New Pyrazolo-Benzimidazole Mannich Bases with Antimicrobial and Antibiofilm Activities
Abstract
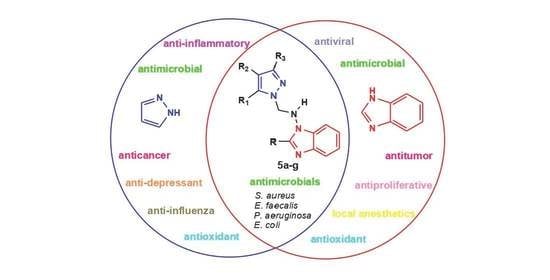
:1. Introduction
2. Resultsand Discussion
2.1. Chemistry
2.1.1. Spectroscopic Characterization of Compounds 5a–g
IR Spectra
Electronic Spectra
NMR Spectral Analysis
MS Spectral Analysis
2.2. Biological Activity
2.2.1. Evaluation of In Vitro Cytotoxicity of the Obtained Compounds
2.2.2. Antimicrobial Activity
Activity against Planktonic Microbial Cells
Antibiofilm Activity
2.2.3. Mechanism of Action
3. Materials and Methods
3.1. Chemistry
3.1.1. General Chemical Characterization Techniques
3.1.2. Mannich Bases 5a–g Synthesis Method
Synthesis of N-[(1H-3,5-dimethylpyrazol-1-yl)methyl]-1-amino-1H-benzimidazole (5a)
Synthesis of N-[(1H-3,5-dimethyl-4-nitropyrazol-1-yl)methyl]-1-amino-1H-benzimidazole (5b)
Synthesis of N-[(1H-3,5-dimethyl-4-iodopyrazol-1yl)methyl]-1-amino-1H-benzimidazole (5c)
Synthesis of N-[(1H-pyrazol-1yl)methyl]-1-amino-2-methyl-1H-benzimidazole (5d)
Synthesis of N-[(1H-3,5-dimetyl-pyrazol-1-yl)methyl]-1-amino-2-methyl-1H-benzimidazole (5e)
Synthesis of N-[(1H-3,5-dimethyl-4-nitropyrazol-1-yl)methyl]-1-amino-2-methyl-1H-benzimidazole (5f)
Synthesis of N-[(1H-3,5-dimethyl-4-iodopyrazol-1yl)methyl]-1-amino-2-methyl-1H-benzimidazole (5g)
3.2. Biological Tests
3.2.1. Cytotoxicity of Samples
3.2.2. Cell Morphology Examination
3.2.3. Statistical Analysis
3.2.4. Quantitative Testing of Antimicrobial Activity on Bacterial Strains
3.2.5. Antibiofilm Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Reddy, T.S.; Kulhari, H.; Reddy, V.G.; Bansal, V.; Kamal, A.; Shukla, R. Design, synthesis and biological evaluation of 1,3-diphenyl-1H-pyrazole derivatives containing benzimidazole skeleton as potential anticancer and apoptosis inducing agents. Eur. J. Med. Chem. 2015, 101, 790–805. [Google Scholar] [CrossRef] [PubMed]
- Marinescu, M.; Zalaru, C.M. Synthesis, Antibacterial and Anti-Tumor Activity of Pyrazole Derivatives. In Recent Trends in Biochemistry; MedDocs eBooks, Ed.; MedDocs Publishers LLC: Reno, NV, USA, 2021; Chapter 3; pp. 18–27. Available online: http://meddocsonline.org/ (accessed on 1 January 2020).
- Kumar, V.; Kaur, K.; Gupta, G.K.; Sharma, A.K. Pyrazole containing natural products: Synthetic preview and biological significance. Eur. J. Med. Chem. 2013, 69, 735–753. [Google Scholar] [CrossRef]
- Caliskan, B.; Yilmaz, A.; Evren, I.; Menevse, S.; Uludag, O.; Banoglu, E. Synthesis and evaluation of analgesic, anti-inflammatory, and anticancer activities of new pyrazole-3(5)-carboxylic acid derivatives. Med. Chem. Res. 2013, 22, 782–793. [Google Scholar] [CrossRef]
- Gokhan-Kelekci, N.; Koyunoglu, S.; Yabanoglu, S.; Yelekci, K.; Ozgen, O.; Ucar, G.; Erol, K.; Kendi, E.; Yesilada, A. New pyrazoline bearing, 4(3H)-quinazolinone inhibitors of monoamine oxidase: Synthesis, biological evaluation, and structural determinants of MAO-A and MAO-B selectivity. Bioorg. Med. Chem. 2009, 17, 675–689. [Google Scholar] [CrossRef] [PubMed]
- Shih, S.R.; Chu, T.Y.; Reddy, G.R.; Tseng, S.N.; Chen, H.L.; Tang, W.F.; Wu, M.S.; Yeh, J.Y.; Chao, Y.S.; Hsu, J.T.; et al. Pyrazole compound BPR1P0034 with potent and selective anti-influenza virus activity. J. Biomed. Sci. 2010, 17, 13. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.F.; Alam, M.M.; Verma, G.; Akhtar, W.G.; Akhter, M.; Shaquiquzzaman, M. The therapeutic voyage of pyrazole and its analogs: A review. Eur. J. Med. Chem. 2016, 120, 170–201. [Google Scholar] [CrossRef]
- Shah, K.; Chhabra, S.; Shrivastava, S.K.; Mishra, P. Benzimidazole, a promising pharmacophore. Med. Chem. Res. 2013, 22, 5077–5104. [Google Scholar] [CrossRef]
- Narasimhan, B.; Sharma, D.; Kumar, P. Benzimidazole: A medicinally important heterocyclic moiety. Med. Chem. Res. 2012, 21, 269–283. [Google Scholar] [CrossRef]
- Yadav, G.; Ganguly, S. Structure activity relationship (SAR) study of benzimidazole scaffold for different biological activities: A mini-review. Eur. J. Med. Chem. 2015, 97, 419–443. [Google Scholar] [CrossRef]
- Zalaru, C.M.; Marinescu, M. Benzimidazole compounds with anti-tumor and antibacterial activities. In Benzimidazole: Preparation and Applications; Vestergaard, A.A., Ed.; Nova Science Publishers: Hauppauge, NY, USA, 2020; pp. 221–250. [Google Scholar]
- Refaat, H.M. Synthesis and anticancer activity of some novel 2-substituted benzimidazole derivatives. Eur. J. Med. Chem. 2010, 45, 2949–2956. [Google Scholar] [CrossRef]
- Castro, A.R.; Rivera, I.L.; Rojas, L.C.; Vazquez, G.N.; Rodríguez, A.N. Synthesis and preliminary evaluation of selected 2-aryl-5(6)-nitro-1H-benzimidazole derivatives as potential anticancer agents. Arch. Pharm. Res. 2011, 34, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Abonia, R.; Cortes, E.; Insuasty, B.; Quiroga, J.; Nogueras, M.; Cobo, J. Synthesis of novel 1,2,5-trisubstituted benzimidazoles as potential antitumor agents. Eur. J. Med. Chem. 2011, 46, 4062–4070. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Mohsen, H.T.A.; Ragab, F.A.F.; Ramla, M.M.; El Diwani, H.I.E. Novel benzimidazole pyrimidine conjugates as potent anti-tumor agents. Eur. J. Med. Chem. 2010, 45, 2336–2344. [Google Scholar] [CrossRef]
- Demirayak, S.; Kayagil, I.; Yurttas, L. Microwave supported synthesis of some novel 1,3-diarylpyrazino [1,2-a]benzimidazole derivatives and investigation of their anticancer activities. Eur. J. Med. Chem. 2011, 46, 411–416. [Google Scholar] [CrossRef]
- Demirayak, S.; Usama, A.; Mohsen, A.C.; Agri, K. Synthesis and anticancer and anti-HIV testing of some pyrazino [12-a]benzimidazole derivatives. Eur. J. Med. Chem. 2002, 37, 255–260. [Google Scholar] [CrossRef]
- Galal, S.A.; Hegab, K.H.; Kassab, A.S.; Rodriguez, M.L.; Kerwin, S.M.; Abdel-Mo’men, A.; El Diwani, H.I. New transition metal ion complexes with benzimidazole-5-carboxylic acid hydrazides with antitumor activity. Eur. J. Med. Chem. 2009, 44, 1500–1508. [Google Scholar] [CrossRef]
- Galal, S.A.; Hegab, K.H.; Hashem, A.M.; Youssef, N.S. Synthesis and antitumor activity of novel benzimidazole-5-carboxylic acid derivatives and their transition metal complexes as topoisomerease II inhibitors. Eur. J. Med. Chem. 2010, 45, 5685–5691. [Google Scholar] [CrossRef]
- Kamal, A.; Kumar, P.P.; Sreekanth, K.; Seshadri, B.N.; Ramulu, P. Synthesis of new benzimidazole linked pyrrolo [2,1-c][1,4] benzodiazepine conjugates with efficient DNA-binding affinity and potent cytotoxicity. Bioorg. Med. Chem. Lett. 2008, 18, 2594–2598. [Google Scholar] [CrossRef]
- Gowda, N.R.; Kavitha, C.V.; Chiruvella, K.K.; Joy, O.; Rangappa, K.S.; Raghavan, S.C. Synthesis and biological evaluation of novel 1-(4-methoxyphenethyl)-1H-benzimidazole-5-carboxylic acid derivatives and their precursors as antileukemic agents. Bioorg. Med. Chem. Lett. 2009, 19, 4594–4600. [Google Scholar] [CrossRef]
- Zalaru, C.; Dumitrascu, F.; Draghici, C.; Iovu, M.; Marinescu, M.; Tarcomnicu, I.; Nitulescu, G.M. Synthesis and biological screening of some novel 2-(1H-pyrazol-1-yl)-acetamides as lidocaine analogue. Ind. J. Chem. B 2014, 53B, 733–739. [Google Scholar]
- Kumar, B.V.S.; Vaidya, S.D.; Kumar, R.V.; Bhirud, S.B.; Mane, R.B. Synthesis and anti-bacterial activity of some novel 2-(6-fluorochroman-2-yl)-1-alkyl/acyl/aroyl-1H-benzimidazoles. Eur. J. Med. Chem. 2006, 41, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Hosamani, K.M.; Seetharamareddy, H.R.; Keri, R.S.; Hanamanthagouda, M.S.; Moloney, M.G. Microwave assisted one-pot synthesis of 5-nitro-2-aryl substituted-1H-benzimidazole libraries: Screening in vitro for antimicrobial activity. J. Enzym. Inhib. Med. Chem. 2009, 24, 1095–1100. [Google Scholar] [CrossRef] [PubMed]
- Kilcig, G.A.; Altanlar, N. Synthesis and antifungal activity of some benzimidazole derivatives. Turk. J. Chem. 2006, 30, 223–228. [Google Scholar]
- Kerimov, I.; Kilcgil, A.G.; Eke, B.C.; Altanlar, N.; Iscan, M.J. Synthesis antifungal and antioxidant screening of some novel benzimidazole derivatives. J. Enzym. Inhib. Med. Chem. 2007, 22, 696–701. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Narasimhan, B.; Kumar, P.; Judge, V.; Narang, R.; De Clercq, E.; Balzarini, J.J. Synthesis antimicrobial and antiviral activity of substituted benzimidazoles. J. Enzym. Inhib. Med. Chem. 2009, 24, 1161. [Google Scholar] [CrossRef]
- Marinescu, M. Synthesis of Antimicrobial Benzimidazole–Pyrazole Compounds and Their Biological Activities. Antibiotics 2021, 10, 1002. [Google Scholar] [CrossRef]
- Marinescu, M.; Tudorache, D.G.; Marton, G.I.; Zalaru, C.M.; Popa, M.; Chifiriuc, M.C.; Stavarache, C.E.; Constantinescu, C. Density functional theory molecular modeling, chemical synthesis, and antimicrobial behaviour of selected benzimidazole derivatives. J. Mol. Struct. 2017, 1130, 463–471. [Google Scholar] [CrossRef]
- Marinescu, M.; Cinteza, L.O.; Marton, G.I.; Chifiriuc, M.C.; Popa, M.; Stanculescu, I.; Zalaru, C.M.; Stavarache, C.E. Synthesis, density functional theory study and in vitro antimicrobial evaluation of new benzimidazole Mannich bases. BMC Chem. 2020, 14, 45. [Google Scholar] [CrossRef]
- Goker, H.; Kus, C.; Boykin, D.W.; Yildiz, S.; Altanlar, N. Synthesis of some new 2-substituted-phenyl-1H-benzimidazole-5-carbonitriles and their potent activity against Candida species. Bioorg. Med. Chem. 2002, 10, 2589–2596. [Google Scholar] [CrossRef]
- Kus, C.; Kilcgil, A.G.; Eke, B.C.; Iscan, M. Synthesis and antioxidant properties of some novel benzimidazole derivatives on lipid peroxidation in the rat liver. Arch. Pharm. Res. 2004, 27, 156–163. [Google Scholar] [CrossRef]
- Kilcgil, G.A.; Altanlar, N. Synthesis and antimicrobial activities of some new benzimidazole derivatives. Il Farm. 2003, 58, 1345–1350. [Google Scholar] [CrossRef]
- Kus, C.; Kilcgil, G.A.; Ozbey, S.; Kaynak, F.B.; Kaya, M.; Coban, T.; Eke, B.C. Synthesis and antioxidant properties of novel N-methyl-1,3,4-thiadiazol-2-amine and 4-methyl-2H-1,2,4-triazole-3(4H)-thione derivatives of benzimidazole class. Bioorg. Med. Chem. 2008, 16, 4294–4303. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, W.; Khan, M.F.; Verma, G.; Shaquiquzzaman, M.; Rizvi, M.A.; Mehdi, S.H.; Akhter, M.; Alam, M.M. Therapeutic evolution of benzimidazole derivatives in the last quinquennial period. Eur. J. Med. Chem. 2017, 126, 705–753. [Google Scholar] [CrossRef] [PubMed]
- Husain, A.; Rashid, M.; Shaharyar, M.; Siddiqui, A.A.; Mishra, R. Benzimidazole clubbed with triazolo-thiadiazoles and triazolo-thiadiazines: New anticancer agents. Eur. J. Med. Chem. 2013, 62, 785–798. [Google Scholar] [CrossRef]
- Paul, K.; Bindal, S.; Luxami, V. Synthesis of new conjugated coumarinbenzimidazole hybrids and their anticancer activity. Bioorg. Med. Chem. 2013, 15, 3667–3672. [Google Scholar] [CrossRef]
- Galal, S.A.; Khairat, S.H.; Ali, H.I.; Shouman, S.A.; Attia, Y.M.; Ali, M.M.; Mahmoud, A.E.; Abdel-Halim, A.H.; Fyiad, A.A.; Tabll, A.; et al. Part II: New candidates of pyrazole-benzimidazole conjugates as checkpoint kinase 2 (Chk2) inhibitors. Eur. J. Med. Chem. 2018, 144, 859–873. [Google Scholar] [CrossRef]
- El-Gohary, N.S.; Shaaban, M.I. Synthesis, antimicrobial, antiquorum-sensing and antitumor activities of new benzimidazole analogs. Eur. J. Med. Chem. 2017, 13, 439–449. [Google Scholar] [CrossRef]
- Mahal, K.; Ahmad, A.; Schmitt, F.; Lockhauserbäumer, J.; Starz, K.; Pradhan, R.; Padhye, S.; Sarkar, F.H.; Koko, W.S.; Schobert, R.; et al. Improved anticancer and antiparasitic activity of new lawsone Mannich bases. Eur. J. Med. Chem. 2017, 126, 421–431. [Google Scholar] [CrossRef]
- Zalaru, C.M.; Dumitrascu, F.; Draghici, C.; Tarcomnicu, I.; Tatia, R.; Moldovan, L.; Chifiriuc, M.C.; Lazar, V.; Marinescu, M.; Nitulescu, M.G.; et al. Synthesis, spectroscopic characterization, DFT study and antimicrobial activity of novel alkylaminopyrazole derivatives. J. Mol. Struct. 2018, 1156, 12–21. [Google Scholar] [CrossRef]
- Bundgaard, H.; Johansen, M. Hydrolysis of N-Mannich bases and its consequences for the biological testing of such agents. Int. J. Pharm. 1981, 9, 7–16. [Google Scholar] [CrossRef]
- Katritzky, A.R.; Boulton, A.J. Advances in Heterocyclic Chemistry; Academic Press: New York, NY, USA; London, UK, 1979; pp. 347–349. [Google Scholar]
- Dvoretzky, I.; Richter, J.H. Formaldehyde condensation in the pyrazole series. J. Org. Chem. 1950, 15, 1285–1288. [Google Scholar] [CrossRef]
- Hüttel, R.; Jochum, P. Die Mannische Reaktion der Pyrazole. Chem. Ber. 1952, 85, 820–826. [Google Scholar] [CrossRef]
- Morgan, G.T.; Ackerman, I. CLII.-Substitution in the pyrazole series. Halogen derivatives of 3:5-dimethylpyrazole. J. Chem.Soc. Trans. 1923, 123, 1308–1318. [Google Scholar] [CrossRef]
- Hüttel, R.; Schäfer, O.; Jochum, P. Die Jodierung der Pyrazole. Justus Liebigs Ann. Der Chem. 1955, 593, 200–207. [Google Scholar] [CrossRef]
- Zalaru, C.M.; Putina, G.; Dumitrascu, F.; Draghici, C. New Mannich bases with pharmacological activity. Rev. Chim. 2007, 58, 773–775. [Google Scholar]
- Phillips, M.A. CCCXVII.—The formation of 2-substituted benziminazoles. J. Chem. Soc. 1928, 2393–2399. [Google Scholar] [CrossRef]
- Wright, J.B. The Chemistry of the Benzimidazoles. Chem. Rev. 1951, 48, 397–541. [Google Scholar] [CrossRef]
- Somei, M.; Matsubara, M.; Kanda, Y.; Natsume, M. A novel N-Amination Method and its Application to the Preparation of N-Aminoheterocycles. Chem. Pharm. Bull. 1978, 26, 2522–2534. [Google Scholar] [CrossRef]
- Wallace, R.G. Hydroxylamine-O-sulfonic acid-a versatile synthetic reagent. Adrichimica Acta 1980, 13, 3–11. [Google Scholar]
- Sączewski, J.; Korcz, M. Synthesis and reactivity of heterocyclic hydroxylamine-O-sulfonates. Heterocycl. Commun. 2014, 20, 133–147. [Google Scholar] [CrossRef]
- Silverstein, R.M.; Bossler, G.C. Spectrometric identification of organic compounds. J. Chem. Educ. 1962, 39, 546–553. [Google Scholar] [CrossRef]
- Santos, L.S.; Padilha, M.C.; Radler de Aquino, F.N.; Dos Pereira, A.S.; Menegatt, R.; Fraga, A.M.C.; Barreiro, E.J.; Eberlin, M.N. Electrospray ionization mass and tandem mass spectra of a series of N-pyrazolylmethyl and N-triazolylmethyl N-phenylpiperazines: New dopaminergic ligands with potential antipsychotic properties. J. Mass Spectrom. 2005, 40, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Tocher, J.H.; Edwards, D.I. The interaction of reduced metronidazole with DNA bases and nucleosides. Int. J. Radiat. Oncol. Biol. Phys. 1992, 22, 661–663. [Google Scholar] [CrossRef]
- Giedraitiene, A.; Pereckaite, L.; Bredelyte-Gruodiene, E.; Virgailis, M.; Ciapiene, I.; Tatarunas, V. CTX-M-producing Escherichia coli strains: Resistance to temocillin, fosfomycin, nitrofurantoin and biofilm formation. Future Microbiol. 2022, 17, 789–802. [Google Scholar] [CrossRef]












| Comp. | Cell Viability (%) | |||||
|---|---|---|---|---|---|---|
| 0.001 µM | 0.01 µM | 0.1 µM | 1 µM | 0.1 mM | 1 mM | |
| Control | 100 ± 0.01 | 100 ± 0.01 | 100 ± 0.02 | 100 ± 0.01 | 100 ± 0.02 | 100 ± 0.01 |
| 5a | 91.6 ± 0.92 | 89.47 ± 2.71 | 89.2 2 ± 2.70 | 87.96 ± 2.11 | 82.2 ± 1.83 | 22.12 ± 3.01 |
| 5b | 100.19 ± 0.54 | 89.79 ± 0.73 | 88.85 ± 1.75 | 85.02 ± 0.61 | 45.99 ± 1.63 | 7.64 ± 0.51 |
| 5c | 85.23 ± 1.02 | 84.37 ± 3.32 | 83.09 ± 2.36 | 80.76 ± 3.60 | 70.89 ± 2.43 | 7.54 ± 0.70 |
| 5d | 109.25 ± 2.08 | 105.41 ± 1.59 | 102.03 ± 1.19 | 93.76 ± 3.15 | 91.35 ± 1.98 | 36.92 ± 0.60 |
| 5e | 102.71 ± 1.92 | 100.07 ± 2.94 | 99.92 ± 2.05 | 94.74 ± 1.26 | 82.86 ± 1.83 | 37.52 ± 2.22 |
| 5f | 100.46 ± 2.79 | 99.39 ± 3.81 | 98.93 ± 1.96 | 98.7 ± 3.17 | 70.78 ± 2.34 | 35.66 ± 2.29 |
| 5g | 93.91 ± 0.57 | 92.23 ± 2.50 | 80.89 ± 2.36 | 81.03 ± 0.82 | 60.39 ± 1.53 | 10.92 ± 0.50 |
 |  |  |  |  |
Control | 0.001 μM | 1 mM | 0.001 μM | 1 mM |
| 5b | 5c | |||
 |  |  |  | |
| 0.001 μM | 1 mM | 0.001 μM | 1 mM | |
| 5d | 5f | |||
| Compound | Gram-Positive Bacteria | Gram-Negative Bacteria | ||
|---|---|---|---|---|
| Staphylococcus aureus ATCC29213 | Enterococcus faecalis ATCC29212 | Pseudomonas aeruginosa ATCC27853 | Escherichia coli ATCC25922 | |
| 5a | 390 ± 3.05 | 390 ± 3.60 | 780 ± 2.31 | 390 ± 4.58 |
| 5b | 190 ± 1.52 | 390 ± 4.7 2 | 780 ± 2.65 | 390 ± 4.72 |
| 5c | 190 ± 3.06 | 390 ± 4.04 | 780 ± 3.21 | 390 ± 4.51 |
| 5d | 780 ± 2.08 | 780 ± 4.51 | 1560 ± 1.15 | 780 ± 2.51 |
| 5e | 190 ± 4.73 | 390 ± 3.79 | 780 ± 2.88 | 390 ± 2.52 |
| 5f | 150 ± 2.64 | 310 ± 1.53 | 310 ±1.73 | 310 ± 2.30 |
| 5g | 1560 ± 1.53 | 1560 ± 1.15 | 3120 ± 2.64 | 1560 ± 2.52 |
| Metronidazole | 1950 ± 2.08 | 970 ± 2.08 | 1562 ± 1.52 | 1950 ± 2.08 |
| Nitrofurantoin | 600 ± 2.52 | 600 ± 1.53 | 3571 ± 4.94 | 600 ± 3.60 |
| Compound | Gram-Positive Bacteria | Gram-Negative Bacteria | ||
|---|---|---|---|---|
| Staphylococcus aureus ATCC29213 | Enterococcus faecalis ATCC29212 | Pseudomonas aeruginosa ATCC27853 | Escherichia coli ATCC25922 | |
| 5a | 390 ± 2.64 | 390 ± 1.53 | 780 ± 3.21 | 1560 ± 1.52 |
| 5b | 190 ± 1.52 | 390 ± 2.52 | 190 ± 2.08 | 390 ± 3.78 |
| 5c | 190 ± 2.30 | 390 ± 2.88 | 1560 ± 3.21 | 780 ± 2.51 |
| 5d | 780± 2.88 | 780 ± 2.64 | 390 ± 2.63 | 1560 ± 1.52 |
| 5e | 390 ± 3.21 | 390 ± 3.21 | 1560 ± 1.52 | 780 ± 3.05 |
| 5f | 310 ± 0.57 | 310 ± 1.52 | 310 ± 2.08 | 310 ± 1.73 |
| 5g | 1560 ± 2.08 | 1560 ± 3.61 | 3120± 2.08 | 1560 ± 2.02 |
| Metronidazole | 7800 ± 2.64 | 3900 ± 2.64 | 970 ± 2.64 | 3900 ± 3.21 |
| Nitrofurantoin | 2230 ± 2.08 | 600 ± 1.53 | 550 ± 3.79 | 600 ± 1.73 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zalaru, C.; Dumitrascu, F.; Draghici, C.; Tarcomnicu, I.; Marinescu, M.; Nitulescu, G.M.; Tatia, R.; Moldovan, L.; Popa, M.; Chifiriuc, M.C. New Pyrazolo-Benzimidazole Mannich Bases with Antimicrobial and Antibiofilm Activities. Antibiotics 2022, 11, 1094. https://doi.org/10.3390/antibiotics11081094
Zalaru C, Dumitrascu F, Draghici C, Tarcomnicu I, Marinescu M, Nitulescu GM, Tatia R, Moldovan L, Popa M, Chifiriuc MC. New Pyrazolo-Benzimidazole Mannich Bases with Antimicrobial and Antibiofilm Activities. Antibiotics. 2022; 11(8):1094. https://doi.org/10.3390/antibiotics11081094
Chicago/Turabian StyleZalaru, Christina, Florea Dumitrascu, Constantin Draghici, Isabela Tarcomnicu, Maria Marinescu, George Mihai Nitulescu, Rodica Tatia, Lucia Moldovan, Marcela Popa, and Mariana Carmen Chifiriuc. 2022. "New Pyrazolo-Benzimidazole Mannich Bases with Antimicrobial and Antibiofilm Activities" Antibiotics 11, no. 8: 1094. https://doi.org/10.3390/antibiotics11081094
APA StyleZalaru, C., Dumitrascu, F., Draghici, C., Tarcomnicu, I., Marinescu, M., Nitulescu, G. M., Tatia, R., Moldovan, L., Popa, M., & Chifiriuc, M. C. (2022). New Pyrazolo-Benzimidazole Mannich Bases with Antimicrobial and Antibiofilm Activities. Antibiotics, 11(8), 1094. https://doi.org/10.3390/antibiotics11081094