Impact of an Antimicrobial Stewardship Program Intervention Associated with the Rapid Identification of Microorganisms by MALDI-TOF and Detection of Resistance Genes in ICU Patients with Gram-Negative Bacteremia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Patient Population
2.2. Laboratory Procedures
2.3. Data Collection
2.4. Outcomes
2.5. Statistical Analysis
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Beganovic, M.; Costello, M.; Wieczorkiewicz, S.M. Effect of Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS) Alone versus MALDI-TOF MS Combined with Real-Time Antimicrobial Stewardship Interventions on Time to Optimal Antimicrobial Therapy in Patients with Positive Blood Cultures. J. Clin. Microbiol. 2017, 55, 1437–1445. [Google Scholar] [CrossRef] [PubMed]
- Timsit, J.F.; Bassetti, M.; Cremer, O.; Daikos, G.; de Waele, J.; Kallil, A.; Kipnis, E.; Kollef, M.; Laupland, K.; Paiva, J.A.; et al. Rationalizing antimicrobial therapy in the ICU: A narrative review. Intensive Care Med. 2019, 45, 172–189. [Google Scholar] [CrossRef]
- Perez, K.K.; Olsen, R.J.; Musick, W.L.; Cernoch, P.L.; Davis, J.R.; Peterson, L.E.; Musser, J.M. Integrating rapid diagnostics and antimicrobial stewardship improves outcomes in patients with antibiotic-resistant Gram-negative bacteremia. J. Infect. 2014, 69, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Barlam, T.F.; Cosgrove, S.E.; Abbo, L.M.; MacDougall, C.; Schuetz, A.N.; Septimus, E.J.; Srinivasan, A.; Dellit, T.H.; Falck-Ytter, Y.T.; Fishman, N.O.; et al. Implementing an Antibiotic Stewardship Program: Guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin. Infect. Dis. 2016, 62, e51–e77. [Google Scholar] [CrossRef] [PubMed]
- Rivard, K.R.; Athans, V.; Lam, S.W.; Gordon, S.M.; Procop, G.W.; Richter, S.S.; Neuner, E. Impact of antimicrobial stewardship and rapid microarray testing on patients with Gram-negative bacteremia. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 1879–1887. [Google Scholar] [CrossRef] [PubMed]
- Timbrook, T.T.; Morton, J.B.; McConeghy, K.W.; Caffrey, A.R.; Mylonakis, E.; LaPlante, K.L. The effect of molecular rapid diagnostic testing on clinical outcomes in bloodstream infections: A systematic review and meta-analysis. Clin. Infect. Dis. 2017, 64, 15–23. [Google Scholar] [CrossRef]
- Huang, A.M.; Newton, D.; Kunapuli, A.; Gandhi, T.N.; Washer, L.L.; Isip, J.; Collins, C.D.; Nagel, J.L. Impact of rapid organism identification via matrix-assisted laser desorption/ionization time-of-flight combined with antimicrobial stewardship team intervention in adult patients with bacteremia and candidemia. Clin. Infect. Dis. 2013, 57, 1237–1245. [Google Scholar] [CrossRef]
- Clerc, O.; Prod’hom, G.; Vogne, C.; Bizzini, A.; Calandra, T.; Greub, G. Impact of matrix-assisted laser desorption ionization time-of-flight mass spectrometry on the clinical management of patients with gram-negative bacteremia: A prospective observational study. Clin. Infect. Dis. 2013, 56, 1101–1107. [Google Scholar] [CrossRef]
- Anton-Vazquez, V.; Hine, P.; Krishna, S.; Chaplin, M.; Planche, T. Rapid versus standard antimicrobial susceptibility testing to guide treatment of bloodstream infection. Cochrane Database Syst. Rev. 2021, 5, CD013235. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute M100-S18. 40. Performance Standards for Antimicrobial Susceptibility Testing. 30th Informational Supplement; CLSI: Wayne, PA, USA, 2020. [Google Scholar]
- Prod’hom, G.; Bizzini, A.; Durussel, C.; Bille, J.; Greub, G. Matrix-assisted laser desorption ionizationtime of flight mass spectrometry for direct bacterial identification from positive blood culture pellets. J. Clin. Microbiol. 2010, 48, 1481–1483. [Google Scholar] [CrossRef] [Green Version]
- Galiana, A.; Coy, J.; Gimeno, A.; Guzman, N.M.; Rosales, F.; Merino, E.; Royo, G.; Rodríguez, J.C. Flow Chip assay for the diagnosis of blood infections. PLoS ONE 2017, 12, e0177627. [Google Scholar] [CrossRef] [PubMed]
- Levin, A.S.S. Guia de Utilização de Anti-Infecciosos e Recomendações Para a Prevenção de Infecções Relacionadas à Assistência à Saúde; 7a edição: Sao Paulo, Brazil, 2018. [Google Scholar]
- Claeys, K.C.; Schlaffer, K.E.; Heil, E.L.; Leekha, S.; Johnson, J.K. Validation of an Antimicrobial Stewardship-Driven Verigene Blood-Culture Gram-Negative Treatment Algorithm to Improve Appropriateness of Antibiotics. Open Forum. Infect. Dis. 2018, 5, ofy233. [Google Scholar] [CrossRef] [PubMed]
- Kollef, M.H.; Morrow, L.E.; Niederman, M.S.; Leeper, K.V.; Anzueto, A.; Benz-Scott, L.; Rodino, F.J. Clinical characteristics, and treatment patterns among patients with ventilator-associated pneumonia. Chest 2006, 129, 1210–1218. [Google Scholar] [CrossRef] [PubMed]
- Garner, J.S.; Jarvis, W.R.; Emori, T.G.; Horan, T.C.; Hughes, J.M. CDC definitions for nosocomial infections, 1988. Am. J. Infect. Control. 1988, 16, 128–140. [Google Scholar] [CrossRef]
- Agência Nacional de Vigilância Sanitária. Diretriz Nacional para o Uso de Antimicrobianos em Serviços de Saúde; ANVISA: Brasília, Brazil, 2016. [Google Scholar]
- Rodrigues, C.; Siciliano, R.F.; Filho, H.C.; Charbel, C.E.; de Carvalho Sarahyba da Silva, L.; Baiardo Redaelli, M.; de Paula Rosa Passetti, A.P.; Franco, M.R.G.; Rossi, F.; Zeigler, R.; et al. The effect of a rapid molecular blood test on the use of antibiotics for nosocomial sepsis: A randomized clinical trial. J. Intensive Care 2019, 7, 37. [Google Scholar] [CrossRef] [PubMed]
- Perez, K.K.; Olsen, R.J.; Musick, W.L.; Cernoch, P.L.; Davis, J.R.; Land, G.A.; Peterson, L.E.; Musser, J.M. Integrating rapid pathogen identification and antimicrobial stewardship significantly decreases hospital costs. Arch. Pathol. Lab. Med. 2013, 137, 1247–1254. [Google Scholar] [CrossRef]
- Musuuza, J.S.; Watson, L.; Parmasad, V.; Putman-Buehler, N.; Christensen, L.; Safdar, N. Prevalence and outcomes of co-infection and superinfection with SARS-CoV-2 and other pathogens: A systematic review and meta-analysis. PLoS ONE 2021, 16, e0251170. [Google Scholar] [CrossRef]
- Polly, M.; de Almeida, B.L.; Lennon, R.P.; Cortês, M.F.; Costa, S.F.; Guimarães, T. Impact of the COVID-19 pandemic on the incidence of multidrug-resistant bacterial infections in an acute care hospital in Brazil. Am. J. Infect. Control. 2022, 50, 32–38. [Google Scholar] [CrossRef]
- Rouzé, A.; Martin-Loeches, I.; Povoa, P.; Makris, D.; Artigas, A.; Bouchereau, M.; Lambiotte, F.; Metzelard, M.; Cuchet, P.; Boulle Geronimi, C.; et al. Relationship between SARS-CoV-2 infection and the incidence of ventilator-associated lower respiratory tract infections: A European multicenter cohort study. Intensive Care Med. 2022, 47, 188–198. [Google Scholar] [CrossRef]
- Coordenadoria de Vigilância em Saúde do Município de São Paulo. Available online: https://www.prefeitura.sp.gov.br/cidade/secretarias/saude/vigilancia_em_saude (accessed on 1 May 2022).
- Porto, A.P.M.; Goossens, H.; Versporten, A.; Costa, S.F.; Brazilian Global-PPS Working Group. Global point prevalence survey of antimicrobial consumption in Brazilian hospitals. J. Hosp. Infect. 2020, 104, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Vlek, A.L.M.; Bonten, M.J.M.; Boel, C.H.E. Direct Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry Improves Appropriateness of Antibiotic Treatment of Bacteremia. PLoS ONE 2012, 7, e32589. [Google Scholar] [CrossRef]
- Munson, E.L.; Diekema, D.J. Detection and treatment of bloodstream infection: Laboratory reporting and antimicrobial management. J. Clin. Microbiol. 2003, 41, 495–497. [Google Scholar] [CrossRef] [PubMed]
- Cattoir, L.; Coorevits, L.; Leroux-Roels, I.; Claeys, G.; Verhasselt, B.; Boelens, J. Improving timelines in reporting results from positive blood cultures: Simulation of impact of rapid identification on therapy on a real-life cohort. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 2253–2260. [Google Scholar] [CrossRef] [PubMed]
- Messacar, K.; Parker, S.K.; Todd, J.K.; Dominguez, S.R. Implementation of Rapid Molecular Infectious Disease Diagnostics: The Role of Diagnostic and Antimicrobial Stewardship. J. Clin. Microbiol. 2017, 55, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Doern, C.D. The Confounding Role of Antimicrobial Stewardship Programs in Understanding the Impact of Technology on Patient Care. J. Clin. Microbiol. 2016, 54, 2420–2423. [Google Scholar] [CrossRef] [Green Version]
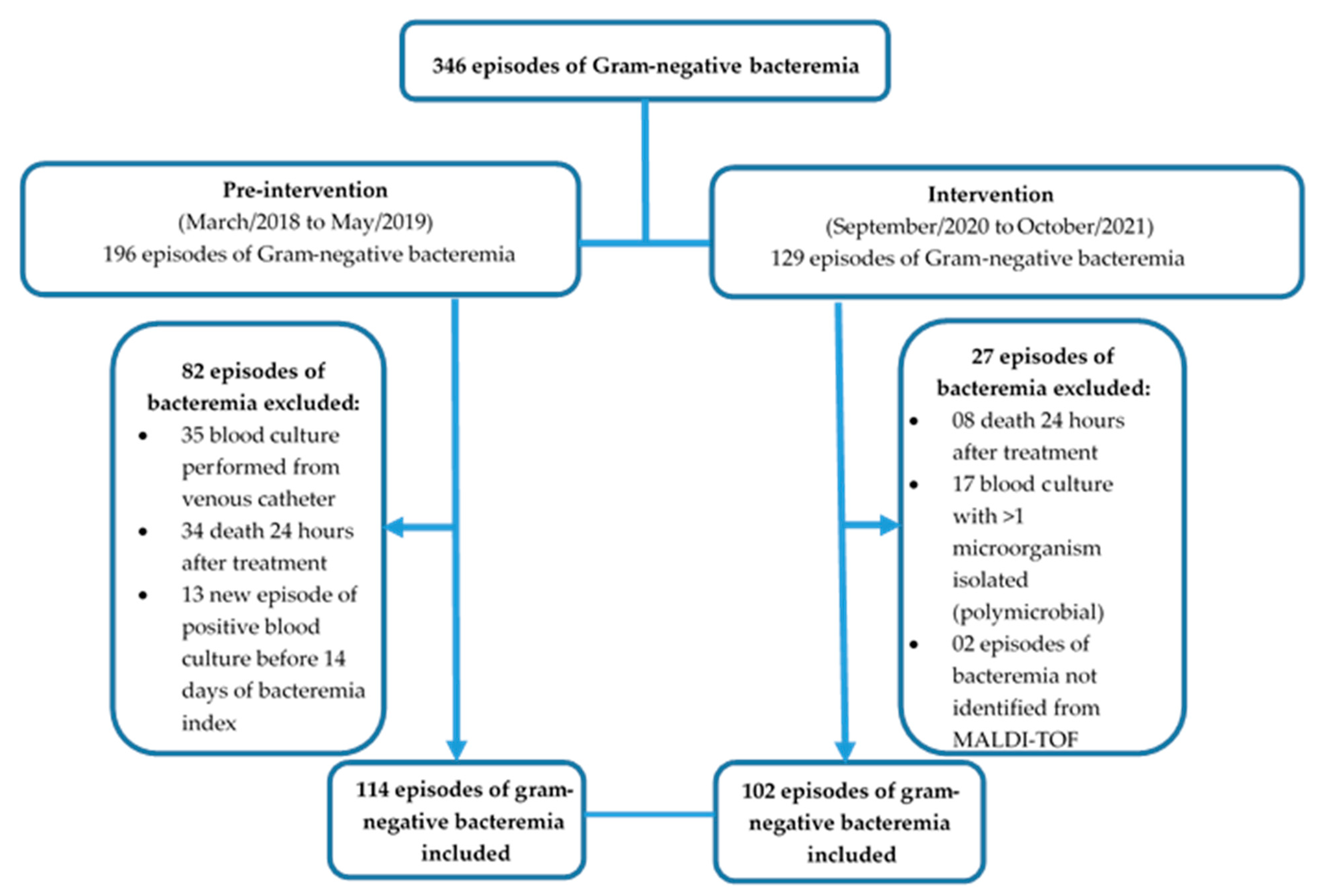
| Variable | Pre-Intervention (N = 114) | Intervention (N = 102) | p-Value |
|---|---|---|---|
| Median age (range in years) | 56 (41–62) | 59 (47–69) | 0.057 a |
| Males (%) | 78 (68.4%) | 65 (63.7%) | 0.466 b |
| End stage renal diseases requiring dialysis, N (%) | 37 (32.5%) | 42 (41.2%) | 0.184 b |
| Central venous catheter, N (%) | 81 (71.1%) | 84 (82.4%) | 0.051 b |
| SAPS3 (median, IQR) | 56 (45–68) | 64 (50–76) | 0.005 a |
| PITT bacteremia score ≥ 6, N (%) | 60 (52.6%) | 62 (60.8%) | 0.228 b |
| ICU admission, N (%) | |||
| Severe acute respiratory syndrome SARS-CoV-2 | 0 (0%) | 45 (44.1%) | <0.0001 b |
| Gastrointestinal diseases | 25 (21.9%) | 5 (4.9%) | 0.0003 b |
| Infectious diseases | 26 (21.9%) | 8 (7.8%) | 0.002 b |
| Cerebrovascular diseases | 16 (14%) | 10 (10%) | 0.34 b |
| Trauma | 13 (11.4%) | 10 (10%) | 0.7 b |
| Kidney diseases | 8 (7%) | 5 (5%) | 0.51 b |
| Cardiovascular diseases | 5 (4.4%) | 5 (5%) | 0.85 b |
| Metabolic diseases | 3 (2.6%) | 1 (1%) | 0.36 b |
| Others | 19 (716.6%) | 13 (13%) | 0.33 b |
| Blood stream infection source, N (%) | |||
| Primary | 58 (50.8%) | 48 (47.1%) | 0.671 b |
| Pulmonary | 13 (11.4%) | 28 (27.5%) | 0.005 b |
| Urinary | 9 (7.9%) | 9 (8.8%) | 1.0 b |
| Intra-abdominal | 17 (14.9%) | 7 (6.9%) | 0.096 b |
| Skin and soft tissue | 7 (6.1%) | 8 (7.8%) | 0.823 b |
| Other sites | 10 (8.7%) | 2 (2.0%) | 0.060 b |
| N (%) of organisms isolated | |||
| K. pneumoniae | 49 (43.0%) | 33 (32.4%) | 0.043 b |
| E. coli | 19 (16.6%) | 13 (12.7%) | 0.536 b |
| A. baumannii | 9 (7.8%) | 17 (16.7%) | 0.077 b |
| P. aeruginosa | 7 (6.1%) | 22 (21.6%) | 0.002 b |
| Enterobacter spp. | 5 (13.1%) | 5 (4.9%) | 0.064 b |
| Others Enterobacterales | 13 (11.4%) | 8 (7.8%) | 0.515 b |
| Others non fermentative | 2 (1.7%) | 4 (3.9%) | 0.580 b |
| N (%) Carbapenem-resistance (N = 76) | |||
| Enterobacterales | 28 (24.6%) | 16 (15.7%) | 0.141 b |
| P. aeruginosa | 3 (2.7%) | 5 (5.0%) | 0.602 b |
| A. baumannii | 8 (7.0%) | 16 (15.7%) | 0.071 b |
| N (%) Genes Resistance (N = 51) | |||
| blaCTX-M | - | 30 (68.6%) | |
| K. pneumoniae | - | 22 | |
| E. coli | - | 4 | |
| Enterobacter spp. | - | 2 | |
| P. aeruginosa | - | 1 | |
| Burkholderia cepacia complex | - | 1 | |
| blaKPC | 14 (27.4%) | ||
| K. pneumoniae | - | 13 | |
| E. coli | - | 1 | |
| blaOXA-23, blaOXA-24, blaOXA-51 | 21 (41.2%) | ||
| A. baumannii | - | 20 | |
| P. aeruginosa | - | 1 | |
| SHV-enzymes | 5 | ||
| K. pneumoniae | - | 5 (9.8%) | |
| blaNDM- | 1 | ||
| K. pneumoniae | - | 1 (2.0%) | |
| Empirical Antimicrobial regimen | |||
| Monotherapy | 56 (49.1%) | 50 (49.0%) | 0.988 b |
| 2 antibiotics | 38 (33.3%) | 48 (47.1%) | 0.040 b |
| 3 antibiotics | 20 (17.5%) | 4 (3.9%) | 0.001 b |
| Duration of antimicrobial therapy (mean; days) | 9.13 | 7.8 | 0.013 a |
| Characteristic Median, IQR | Pre-Intervention N = 114 | Intervention N = 102 | p-Value |
|---|---|---|---|
| 30-day mortality | 29 (25%) | 36 (35%) | 0.115 b |
| Hospital LOS | 44 (20–59) | 39 (14–48) | 0.005 a |
| ICU LOS | 17 (7–22) | 13 (5–16) | 0.033 a |
| Univariate Analysis | ||||
|---|---|---|---|---|
| Variable | Death | |||
| No | Yes | HR (CI 95%) | p-Value | |
| (N = 151) | (N = 65) | |||
| Pre intervention, N | 49 (43%) | 65 (57%) | ||
| Intervention, N | 34 (33.3%) | 68 (66.6%) | 1.38 (0.97–1.95) | 0.072 |
| SARS-CoV-2, N | 9 (20%) | 36 (80%) | 1.74 (1.17–2.59) | 0.006 |
| SAPS3 (median, IQR) | 51 (45–67) | 62 (50–74) | 1.01 (1.00–1.02) | 0.083 |
| Gastrointestinal diseases, N | 17 (56.6%) | 13 (43.3%) | 1.18 (0.85–1.64) | 0.333 |
| Infectious diseases, N | 12 (14.5%) | 22 (16.5%) | 0.83 (0.52–1.32) | 0.432 |
| Blood stream infection source: pulmonary, N | 18 (43.9%) | 23 (56.1%) | 1.39 (1.07–1.80) | 0.036 |
| Organism isolated: P. aeruginosa | 11 (37.9%) | 18 (62.1%) | 1.38 (1.09–1.74) | 0.002 |
| Multivariate analysis | ||||
| OR | CI95% | p-value | ||
| SARS-CoV-2 | 1.54 | (1.02–2.29) | 0.036 | |
| Blood stream infection source: pulmonary | 1.75 | (0.87–3.51) | 0.113 | |
| Organism isolated: P. aeruginosa | 1.26 | (0.56–2.83) | 0.573 | |
| Antimicrobial Consumption and Cost | Pre-Intervention | Intervention | p-Value | Percent of Change |
|---|---|---|---|---|
| Consumption (DOT/1000 days present, median, IQR) | ||||
| All antimicrobials | N = 114 1.381 (1.103–2.251) | N = 102 1.262 (1.063–1.662) | 0.032 a | |
| Antimicrobial for gram-negative bacteria | N = 114 1.281 (1.004–1.775) | N = 102 1.172 (1.006–1.427) | 0.067 a | |
| Antimicrobial for gram-positive bacteria | N = 52 475 (238–761) | N = 43 270 (139–467) | 0.004 a | |
| Carbapenems | N = 76 836 (504–1056) | N = 72 543 (301–991) | 0.040 a | |
| Colistin/Polymyxin B | N = 45 722 (390–932) | N = 40 881 (462–1001) | 0.299 a | |
| Ceftazidime/avibactam | N = 3 931 (725–1022) | N = 8 911 (823–940) | 0.865 a | |
| Costs (Sum USD) | ||||
| All antimicrobials | N = 114 $23,937.60 | N = 102 $31,126.05 | 0.30 | +30% |
| Gram-negative antimicrobials | N = 114 $23,331.30 | N = 102 $30,332.39 | 0.283 | +33% |
| Gram-positive antimicrobials | N = 52 $627.00 | N = 43 $202.68 | 0.008 | −78% |
| Carbapenems | N = 76 $6003.20 | N = 72 $3938.63 | 0.039 | −34% |
| Colistin/Polymyxin B | N = 45 $5450.26 | N = 40 $3030.88 | 0.067 | −47% |
| Ceftazidime/avibactam | N = 3 $4471.50 | N = 8 $19,949.77 | 0.008 | +346% |
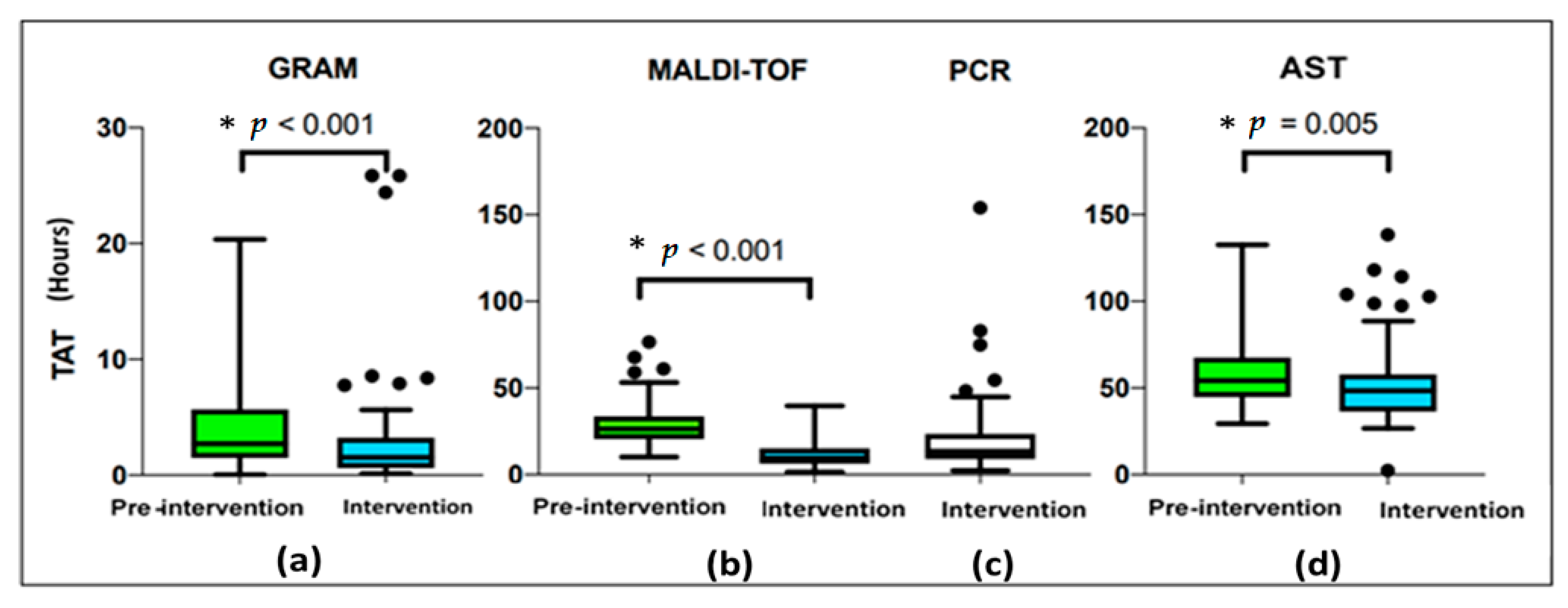
| Turnaround Time (TAT) (Hours, Minutes) | Pre-Intervention N = 114 | Intervention N = 102 | p-Value |
|---|---|---|---|
| Time to positivity (TTP), median, (IQR) | 11 h 53 min (9 h 47 min–17 h 26 min) | 12 h 29 min (10 h 07 min–18 h 18 mim) | 0.302 a |
| Time between TTP and Gram stain, median, (IQR) | 2 h 43 min (1 h 30 min–5 h 40 min) | 01 h 32 min (37 min–3 h 11 min) | <0.001 a |
| Time between TTP and MALDI-TOF, median, (IQR) | 26 h 31 min (20 h 33 min–33 h 42 min) | 9 h 31 min (6 h 28 min–15 h 10 min) | <0.001 a |
| Time between TTP and PCR, median, (IQR) | 8 h 49 min (7 h 57 min–13 h 33 min) | ||
| Time between TTP and AST, median, (IQR) | 54 h 14 min (44 h 49 min–67 h 12 min) | 48 h 28 min (36 h 33 min–57 h 47 min) | 0.005 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campos, A.F.; Arantes, T.; Cambiais, A.M.V.B.; Cury, A.P.; Tiroli, C.G.; Rossi, F.; Malbouisson, L.M.S.; Costa, S.F.; Guimarães, T. Impact of an Antimicrobial Stewardship Program Intervention Associated with the Rapid Identification of Microorganisms by MALDI-TOF and Detection of Resistance Genes in ICU Patients with Gram-Negative Bacteremia. Antibiotics 2022, 11, 1226. https://doi.org/10.3390/antibiotics11091226
Campos AF, Arantes T, Cambiais AMVB, Cury AP, Tiroli CG, Rossi F, Malbouisson LMS, Costa SF, Guimarães T. Impact of an Antimicrobial Stewardship Program Intervention Associated with the Rapid Identification of Microorganisms by MALDI-TOF and Detection of Resistance Genes in ICU Patients with Gram-Negative Bacteremia. Antibiotics. 2022; 11(9):1226. https://doi.org/10.3390/antibiotics11091226
Chicago/Turabian StyleCampos, Aléia Faustina, Tiago Arantes, Amanda Magalhães Vilas Boas Cambiais, Ana Paula Cury, Camila Guimarães Tiroli, Flávia Rossi, Luiz Marcelo Sa Malbouisson, Silvia Figueiredo Costa, and Thaís Guimarães. 2022. "Impact of an Antimicrobial Stewardship Program Intervention Associated with the Rapid Identification of Microorganisms by MALDI-TOF and Detection of Resistance Genes in ICU Patients with Gram-Negative Bacteremia" Antibiotics 11, no. 9: 1226. https://doi.org/10.3390/antibiotics11091226
APA StyleCampos, A. F., Arantes, T., Cambiais, A. M. V. B., Cury, A. P., Tiroli, C. G., Rossi, F., Malbouisson, L. M. S., Costa, S. F., & Guimarães, T. (2022). Impact of an Antimicrobial Stewardship Program Intervention Associated with the Rapid Identification of Microorganisms by MALDI-TOF and Detection of Resistance Genes in ICU Patients with Gram-Negative Bacteremia. Antibiotics, 11(9), 1226. https://doi.org/10.3390/antibiotics11091226







