Resistance of Streptococcus suis Isolates from the Czech Republic during 2018–2022
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Isolates
4.2. Serotyping
4.3. Antimicrobial Susceptibility Testing
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tan, M.F.; Tan, J.; Zeng, Y.B.; Li, H.Q.; Yang, Q.; Zhou, R. Antimicrobial resistance phenotypes and genotypes of Streptococcus suis isolated from clinically healthy pigs from 2017 to 2019 in Jiangchi Province, China. J. Appl. Microbiol. 2020, 130, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Goyette-Desjardins, G.; Auger, J.P.; Xu, J.; Segura, M.; Gottschalk, M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microb. Infect. 2014, 3, e45. [Google Scholar] [CrossRef] [PubMed]
- Staats, J.J.; Feder, I.; Okwumabua, O.; Chengappa, M.M. Streptococcus suis: Past and present. Vet. Res. Commun. 1997, 21, 381–407. [Google Scholar] [CrossRef]
- Varela, N.P.; Gadbois, P.; Thibault, C.; Gottschalk, M.; Dick, P.; Wilson, J. Antimicrobial resistance and prudent drug use for Streptococcus suis. Anim. Health Res. Rev. 2013, 14, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Devi, M.; Dutta, J.B.; Rajkhowa, S.; Kalita, D.; Saikia, G.K.; Das, B.C.; Hazarika, R.A.; Mahato, G. Prevalence of multiple drug resistant Streptococcus suis in and around Guwahati. India Vet. World 2017, 10, 556–561. [Google Scholar] [CrossRef] [PubMed]
- Yongkiettrakul, S.; Maneerat, K.; Arechanajan, B.; Malila, Y.; Srimanote, P.; Gottschalk, M.; Visessanguan, W. Antimicrobial susceptibility of Streptococcus suis isolated from diseased pigs, asymptomatic pigs, and human patients in Thailand. BMC Vet. Res. 2019, 15, 5. [Google Scholar] [CrossRef]
- El Garch, F.; de Jong, A.; Simjee, S.; Moyaert, H.; Klein, U.; Ludwig, C.; Marion, H.; Haag-Diergarten, S.; Richard-Mazet, A.; Thomas, V.; et al. Monitoring of antimicrobial susceptibility of respiratory tract pathogens isolated from diseased cattle and pigs across Europe. 2009–2012: VetPath results. Vet. Microbiol. 2016, 194, 11–22. [Google Scholar] [CrossRef]
- Perch, B.; Pedersen, K.B.; Heinrichsen, J. Serology of capsulated streptococci pathogenic for pigs: Six new serotypes of Streptococcus suis. J. Clin. Microbiol. 1983, 17, 993–996. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, M.; Higgins, R.; Jacques, M.; Mittal, K.R.; Henrichsen, J. Description of 14 new capsular types of Streptococcus suis. J. Clin. Microbiol. 1989, 27, 2633–2635. [Google Scholar] [CrossRef]
- Gottschalk, M.; Higgins, R.; Jacques, M.; Beaudoin, M.; Henrichsen, J. Characterization of six new capsular types (23 through 28) of Streptococcus suis. J. Clin. Microbiol. 1991, 29, 2590–2594. [Google Scholar] [CrossRef] [Green Version]
- Higgins, R.; Gottschalk, M.; Boudreau, M.; Lebrun, A.; Henrichsen, J. Description of six new capsular types (29–34) of Streptococcus suis. J. Vet. Diagn. Investig. 1995, 7, 405–406. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.E.; Gottschalk, M.; Brousseau, R.; Harel, J.; Hemmingsen, S.M.; Goh, S.H. Biochemical analysis, cpn60 and 16S rDNA sequence data indicate that Streptococcus suis serotypes 32 and 34. isolated from pigs. are Streptococcus orisratti. Vet. Microbiol. 2005, 107, 63–69. [Google Scholar] [CrossRef]
- Le Tien, H.T.; Nishibori, T.; Nishitani, Y.; Nomoto, R.; Osawa, R. Reappraisal of the taxonomy of Streptococcus suis serotypes 20, 22, 26, and 33 based on DNA-DNA homology and sodA and recN phylogenies. Vet. Microbiol. 2013, 162, 842–849. [Google Scholar] [CrossRef]
- Okura, M.; Lachance, C.; Osaki, M.; Sekizaki, T.; Maruyama, F.; Nozawa, T.; Nakagawa, I.; Hamada, S.; Rossignol, C.; Gottschalk, M.; et al. Development of a two-step multiplex PCR assay for typing of capsular polysaccharide synthesis gene clusters of Streptococcus suis. J. Clin. Microbiol. 2014, 52, 1714–1719. [Google Scholar] [CrossRef]
- Higgins, R.; Gottschalk, M. An update on Streptococcus suis identification. J. Vet. Diagn. Investig. 1990, 2, 249–252. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zheng, H.; Gottschalk, M.; Bai, X.; Lan, R.; Ji, S.; Liu, H.; Xu, J. Development of multiplex PCR assays for the identification of the 33 serotypes of Streptococcus suis. PLoS ONE 2013, 8, e72070. [Google Scholar] [CrossRef]
- Gottschalk, M.; Higgins, R.; Boudreau, M. Use of polyvalent coagglutination reagents for serotyping of Streptococcus suis. J. Clin. Microbiol. 1993, 31, 2192–2194. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, M.; Segura, M.; Xu, J. Streptococcus suis infections in humans: The Chinese experience and the situation in North America. Anim. Health Res. Rev. 2007, 8, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Athey, T.B.; Teatero, S.; Lacouture, S.; Takamatsu, D.; Gottschalk, M.; Fittipaldi, N. Determining Streptococcus suis serotype from short-read whole-genome sequencing data. BMC Microbiol. 2016, 16, 162. [Google Scholar] [CrossRef]
- Matiasovic, J.; Zouharova, M.; Nedbalcova, K.; Kralova, N.; Matiaskova, K.; Simek, B.; Kucharovicova, I.; Gottschalk, M. Resolution of Streptococcus suis serotypes ½ versus 2 and 1 versus 14 by PCR-restriction fragment lenght polymorphism method. J. Clin. Microbiol. 2020, 58, e00480-20. [Google Scholar] [CrossRef]
- Vela, A.I.; Moreno, M.A.; Cebolla, J.A.; Gonzales, S.; Latre, M.V.; Dominguez, L.; Fernandez-Garayzabal, J.F. Antimicrobial susceptibility of clinical strains of Streptococcus suis isolated from pigs in Spain. Vet. Microbiol. 2005, 105, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Hernandes-Garcia, J.; Wang, J.; Restif, O.; Holmes, M.A.; Mather, A.E.; Weinert, L.A.; Wileman, T.M.; Thomson, J.R.; Langford, P.R.; Wren, B.W.; et al. Patterns of antimicrobial resistance in Streptococcus suis isolates from pigs with or without streptococcal disease in England between 2009 and 2014. Vet. Microbiol. 2017, 207, 117–124. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, P.; Wang, Y.; Fu, L.; Liu, L.; Xu, D.; Hou, Y.; Li, Y.; Fu, M.; Wang, X.; et al. Capsular serotypes, antimicrobial susceptibility, and the presence of transferable oxazolidinone resistance genes in Streptococcus suis isolated from healthy pigs in China. Vet. Microbiol. 2020, 247, 108750. [Google Scholar] [CrossRef]
- Lunha, K.; Chumpol, W.; Samngammim, S.; Jiemsup, S.; Assavacheep, P.; Yongkiettrakul, S. Antimicrobial susceptibility of Streptococcus suis isolated from diseased pigs in Thailand, 2018–2020. Antibiotics 2022, 11, 410. [Google Scholar] [CrossRef]
- Burch, D.G.S.; Duran, C.O.; Aarestrup, F.M. Guidelines for antimicrobial use in swine. In Guide to Antimicrobial Use in Animals; Guardabassi, L., Jensen, L.B., Kruse, H., Eds.; Blackwell Publishing: Oxford, UK, 2008; pp. 102–125. [Google Scholar]
- Charpentier, X.; Polard, P.; Claverys, J.P. Induction of competence for genetic transformation by antibiotics: Convergent evolution of stress responses in distant bacterial species lacking SOS? Curr. Opin. Microbiol. 2012, 15, 570–576. [Google Scholar] [CrossRef] [PubMed]
- Lerminiaux, N.A.; Cameron, A.D.S. Horizontal transfeer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 2019, 65, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, M.; Xu, J.; Calzas, C.; Segura, M. Streptococcus suis: A new emerging or an old neglected zoonotic pathogen? Future Microbiol. 2010, 5, 371–391. [Google Scholar] [CrossRef]
- Correa-Fiz, F.; Neila-Ibanez, C.; Lopez-Soria, S.; Napp, S.; Martinez, B.; Sobervia, L.; Tibble, S.; Aragon, V.; Migura-Garcia, L. Feed additives for the control of post-weaning Streptococcus suis disease and the effect on the faecal and nasal microbiota. Sci. Rep. 2020, 10, 20354. [Google Scholar]
- Wisselink, H.J.; Veldman, K.T.; van den Eede, C.; Salmon, S.A.; Mevius, D.J. Quantitative susceptibility of Streptococcus suis strains isolated from diseased pigs in seven European countries to antimicrobial agents licenced in veterinary medicine. Vet. Microbiol. 2006, 113, 73–82. [Google Scholar] [CrossRef]
- De Jong, A.; Thomas, V.; Simjee, S.; Moyaert, H.; El Garch, F.; Mahler, K.; Morrissey, I.; Butty, P.; Klein, U.; Marion, H.; et al. Antimicrobial susceptibility monitoring of respiratory tract pathogens isolated from diseased cattle and pigs across Europe: The VetPath study. Vet. Microbiol. 2014, 172, 202–215. [Google Scholar] [CrossRef]
- Heuvelink, A.E.; van Hout, A.J.; Gonggrijp, M. Monitoring of antimicrobialsusceptibility of swine respiratory pathogens in The Netherlands, 2012–2014. In Proceedings of the 6th Symposium on Antimicrobial Resistance in Animals and the Environment (ARAE), Tours, France, 29 June–1 July 2015; Poster P34. p. 98. [Google Scholar]
- Hendriksen, R.S.; Mevius, D.J.; Schroeter, A.; Teale, C.; Jouy, E.; Butaye, P.; Franco, A.; Utinane, A.; Amado, A.; Moreno, M.; et al. Occurence of antimicrobial resistance among bacterial pathogens and indicator bacteria in pigs in different European countries from year 2002–2004: The ARBAO-II study. Acta Vet. Scand. 2008, 50, 19. [Google Scholar]
- Riley, B.R.; Chidgey, K.L.; Bridges, J.P.; Gordon, E.; Lawrence, K.E. Isolates, antimicrobial susceptibility profiles and multidrug resistance of bacteria cultured from pig submissions in New Zealand. Animals 2020, 10, 1427. [Google Scholar] [CrossRef] [PubMed]
- Werinder, A.; Aspan, A.; Backhans, A.; Sjolund, M.; Guss, B.; Jacobson, B. Streptococcus suis in Swedish grower pigs: Occurence, serotypes, and antimicrobial susceptibility. Acta Vet. Scand. 2020, 62, 36. [Google Scholar] [CrossRef]
- Kerdsin, A.; Akeda, Y.; Hatrongjit, R.; Detchawna, U.; Sekizaki, T.; Hamada, S.; Gottschalk, M.; Oishi, K. Streptococcus suis serotyping by a new multiplex PCR. J. Med. Microbiol. 2014, 63, 824–830. [Google Scholar] [CrossRef]
- Ishida, S.; Tien Le, H.T.; Osawa, R.; Tohya, M.; Nomoto, R.; Kawamura, Y.; Takahashi, T.; Kikuchi, N.; Kikuchi, K.; Sekizaki, T. Development of an appropriate PCR system for the reclassification of Streptococcus suis. J. Microbiol. Methods 2014, 107, 66–70. [Google Scholar] [CrossRef]
- Lakkitjaroen, N.; Takamatsu, D.; Okura, M.; Sato, M.; Osaki, M.; Sekizaki, T. Loss of capsule among Streptococcus suis isolates from porcine endocarditis and its biological significance. J. Med. Microbiol. 2011, 60, 1669–1676. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, Z.; Song, L.; Fan, X.; Wen, F.; Xu, S.; Ning, Y. Antimicrobial resistance profile and genotypic characteristics of Streptococcus suis capsular type 2 isolated from clinical carrier sows and diseased pigs in China. Biomed. Res. Int. 2015, 2015, 284303. [Google Scholar]
- Prufer, T.L.; Rohde, J.; Verspohl, J.; Rohde, M.; de Greeff, A.; Willenborg, J.; Valentin-Weigand, P. Molecular typing of Streptococcus suis strains isolated from diseased and healthy pigs between 1996–2016. PLoS ONE 2019, 14, e0210801. [Google Scholar] [CrossRef]
- Segura, M.; Aragon, V.; Brockmeier, S.L.; Gebhart, C.; Greeff, A.; Kerdsin, A.; O’Dea, M.A.; Okura, M.; Saléry, M.; Schultsz, C.; et al. Update on Streptococcus suis research and prevention in the era of antimicrobial restriction: 4th International Workshop on S. suis. Pathogens 2020, 9, 374. [Google Scholar] [CrossRef]
- Kerdsin, A.; Takeuchi, D.; Nuangmek, A.; Akeda, Y.; Gottschalk, M.; Oishi, K. Genotypic comparison between Streptococcus suis isolated from pigs and humans in Thailand. Pathogens 2020, 9, 50. [Google Scholar] [CrossRef] [PubMed]
- Mittal, K.R.; Higgins, R.; Larivière, S. Identification and serotyping of Haemophilus pleuropneumoniae by coagglutination test. J. Clin. Microbiol. 1983, 18, 1351–1354. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. In CLSI Supplement VET08, 4th ed.; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2018; 170p. [Google Scholar]
- Jones, R.N.; Pfaller, M.A.; Rhomberg, P.R.; Walter, D.H. Tiamulin activity against fastidious and nonfastidious veterinary and human bacterial isolates: Initial development of in vitro susceptibility test methods. J. Clin. Microbiol. 2002, 40, 461–465. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. In CLSI Document VET01-A4—Approved Standard, 4th ed.; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2013; 70p. [Google Scholar]
- Schwarz, S.; Silley, P.; Simjee, S.; Woodford, N.; van Duijkeren, E.; Johnson, A.P.; Gaastra, W. Assessing the antimicrobial susceptibility of bacteria obtained from animals. Vet. Microbiol. 2010, 141, 601–604. [Google Scholar] [CrossRef] [PubMed]




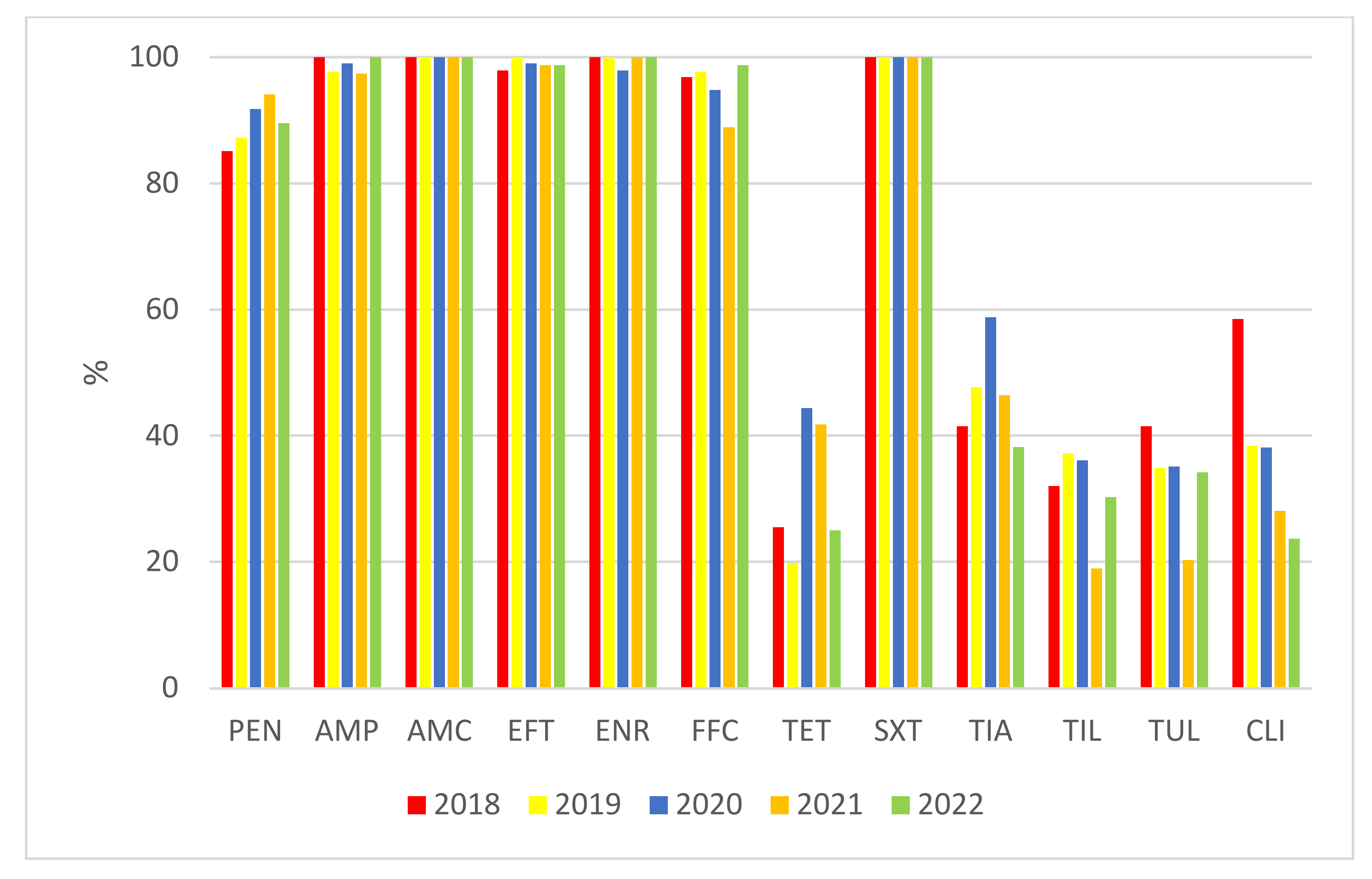








| MIC (mg/L) | S (%) | I (%) | R (%) | MIC50 (mg/L) | MIC90 (mg/L) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.03 | 0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | ||||||
| 2018: n = 94 | |||||||||||||||||||
| PEN | 27 | 34 | 10 | 9 | 4 | 7 | 3 | 85.1 | 4.3 | 10.6 | 0.06 | 1 | |||||||
| AMP | 43 | 30 | 16 | 5 | 100 | 0 | 0 | 0.06 | 0.125 | ||||||||||
| AMC | 93 | 1 | 100 | 0 | 0 | ≤0.25 | ≤0.25 | ||||||||||||
| EFT | 37 | 25 | 9 | 13 | 8 | 2 | 97.9 | 2.1 | 0 | 0.25 | 2 | ||||||||
| ENR | 5 | 9 | 49 | 31 | 100 | 0 | 0 | 0.25 | 0.5 | ||||||||||
| FFC | 1 | 16 | 74 | 3 | 96.8 | 3.2 | 0 | 2 | 2 | ||||||||||
| TET | 20 | 4 | 8 | 4 | 4 | 5 | 7 | 37 | 5 | 25.5 | 8.5 | 66 | 16 | 32 | |||||
| SXT | 66 | 17 | 9 | 2 | 100 | 0 | 0 | ≤0.06 | 0.25 | ||||||||||
| TIA | 2 | 5 | 6 | 5 | 21 | 12 | 7 | 9 | 27 | 41.5 | 21.2 | 38.3 | 8 | >32 | |||||
| TIL | 2 | 9 | 20 | 22 | 6 | 10 | 25 | 32 | - | 68 | 64 | >128 | |||||||
| TUL | 3 | 10 | 14 | 12 | 6 | 5 | 1 | 43 | 41.5 | 6.4 | 52.1 | 64 | >128 | ||||||
| CLI | 46 | 9 | 1 | 1 | 1 | 5 | 4 | 3 | 24 | 58.5 | 1.1 | 40.4 | 0.25 | >16 | |||||
| 2019: n = 86 | |||||||||||||||||||
| PEN | 30 | 34 | 4 | 7 | 4 | 2 | 4 | 1 | 87.2 | 4 | 7 | 0.06 | 0.5 | ||||||
| AMP | 37 | 28 | 16 | 1 | 2 | 2 | 97.7 | 2.3 | 0 | 0.06 | 0.125 | ||||||||
| AMC | 85 | 1 | 100 | 0 | 0 | ≤0.25 | ≤0.25 | ||||||||||||
| EFT | 34 | 32 | 7 | 1 | 100 | 0 | 0 | 0.25 | 1 | ||||||||||
| ENR | 3 | 25 | 45 | 13 | 100 | 0 | 0 | 0.25 | 0.5 | ||||||||||
| FFC | 1 | 20 | 63 | 1 | 1 | 97.7 | 1.15 | 1.15 | 2 | 2 | |||||||||
| TET | 11 | 6 | 13 | 1 | 5 | 14 | 34 | 2 | 19.8 | 15.1 | 65.1 | 16 | 32 | ||||||
| SXT | 61 | 15 | 8 | 2 | 100 | 0 | 0 | ≤0.06 | 0.25 | ||||||||||
| TIA | 2 | 6 | 2 | 10 | 21 | 11 | 6 | 5 | 23 | 47.7 | 19.7 | 32.6 | 8 | >32 | |||||
| TIL | 1 | 2 | 29 | 14 | 1 | 39 | 37.2 | - | 62.8 | 32 | >128 | ||||||||
| TUL | 1 | 2 | 3 | 13 | 11 | 9 | 6 | 2 | 39 | 34.9 | 10.4 | 54.7 | 64 | >128 | |||||
| CLI | 26 | 7 | 2 | 2 | 8 | 3 | 1 | 37 | 38.4 | 2.3 | 59.3 | 4 | >16 | ||||||
| 2020: n = 97 | |||||||||||||||||||
| PEN | 43 | 37 | 7 | 2 | 5 | 1 | 2 | 91.8 | 5.1 | 3.1 | 0.06 | 0.25 | |||||||
| AMP | 52 | 37 | 2 | 4 | 1 | 1 | 99 | 1 | 0 | ≤0.03 | 0.125 | ||||||||
| AMC | 95 | 2 | 100 | 0 | 0 | ≤0.25 | ≤0.25 | ||||||||||||
| EFT | 63 | 22 | 4 | 5 | 2 | 1 | 99 | 1 | 0 | ≤0.125 | 0.5 | ||||||||
| ENR | 2 | 16 | 59 | 18 | 2 | 97.9 | 2.1 | 0 | 0.25 | 0.5 | |||||||||
| FFC | 1 | 25 | 66 | 5 | 94.8 | 5.2 | 0 | 2 | 2 | ||||||||||
| TET | 35 | 8 | 11 | 6 | 1 | 4 | 9 | 18 | 5 | 44.35 | 11.3 | 44.35 | 1 | 32 | |||||
| SXT | 78 | 12 | 6 | 1 | 100 | 0 | 0 | ≤0.06 | 0.125 | ||||||||||
| TIA | 11 | 6 | 9 | 27 | 7 | 5 | 12 | 16 | 58.8 | 12.4 | 28.8 | 4 | >32 | ||||||
| TIL | 6 | 29 | 20 | 42 | 36.1 | - | 63.9 | 128 | >128 | ||||||||||
| TUL | 1 | 1 | 5 | 9 | 18 | 11 | 10 | 1 | 41 | 35.1 | 11.3 | 53.6 | 64 | >128 | |||||
| CLI | 33 | 4 | 3 | 1 | 6 | 1 | 5 | 44 | 38.1 | 3.1 | 58.8 | 16 | >16 | ||||||
| 2021 (n = 153) | |||||||||||||||||||
| PEN | 79 | 59 | 4 | 2 | 4 | 2 | 1 | 2 | 94.1 | 2.6 | 3.3 | ≤0.03 | 0.06 | ||||||
| AMP | 84 | 57 | 4 | 3 | 1 | 3 | 1 | 97.4 | 2 | 0.6 | ≤0.03 | 0.06 | |||||||
| AMC | 151 | 2 | 100 | 0 | 0 | ≤0.25 | ≤0.25 | ||||||||||||
| EFT | 111 | 23 | 6 | 6 | 5 | 2 | 98.7 | 1.3 | 0 | ≤0.125 | 0.5 | ||||||||
| ENR | 2 | 37 | 77 | 37 | 100 | 0 | 0 | 0.25 | 0.5 | ||||||||||
| FFC | 42 | 94 | 17 | 88.9 | 11.1 | 0 | 2 | 4 | |||||||||||
| TET | 52 | 12 | 18 | 11 | 1 | 11 | 35 | 13 | 41.8 | 11.8 | 46.4 | 1 | 32 | ||||||
| SXT | 123 | 19 | 6 | 3 | 2 | 100 | 0 | 0 | ≤0.06 | 0.125 | |||||||||
| TIA | 5 | 16 | 10 | 17 | 23 | 15 | 14 | 12 | 41 | 46.4 | 19 | 34.6 | 8 | >32 | |||||
| TIL | 4 | 8 | 1 | 16 | 37 | 2 | 2 | 83 | 19 | - | 81 | >128 | >128 | ||||||
| TUL | 10 | 4 | 3 | 8 | 6 | 15 | 21 | 4 | 82 | 20.3 | 9.8 | 69.9 | >128 | >128 | |||||
| CLI | 35 | 8 | 5 | 1 | 2 | 12 | 7 | 1 | 82 | 28.1 | 3.3 | 68.6 | >16 | >16 | |||||
| 2022 (n = 76) | |||||||||||||||||||
| PEN | 32 | 26 | 5 | 5 | 5 | 2 | 1 | 89.5 | 6.6 | 3.9 | 0.06 | 0.5 | |||||||
| AMP | 36 | 29 | 4 | 6 | 1 | 100 | 0 | 0.4 | 0.06 | 0.125 | |||||||||
| AMC | 76 | 100 | 0 | 0 | ≤0.25 | ≤0.25 | |||||||||||||
| EFT | 57 | 6 | 7 | 3 | 2 | 1 | 98.7 | 1.3 | 0 | ≤0.125 | 0.5 | ||||||||
| ENR | 1 | 8 | 49 | 18 | 100 | 0 | 0 | 0.25 | 0.5 | ||||||||||
| FFC | 14 | 61 | 1 | 98.7 | 0 | 1.3 | 2 | 2 | |||||||||||
| TET | 14 | 5 | 16 | 2 | 2 | 7 | 28 | 2 | 25 | 21 | 54 | 8 | 32 | ||||||
| SXT | 61 | 10 | 3 | 1 | 1 | 100 | 0 | 0 | ≤0.06 | 0.125 | |||||||||
| TIA | 2 | 3 | 7 | 17 | 4 | 6 | 9 | 28 | 38.2 | 13.1 | 48.7 | 16 | >32 | ||||||
| TIL | 1 | 1 | 21 | 21 | 32 | 30.3 | - | 69.7 | >128 | >128 | |||||||||
| TUL | 5 | 9 | 2 | 10 | 9 | 8 | 1 | 32 | 34.2 | 11.8 | 54 | >128 | >128 | ||||||
| CLI | 17 | 1 | 7 | 1 | 10 | 4 | 2 | 34 | 23.7 | 9.2 | 67.1 | >16 | >16 | ||||||
| 2018–2022 (n = 506) | |||||||||||||||||||
| PEN | 211 | 190 | 30 | 25 | 22 | 14 | 11 | 3 | 90.1 | 4.3 | 5.6 | 0.06 | 0.25 | ||||||
| AMP | 252 | 181 | 42 | 19 | 5 | 5 | 2 | 98.6 | 1 | 0.4 | 0.06 | 0.125 | |||||||
| AMC | 500 | 6 | 100 | 0 | 0 | ≤0.25 | ≤0.25 | ||||||||||||
| EFT | 302 | 108 | 33 | 40 | 17 | 6 | 98.8 | 0.2 | 0 | ≤0.125 | 1 | ||||||||
| ENR | 13 | 95 | 279 | 117 | 2 | 99.6 | 0.4 | 0 | 0.25 | 0.5 | |||||||||
| FFC | 3 | 117 | 358 | 26 | 2 | 94.5 | 5.1 | 0.4 | 2 | 2 | |||||||||
| TET | 132 | 35 | 66 | 24 | 5 | 17 | 48 | 152 | 27 | 33 | 13 | 54 | 2 | 32 | |||||
| SXT | 389 | 73 | 32 | 9 | 3 | 100 | 0 | 0 | ≤0.06 | 0.125 | |||||||||
| TIA | 13 | 40 | 27 | 48 | 109 | 49 | 38 | 47 | 135 | 46.8 | 17.2 | 36 | 8 | >32 | |||||
| TIL | 1 | 4 | 11 | 19 | 115 | 114 | 8 | 13 | 221 | 29.6 | - | 70.4 | 32 | >128 | |||||
| TUL | 12 | 15 | 30 | 46 | 57 | 50 | 50 | 9 | 237 | 35.2 | 9.9 | 54.9 | 64 | >128 | |||||
| CLI | 157 | 29 | 18 | 5 | 4 | 41 | 19 | 12 | 221 | 36.8 | 3.5 | 59.7 | 4 | >16 | |||||
| Body Site of Isolation | Number of Isolates | |||||
|---|---|---|---|---|---|---|
| 2018 | 2019 | 2020 | 2021 | 2022 | Total | |
| nasal swabs | 13 | 8 | 10 | 17 | 4 | 52 |
| tonsils | - | 9 | - | - | - | 9 |
| lower respiratory tract | 51 | 31 | 43 | 54 | 41 | 220 |
| lymph nodes | 11 | 16 | 17 | 19 | 14 | 77 |
| joint | 3 | 4 | 5 | 10 | 5 | 27 |
| brain | 9 | 12 | 13 | 26 | 8 | 68 |
| digestive system | 2 | 2 | 6 | 6 | 3 | 19 |
| urogenital tract | 2 | 1 | 2 | 2 | 1 | 8 |
| skin | 2 | 2 | - | - | - | 4 |
| not specified | 1 | 1 | 1 | 19 | - | 22 |
| PEN | AMP | AMC * | EFT | ENR | FFC | CLI | TIA | TIL | TUL | TET | SXT ** |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 4 | 32 | 16 | 8 | 64 | 16 | 32 | 128 | 128 | 32 | PGC |
| 2 | 2 | 16 | 8 | 4 | 32 | 8 | 16 | 64 | 64 | 16 | 4 |
| 1 | 1 | 8 | 4 | 2 | 16 | 4 | 8 | 32 | 32 | 8 | 2 |
| 0.5 | 0.5 | 4 | 2 | 1 | 8 | 2 | 4 | 16 | 16 | 4 | 1 |
| 0.25 | 0.25 | 2 | 1 | 0.5 | 4 | 1 | 2 | 8 | 8 | 2 | 0.5 |
| 0.125 | 0.125 | 1 | 0.5 | 0.25 | 2 | 0.5 | 1 | 4 | 4 | 1 | 0.25 |
| 0.06 | 0.06 | 0.5 | 0.25 | 0.125 | 1 | 0.25 | 0.5 | 2 | 2 | 0.5 | 0.125 |
| 0.03 | 0.03 | 0.25 | 0.125 | 0.06 | 0.5 | 0.125 | 0.25 | 1 | 1 | 0.25 | 0.06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nedbalcova, K.; Kucharovicova, I.; Zouharova, M.; Matiaskova, K.; Kralova, N.; Brychta, M.; Simek, B.; Pecha, T.; Plodkova, H.; Matiasovic, J. Resistance of Streptococcus suis Isolates from the Czech Republic during 2018–2022. Antibiotics 2022, 11, 1214. https://doi.org/10.3390/antibiotics11091214
Nedbalcova K, Kucharovicova I, Zouharova M, Matiaskova K, Kralova N, Brychta M, Simek B, Pecha T, Plodkova H, Matiasovic J. Resistance of Streptococcus suis Isolates from the Czech Republic during 2018–2022. Antibiotics. 2022; 11(9):1214. https://doi.org/10.3390/antibiotics11091214
Chicago/Turabian StyleNedbalcova, Katerina, Ivana Kucharovicova, Monika Zouharova, Katarina Matiaskova, Natalie Kralova, Marek Brychta, Bronislav Simek, Tomas Pecha, Hana Plodkova, and Jan Matiasovic. 2022. "Resistance of Streptococcus suis Isolates from the Czech Republic during 2018–2022" Antibiotics 11, no. 9: 1214. https://doi.org/10.3390/antibiotics11091214





