28.48 | 50.77 | |
| Minor tail protein (280) | 11.07 | 13.21 | 39.64 | 36.07 | |
| Minor tail protein (87) | 10.34 | 12.64 | 32.18 | 44.83 | |
| Minor tail protein (272) | 9.93 | 9.19 | 34.93 | 45.96 | |
| Minor tail subunit (315) | 14.6 | 10.48 | 29.84 | 45.08 | |
| Major tail protein (210) | 14.29 | 12.38 | 32.38 | 40.95 | |
| Head scaffold protein (183) | 21.31 | 16.94 | 28.42 | 33.33 | |
| Portal protein (441) | 14.06 | 10.66 | 30.61 | 44.67 | |
| Escherichia coli ADB-2 phage (NC_019725) | Tape measure protein | 13.06 | 12.75 | 33.54 | 40.65 |
| Capsid protein | 17.03 | 15.14 | 25.14 | 42.7 | |
| Minor tail protein | 9.4 | 8.55 | 42.74 | 39.32 | |
| Tail component 1 | 12.93 | 9.64 | 38.19 | 39.25 | |
| Tail component 2 | 7.04 | 10.05 | 37.69 | 45.23 | |
| Tail fiber | 14.52 | 11.45 | 33.87 | 40.16 |
| Tape Measure Protein of Bacteriophage (Number of Amino Acids) | Acidic Amino Acids (%) | Basic Amino Acids (%) | Neutral Amino Acids (%) | Hydrophobic Amino Acids (%) | Accession Number |
|---|---|---|---|---|---|
| Cutibacterium acnes phage 2012-15 (921) | 3.58 | 8.9 | 31.81 | 55.7 | YP_009151455.1 |
| Escherichia coli phage Lambda (853) | 11.84 | 13.25 | 35.05 | 39.86 | NP_040595.1 |
| Escherichia coli phage T5 (1226) | 12.23 | 11.83 | 36.62 | 39.31 | VUF55695.1 |
| Bacillus phage SPP1 (1032) | 4.94 | 12.79 | 31.88 | 50.39 | Q0PDK7.1 |
| Lactococcus phage c2 (624) | 8.97 | 12.34 | 41.51 | 37.18 | ASZ70817.1 |
| Lactococcus phage P2 (999) | 7.21 | 7.71 | 39.74 | 45.35 | D3WAD2.1 |
| Lactococcus phage 1358 (690 AAs) | 7.39 | 8.84 | 35.65 | 48.12 | YP_009140403.1 |
| Lactococcus phage TP901-1 (937 AAs) | 8.75 | 9.71 | 34.79 | 46.74 | AAG32164.1 |
| Lactococcus phage Tuc2009 (1025 AAs) | 8.68 | 9.56 | 34.93 | 46.83 | NP_108725.1 |
| Listeria phage PSA (1026 AAs) | 13.16 | 11.4 | 30.31 | 45.13 | NP_510993.1 |
| Streptococcus phage STP1 (1656 AAs) | 11.11 | 15.22 | 35.14 | 38.53 | ATI20023.1 |
| Staphylococcus phage 80α (1154 AAs) | 5.89 | 10.23 | 34.06 | 49.83 | ABF71627.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.; Song, H.; Jin, J.S.; Lee, W.J.; Kim, J. Genomic and Phenotypic Characterization of Cutibacterium acnes Bacteriophages Isolated from Acne Patients. Antibiotics 2022, 11, 1041. https://doi.org/10.3390/antibiotics11081041
Kim S, Song H, Jin JS, Lee WJ, Kim J. Genomic and Phenotypic Characterization of Cutibacterium acnes Bacteriophages Isolated from Acne Patients. Antibiotics. 2022; 11(8):1041. https://doi.org/10.3390/antibiotics11081041
Chicago/Turabian StyleKim, Shukho, Hyesoon Song, Jong Sook Jin, Weon Ju Lee, and Jungmin Kim. 2022. "Genomic and Phenotypic Characterization of Cutibacterium acnes Bacteriophages Isolated from Acne Patients" Antibiotics 11, no. 8: 1041. https://doi.org/10.3390/antibiotics11081041
APA StyleKim, S., Song, H., Jin, J. S., Lee, W. J., & Kim, J. (2022). Genomic and Phenotypic Characterization of Cutibacterium acnes Bacteriophages Isolated from Acne Patients. Antibiotics, 11(8), 1041. https://doi.org/10.3390/antibiotics11081041






