Molecular Characterization of Extended Spectrum β-Lactamase (ESBL) and Virulence Gene-Factors in Uropathogenic Escherichia coli (UPEC) in Children in Duhok City, Kurdistan Region, Iraq
Abstract
1. Introduction
2. Results
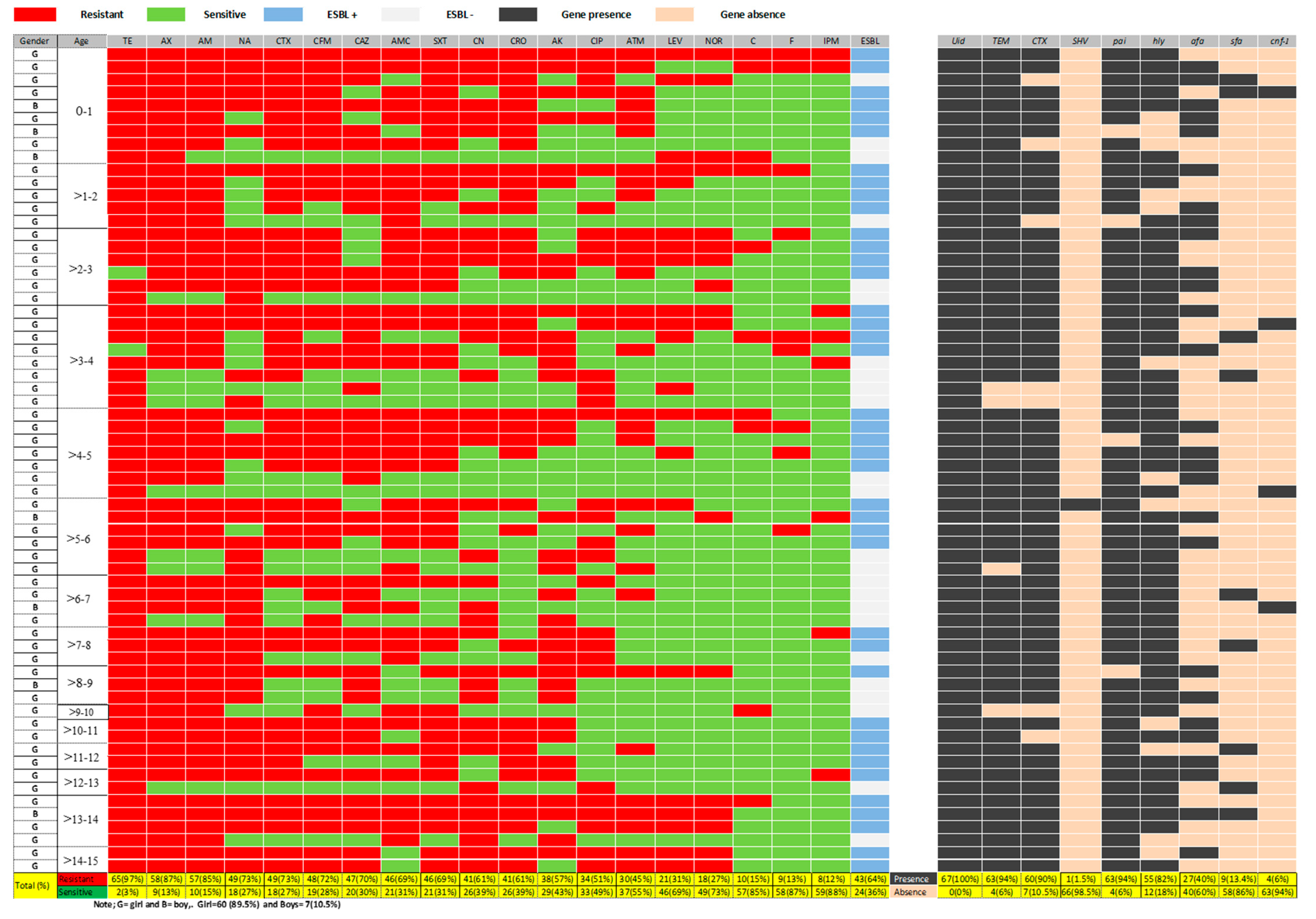
2.1. E. coli Isolation with Phenotypic and Genotypic Detection
2.2. Antimicrobial Susceptibility Pattern
2.3. Phenotypic and Molecular Detection of ESBL Production
2.4. Molecular Detection of Virulence Factors Genes
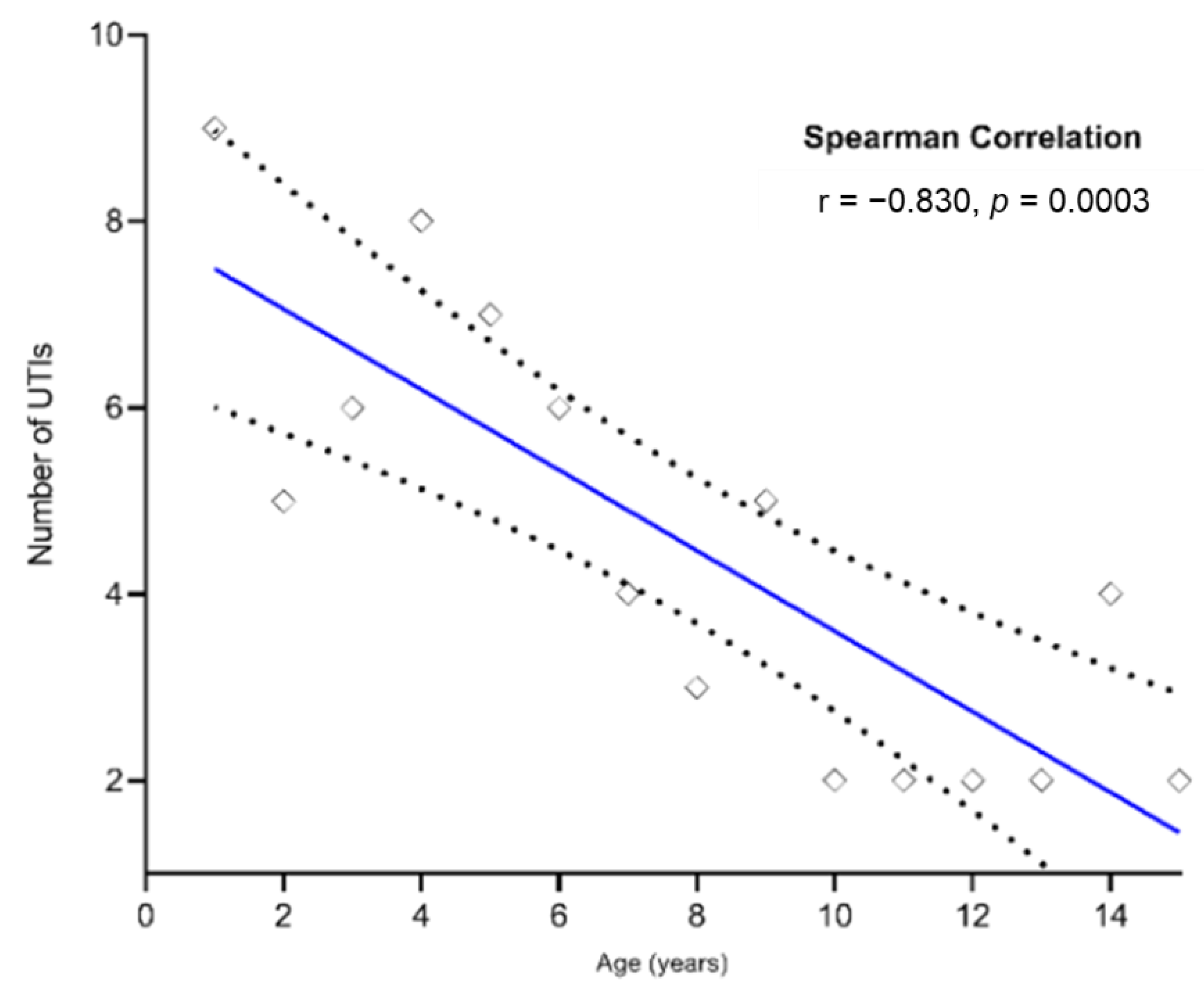
2.5. Relationship between UTIs by E. coli and Children Age
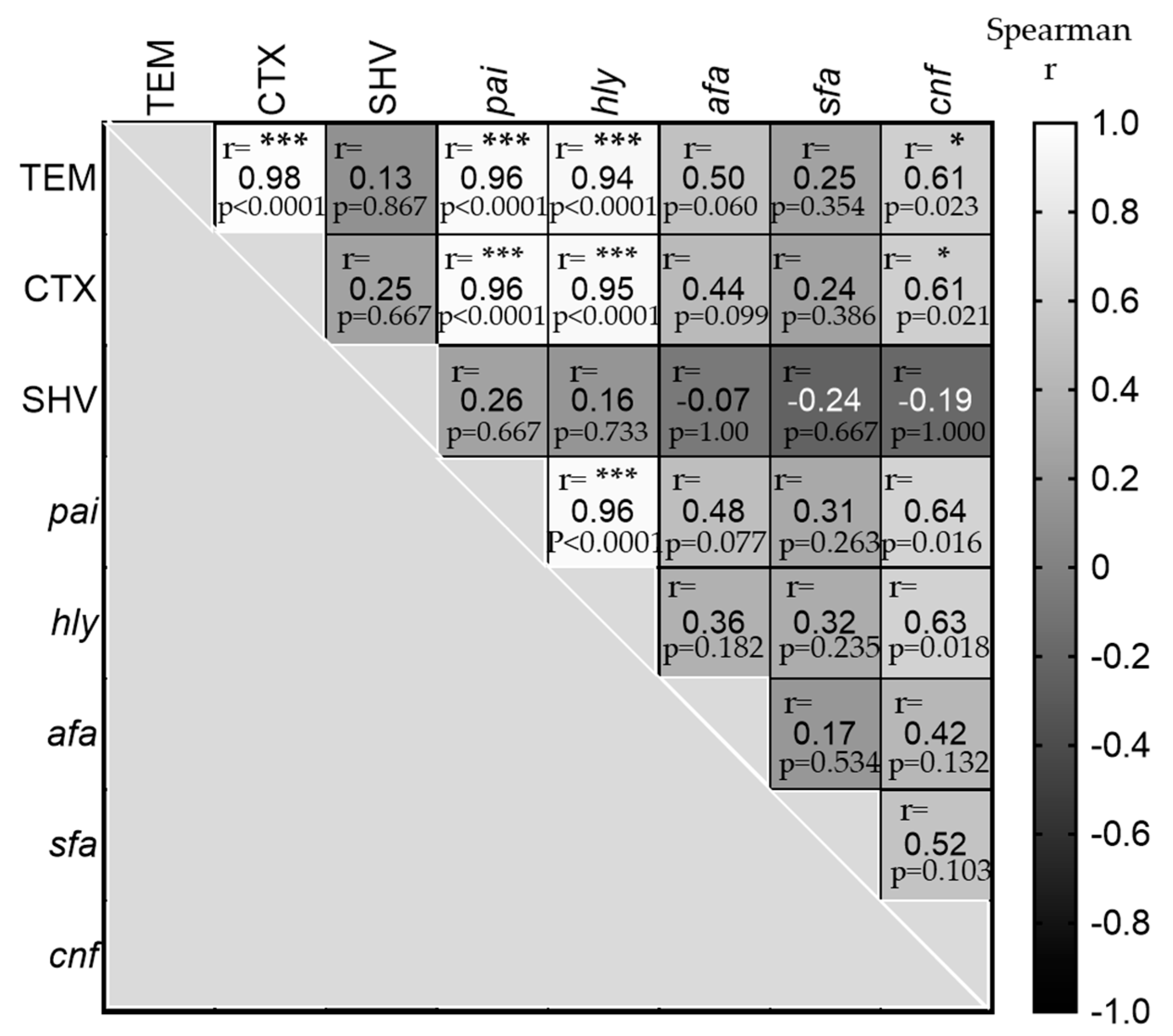
2.6. Relationship between Antibiotic Resistance and Marker Genes of E. coli and Children Ages Having UTIs
2.7. Relationship between Antibiotics Resistance and Virulence Factors Genes
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. E. coli Isolation and Identification
4.3. Antimicrobial Susceptibility Test and ESBLs Detection
4.4. Bacterial DNA Extraction from E. coli
4.5. Statistical Analysis
4.6. Ethical Approval
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Núñez-Samudio, V.; Pecchio, M.; Pimentel-Peralta, G.; Quintero, Y.; Herrera, M.; Landires, I. Molecular Epidemiology of Escherichia coli Clinical Isolates from Central Panama. Antibiotics 2021, 10, 899. [Google Scholar] [CrossRef] [PubMed]
- Kresken, M.; Pfeifer, Y.; Wagenlehner, F.; Werner, G.; Wohlfarth, E.; Study Group ‘Antimicrobial Resistance’ of the Paul Ehrlich Society for Infection Therapy. Resistance to Mecillinam and Nine Other Antibiotics for Oral Use in Escherichia coli Isolated from Urine Specimens of Primary Care Patients in Germany, 2019/20. Antibiotics 2022, 11, 751. [Google Scholar] [CrossRef] [PubMed]
- Sahu, R.; Sahoo, R.K.; Prusty, S.K.; Sahu, P.K. Urinary Tract Infection and Its Management. Syst. Rev. Pharm. 2019, 10, 42–48. [Google Scholar] [CrossRef]
- Stamm, W.E.; Norrby, S.R. Urinary Tract Infections: Disease Panorama and Changes. J. Infect. Dis. 2001, 183, S1–S4. [Google Scholar] [CrossRef]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary Tract Infections: Epidemiology, Mechanisms of Infection and Treatment Options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef]
- Millner, R.; Becknell, B. Urinary Tract Infections. Pediatr. Clin. N. Am. 2019, 66, 1–13. [Google Scholar] [CrossRef]
- Schmidt, B.; Copp, H.L. Work-up of Pediatric Urinary Tract Infection. Urol. Clin. 2015, 42, 519–526. [Google Scholar] [CrossRef]
- Freedman, A.L.; Urologic Diseases in America Project. Urologic Diseases in North America Project: Trends in Resource Utilization for Urinary Tract Infections in Children. J. Urol. 2005, 173, 949–954. [Google Scholar] [CrossRef]
- Nibhanipudi, K.V. A Study to Determine the Incidence of Urinary Tract Infections in Infants and Children Ages 4 Months to 6 Years with Febrile Diarrhea. Glob. Pediatr. Health 2016, 3, 2333794X16667175. [Google Scholar] [CrossRef]
- Tullus, K.; Shaikh, N. Urinary Tract Infections in Children. Lancet 2020, 395, 1659–1668. [Google Scholar] [CrossRef]
- Ismaili, K.; Wissing, K.M.; Lolin, K.; Le, P.Q.; Christophe, C.; Lepage, P.; Hall, M. Characteristics of First Urinary Tract Infection with Fever in Children: A Prospective Clinical and Imaging Study. Pediatr. Infect. Dis. J. 2011, 30, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Montini, G.; Tullus, K.; Hewitt, I. Febrile Urinary Tract Infections in Children. N. Engl. J. Med. 2011, 365, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, N.; Morone, N.E.; Bost, J.E.; Farrell, M.H. Prevalence of Urinary Tract Infection in Childhood: A Meta-Analysis. Pediatr. Infect. Dis. J. 2008, 27, 302–308. [Google Scholar] [CrossRef]
- Johnson, J.R.; Kuskowski, M.A.; O’bryan, T.T.; Colodner, R.; Raz, R. Virulence Genotype and Phylogenetic Origin in Relation to Antibiotic Resistance Profile among Escherichia Coli Urine Sample Isolates from Israeli Women with Acute Uncomplicated Cystitis. Antimicrob. Agents Chemother. 2005, 49, 26–31. [Google Scholar] [CrossRef]
- Wiles, T.J.; Kulesus, R.R.; Mulvey, M.A. Origins and Virulence Mechanisms of Uropathogenic Escherichia coli. Exp. Mol. Pathol. 2008, 85, 11–19. [Google Scholar] [CrossRef]
- Mulvey, M.A.; Schilling, J.D.; Martinez, J.J.; Hultgren, S.J. Bad Bugs and Beleaguered Bladders: Interplay between Uropathogenic Escherichia coli and Innate Host Defenses. Proc. Natl. Acad. Sci. USA 2000, 97, 8829–8835. [Google Scholar] [CrossRef]
- Marrs, C.F.; Zhang, L.; Foxman, B. Escherichia coli Mediated Urinary Tract Infections: Are There Distinct Uropathogenic E. coli (UPEC) Pathotypes? FEMS Microbiol. Lett. 2005, 252, 183–190. [Google Scholar] [CrossRef]
- Bien, J.; Sokolova, O.; Bozko, P. Role of Uropathogenic Escherichia coli Virulence Factors in Development of Urinary Tract Infection and Kidney Damage. Int. J. Nephrol. 2012, 2012, 681473. [Google Scholar] [CrossRef]
- Rupp, M.E.; Fey, P.D. Extended Spectrum β-Lactamase (ESBL)-Producing Enterobacteriaceae. Drugs 2003, 63, 353–365. [Google Scholar] [CrossRef]
- Toussaint, K.A.; Gallagher, J.C. β-Lactam/β-Lactamase Inhibitor Combinations: From Then to Now. Ann. Pharmacother. 2015, 49, 86–98. [Google Scholar] [CrossRef]
- Paterson, D.L. Recommendation for Treatment of Severe Infections Caused by Enterobacteriaceae Producing Extended-Spectrum β-Lactamases (ESBLs). Clin. Microbiol. Infect. 2000, 6, 460–463. [Google Scholar] [CrossRef] [PubMed]
- Paterson, D.L.; Bonomo, R.A. Extended-Spectrum β-Lactamases: A Clinical Update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef] [PubMed]
- Ngoi, S.T.; Teh, C.S.J.; Chong, C.W.; Abdul Jabar, K.; Tan, S.C.; Yu, L.H.; Leong, K.C.; Tee, L.H.; AbuBakar, S. In Vitro Efficacy of Flomoxef against Extended-Spectrum Beta-Lactamase-Producing Escherichia coli and Klebsiella Pneumoniae Associated with Urinary Tract Infections in Malaysia. Antibiotics 2021, 10, 181. [Google Scholar] [CrossRef]
- World Health Organization. Global Priority Pathogens List of Antibiotic-Resistant Bacteria; WHO: Geneva, Switzerland, 2021; Volume 500. Available online: https://www.doherty.edu.au/news-events/news/who-global-priority-pathogens-list-of-antibiotic-501resistant-bacteria (accessed on 30 June 2022).
- Raphael, E.; Glymour, M.M.; Chambers, H.F. Trends in Prevalence of Extended-Spectrum Beta-Lactamase-Producing Escherichia coli Isolated from Patients with Community-and Healthcare-Associated Bacteriuria: Results from 2014 to 2020 in an Urban Safety-Net Healthcare System. Antimicrob. Resist. Infect. Control 2021, 10, 118. [Google Scholar] [CrossRef] [PubMed]
- Yasir, M.; Ajlan, A.M.; Shakil, S.; Jiman-Fatani, A.A.; Almasaudi, S.B.; Farman, M.; Baazeem, Z.M.; Baabdullah, R.; Alawi, M.; Al-Abdullah, N. Molecular Characterization, Antimicrobial Resistance and Clinico-Bioinformatics Approaches to Address the Problem of Extended-Spectrum β-Lactamase-Producing Escherichia coli in Western Saudi Arabia. Sci. Rep. 2018, 8, 14847. [Google Scholar] [CrossRef]
- Awean, G.Z.A.; Salameh, K.; Elmohamed, H.; Alshmayt, H.; Omer, M.R.B. Prevalence of ESBL Urinary Tract Infection in Children. J Adv. Pediatr. Child Health 2019, 2, 4–7. [Google Scholar] [CrossRef]
- Rezai, M.S.; Salehifar, E.; Rafiei, A.; Langaee, T.; Rafati, M.; Shafahi, K.; Eslami, G. Characterization of Multidrug Resistant Extended-Spectrum Beta-Lactamase-Producing Escherichia coli among Uropathogens of Pediatrics in North of Iran. Biomed Res. Int. 2015, 2015, 309478. [Google Scholar] [CrossRef]
- Kizilca, O.; Siraneci, R.; Yilmaz, A.; Hatipoglu, N.; Ozturk, E.; Kiyak, A.; Ozkok, D. Risk Factors for Community-acquired Urinary Tract Infection Caused by ESBL-producing Bacteria in Children. Pediatr. Int. 2012, 54, 858–862. [Google Scholar] [CrossRef]
- Rodríguez-Bano, J.; Navarro, M.D.; Romero, L.; Martínez-Martínez, L.; Muniain, M.A.; Perea, E.J.; Pérez-Cano, R.; Pascual, A. Epidemiology and Clinical Features of Infections Caused by Extended-Spectrum Beta-Lactamase-Producing Escherichia coli in Nonhospitalized Patients. J. Clin. Microbiol. 2004, 42, 1089–1094. [Google Scholar] [CrossRef]
- Kaarme, J.; Riedel, H.; Schaal, W.; Yin, H.; Nevéus, T.; Melhus, Å. Rapid Increase in Carriage Rates of Enterobacteriaceae Producing Extended-Spectrum β-Lactamases in Healthy Preschool Children, Sweden. Emerg. Infect. Dis. 2018, 24, 1874. [Google Scholar] [CrossRef]
- Ibrahim, M.; Khalid, H.M.; Mer, W.M.S. The Prevalence of Uropathogenic Escherichia coli Strains among Outpatients with Urinary Tract Infection in Zakho Hospitals-Zakho City, Duhok Province/Iraq. Al-Qadisiyah J. Pure Sci. 2021, 26, 26–40. [Google Scholar] [CrossRef]
- Merza, N.S.; Jubrael, J.M.S. The Prevalence of Virulence Factors among Uropathogenic Escherichia coli Strains Isolated from Different Hospitals in Kurdistan Region-Iraq. Int. J. Bioinforma Biomed. Eng. 2015, 1, 338–343. [Google Scholar]
- Naqid, I.A.; Balatay, A.A.; Hussein, N.R.; Ahmed, H.A.; Saeed, K.A.; Abdi, S.A. Bacterial Strains and Antimicrobial Susceptibility Patterns in Male Urinary Tract Infections in Duhok Province, Iraq. Middle East J. Rehabil. Health Stud. 2020, 7, e103529. [Google Scholar] [CrossRef]
- Ibrahim, S.A.; Mohamed, D.A.; Suleman, S.K. Microbial Causes of Urinary Tract Infection and Its Sensitivity to Antibiotics at Heevi Pediatric Teaching Hospital/Duhok City. Med. J. Babylon 2020, 17, 109. [Google Scholar]
- Saeed, C.H.; AL-Otraqchi, K.I.B.; Mansoor, I.Y. Prevalence of Urinary Tract Infections and Antibiotics Susceptibility Pattern among Infants and Young Children in Erbil City. Zanco J. Med. Sci. 2015, 19, 915–922. [Google Scholar] [CrossRef]
- Alkhateeb, N.E.R. Antibiotic Resistance of Urinary Tract Pathogens and Rationale for Empirical Antibiotic Therapy in Children with Urinary Tract Infection. Zanco J. Med. Sci. 2016, 20, 1458_1466. [Google Scholar]
- Hussein, N.H. Prevalence and Antimicrobial Susceptibility Patterns of Bacteria Isolated from Urinary Tract Infections (UTIs) in Children at Children Hospital in Baghdad. Al-Kindy Coll. Med. J. 2017, 13, 102–107. [Google Scholar] [CrossRef]
- Chen, H.E.; Tain, Y.L.; Kuo, H.C.; Hsu, C.N. Trends in Antimicrobial Susceptibility of Escherichia coli Isolates in a Taiwanese Child Cohort with Urinary Tract Infections between 2004 and 2018. Antibiotics 2020, 9, 501. [Google Scholar] [CrossRef]
- Foxman, B. Urinary Tract Infection Syndromes: Occurrence, Recurrence, Bacteriology, Risk Factors, and Disease Burden. Infect. Dis. Clin. 2014, 28, 1–13. [Google Scholar] [CrossRef]
- Moreno, E.; Andreu, A.; Perez, T.; Sabaté, M.; Johnson, J.R.; Prats, G. Relationship between Escherichia coli Strains Causing Urinary Tract Infection in Women and the Dominant Faecal Flora of the Same Hosts. Epidemiol. Infect. 2006, 134, 1015–1023. [Google Scholar] [CrossRef]
- Morand, A.; Cornu, F.; Dufour, J.-C.; Tsimaratos, M.; Lagier, J.-C.; Raoult, D. Human Bacterial Repertoire of the Urinary Tract: A Potential Paradigm Shift. J. Clin. Microbiol. 2019, 57, e00675-18. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.K.C.; Wong, A.H.C.; Leung, A.A.M.; Hon, K.L. Urinary Tract Infection in Children. Recent Pat. Inflamm. Allergy Drug Discov. 2019, 13, 2–18. [Google Scholar] [CrossRef] [PubMed]
- Fahimzad, A.; Taherian, M.; Dalirani, R.; Shamshiri, A. Diaper Type as a Risk Factor in Urinary Tract Infection of Children. Iran. J. Pediatr. 2010, 20, 97–100. [Google Scholar] [PubMed]
- Banieghbal, B. Optimal Time for Neonatal Circumcision: An Observation-Based Study. J. Pediatr. Urol. 2009, 5, 359–362. [Google Scholar] [CrossRef] [PubMed]
- To, T.; Agha, M.; Dick, P.T.; Feldman, W. Cohort Study on Circumcision of Newborn Boys and Subsequent Risk of Urinary-Tract Infection. Lancet 1998, 352, 1813–1816. [Google Scholar] [CrossRef]
- Singh-Grewal, D.; Macdessi, J.; Craig, J. Circumcision for the Prevention of Urinary Tract Infection in Boys: A Systematic Review of Randomised Trials and Observational Studies. Arch. Dis. Child. 2005, 90, 853–858. [Google Scholar] [CrossRef]
- Ibrahim, M.S.; Khalid, H.M.; Mero, W.M. Molecular Characterization of Some Virulence Genes and Antibiotics Resistance of Uropathogenic Escherichia coli Isolated from Patients in Zakho City, Kurdistan Region/Iraq. Zanco J. of Pure and Appl. Sci. 2020, 32, 167–177. [Google Scholar] [CrossRef]
- Al- Barzinji, R.; Esmahil, S.; Sulaiman, S.; Raheem, S. Effect of Some Antimicrobial Agents on Isolated Bacteria from Patients with Urinary Tract Infection in Kurdistan Region. Zanco J. Med. Sci. 2010, 14, 61–67. [Google Scholar] [CrossRef]
- Murray, P.R.; Rosenthal, K.S.; Pfaller, M.A. Antimicrobial Agents; Elsevier: Amsterdam, The Netherlands, 2016; pp. 183–190. [Google Scholar]
- Nicolle, L.E. Complicated Urinary Tract Infection in Adults. Can. J. Infect. Dis. Med. Microbiol. 2005, 16, 349–360. [Google Scholar] [CrossRef]
- Demir, M.; Kazanasmaz, H. Uropathogens and Antibiotic Resistance in the Community and Hospital-Induced Urinary Tract Infected Children. J. Glob. Antimicrob. Resist. 2020, 20, 68–73. [Google Scholar] [CrossRef]
- Kot, B.; Grużewska, A.; Szweda, P.; Wicha, J.; Parulska, U. Antibiotic Resistance of Uropathogens Isolated from Patients Hospitalized in District Hospital in Central Poland in 2020. Antibiotics 2021, 10, 447. [Google Scholar] [CrossRef] [PubMed]
- Paterson, D.L. Resistance in Gram-Negative Bacteria: Enterobacteriaceae. Am. J. Infect. Control 2006, 34, S20–S28. [Google Scholar] [CrossRef] [PubMed]
- Sharif, M.; Nori, S. The Frequency and Antibiotic Resistance of Urinary Tract Infection Organisms in Hospitalized Children Shahid Beheshti Hospital, Kashan 2012–2013. Iran. J. Infect. Dis. Trop. Med. 2014, 65, 47–51. [Google Scholar]
- Assafi, M.S.A.; Ibrahim, N.M.R.; Hussein, N.R.; Taha, A.A.; Balatay, A.A. Urinary Bacterial Profile and Antibiotic Susceptibility Pattern among Patients with Urinary Tract Infection in Duhok City, Kurdistan Region, Iraq. Int. J. Pure Appl. Sci. Technol. 2015, 30, 54. [Google Scholar]
- Aka, S.T.; Haji, S.H. Evaluation of Multi Drug Resistance among Extended Spectrum β-Lactamase-Producing Escherichia coli Causing Urinary Tract Infection in Erbil City. Zanco J. Med. Sci. 2015, 19, 998–1004. [Google Scholar] [CrossRef][Green Version]
- Alqasim, A.; Abu Jaffal, A.; Alyousef, A.A. Prevalence of Multidrug Resistance and Extended-Spectrum β-Lactamase Carriage of Clinical Uropathogenic Escherichia coli Isolates in Riyadh, Saudi Arabia. Int. J. Microbiol. 2018, 2018, 3026851. [Google Scholar] [CrossRef]
- Agegnehu, A.; Worku, M.; Nigussie, D.; Lulu, B.; Tadesse, B.T. Pediatric Febrile Urinary Tract Infection Caused by ESBL Producing Enterobacteriaceae Species. Biomed Res. Int. 2020, 2020, 6679029. [Google Scholar] [CrossRef]
- Mortazavi-Tabatabaei, S.A.R.; Ghaderkhani, J.; Nazari, A.; Sayehmiri, K.; Sayehmiri, F.; Pakzad, I. Pattern of Antibacterial Resistance in Urinary Tract Infections: A Systematic Review and Meta-Analysis. Int. J. Prev. Med. 2019, 10, 169. [Google Scholar]
- Calbo, E.; Romaní, V.; Xercavins, M.; Gómez, L.; Vidal, C.G.; Quintana, S.; Vila, J.; Garau, J. Risk Factors for Community-Onset Urinary Tract Infections Due to Escherichia coli Harbouring Extended-Spectrum β-Lactamases. J. Antimicrob. Chemother. 2006, 57, 780–783. [Google Scholar] [CrossRef]
- Rawat, D.; Nair, D. Extended-Spectrum β-Lactamases in Gram Negative Bacteria. J. Glob. Infect. Dis. 2010, 2, 263. [Google Scholar] [CrossRef]
- Cleuziat, P.; Robert-Baudouy, J. Specific Detection of Escherichia coli and Shigella Species Using Fragments of Genes Coding for β-Glucuronidase. FEMS Microbiol. Lett. 1990, 72, 315–322. [Google Scholar] [CrossRef]
- Feng, P.; Lum, R.; Chang, G.W. Identification of UidA Gene Sequences in Beta-D-Glucuronidase-Negative Escherichia coli. Appl. Environ. Microbiol. 1991, 57, 320–323. [Google Scholar] [CrossRef]
- Farnleitner, A.H.; Kreuzinger, N.; Kavka, G.G.; Grillenberger, S.; Rath, J.; Mach, R.L. Simultaneous Detection and Differentiation of Escherichia coli Populations from Environmental Freshwaters by Means of Sequence Variations in a Fragment of the β-D-Glucuronidase Gene. Appl. Environ. Microbiol. 2000, 66, 1340–1346. [Google Scholar] [CrossRef] [PubMed]
- Brons, J.K.; Vink, S.N.; de Vos, M.G.J.; Reuter, S.; Dobrindt, U.; van Elsas, J.D. Fast Identification of Escherichia coli in Urinary Tract Infections Using a Virulence Gene Based PCR Approach in a Novel Thermal Cycler. J. Microbiol. Methods 2020, 169, 105799. [Google Scholar] [CrossRef] [PubMed]
- Karaolis, D.K.R. Pathogenicity Islands. In Encyclopedia of Genetics; Academic Press (Elsevier): Amsterdam, The Netherlands, 2001; pp. 1422–1424. [Google Scholar]
- Tarchouna, M.; Ferjani, A.; Ben-Selma, W.; Boukadida, J. Distribution of Uropathogenic Virulence Genes in Escherichia coli Isolated from Patients with Urinary Tract Infection. Int. J. Infect. Dis. 2013, 17, 10–13. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, M.; Brooks, B.D.; Brooks, A.E. The Complex Relationship between Virulence and Antibiotic Resistance. Genes 2017, 8, 39. [Google Scholar] [CrossRef]
- Giaouris, E.; Heir, E.; Desvaux, M.; Hébraud, M.; Møretrø, T.; Langsrud, S.; Doulgeraki, A.; Nychas, G.-J.; Kačániová, M.; Czaczyk, K. Intra-and Inter-Species Interactions within Biofilms of Important Foodborne Bacterial Pathogens. Front. Microbiol. 2015, 6, 841. [Google Scholar] [CrossRef]
- Srivastava, N.; Srivastava, M.; Mishra, P.K.; Ramteke, P.W.; Singh, R.L. New and Future Developments in Microbial Biotechnology and Bioengineering: From Cellulose to Cellulase: Strategies to Improve Biofuel Production; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Abd El-Baky, R.M.; Ibrahim, R.A.; Mohamed, D.S.; Ahmed, E.F.; Hashem, Z.S. Prevalence of Virulence Genes and Their Association with Antimicrobial Resistance among Pathogenic E. coli Isolated from Egyptian Patients with Different Clinical Infections. Infect. Drug Resist. 2020, 13, 1221. [Google Scholar] [CrossRef]
- Pärnänen, K.; Karkman, A.; Hultman, J.; Lyra, C.; Bengtsson-Palme, J.; Larsson, D.G.; Rautava, S.; Isolauri, E.; Salminen, S.; Kumar, H. Maternal Gut and Breast Milk Microbiota Affect Infant Gut Antibiotic Resistome and Mobile Genetic Elements. Nat. Commun. 2018, 9, 3891. [Google Scholar] [CrossRef]
- Zwane, T.; Shuping, L.; Perovic, O. Etiology and Antimicrobial Susceptibility of Pathogens Associated with Urinary Tract Infections among Women Attending Antenatal Care in Four South African Tertiary-Level Facilities, 2015–2019. Antibiotics 2021, 10, 669. [Google Scholar] [CrossRef]
- Roberts, K.B.; Downs, S.M.; Finnell, S.M.E.; Hellerstein, S.; Shortliffe, L.D.; Wald, E.R.; Zerin, J.M. Urinary Tract Infection: Clinical Practice Guideline for the Diagnosis and Management of the Initial UTI in Febrile Infants and Children 2 to 24 Months. Pediatrics 2011, 128, 595–610. [Google Scholar] [CrossRef] [PubMed]
- Larcombe, J. Urinary tract infection in children. Am. Fam. Physician 2010, 82, 1252–1256. [Google Scholar] [PubMed]
- Leboffe, M.J.; Pierce, B.E. A Photographic Atlas for the Microbiology Laboratory, 4th ed.; Morton Pub. Co: Englewood, CO, USA, 2011. [Google Scholar]
- Hudzicki, J. Kirby-Bauer Disk Diffusion Susceptibility Test Protocol. Am. Soc. Microbiol. 2009, 15, 55–63. [Google Scholar]
- CLSI (Clinical and Laboratory Standards Institute). Performanc Standards for Antimicrobial 409 Susceptibility testing; Seventeeth Informational Supplement. M100-17; Clinical and Laboratory Standards 410 Institute: Wayne, PA, USA, 2007. [Google Scholar]
- Jarlier, V.; Nicolas, M.-H.; Fournier, G.; Philippon, A. Extended Broad-Spectrum β-Lactamases Conferring Transferable Resistance to Newer β-Lactam Agents in Enterobacteriaceae: Hospital Prevalence and Susceptibility Patterns. Clin. Infect. Dis. 1988, 10, 867–878. [Google Scholar] [CrossRef]
- Adamus-Bialek, W.; Wojtasik, A.; Majchrzak, M.; Sosnowski, M.; Parniewski, P. (CGG) 4-Based PCR as a Novel Tool for Discrimination of Uropathogenic Escherichia coli Strains: Comparison with Enterobacterial Repetitive Intergenic Consensus-PCR. J. Clin. Microbiol. 2009, 47, 3937–3944. [Google Scholar] [CrossRef]
- Oliveira, F.A.; Paludo, K.S.; Arend, L.; Farah, S.; Pedrosa, F.O.; Souza, E.M.; Surek, M.; Picheth, G.; Fadel-Picheth, C.M. Virulence Characteristics and Antimicrobial Susceptibility of Uropathogenic Escherichia coli Strains. Genet. Mol. Res. 2011, 10, 4114–4125. [Google Scholar] [CrossRef]
- Le Bouguenec, C.; Archambaud, M.; Labigne, A. Rapid and Specific Detection of the Pap, Afa, and Sfa Adhesin-Encoding Operons in Uropathogenic Escherichia coli Strains by Polymerase Chain Reaction. J. Clin. Microbiol. 1992, 30, 1189–1193. [Google Scholar] [CrossRef]
- Mladin, C.; Usein, C.-R.; Chifiriuc, M.-C.; Palade, A.; Slavu, C.L.; Negut, M.; Damian, M. Genetic Analysis of Virulence and Pathogenicity Features of Uropathogenic Escherichia coli Isolated from Patients with Neurogenic Bladder. Rom. Biotechnol. Lett. 2009, 14, 4906–4911. [Google Scholar]
- Chapman, T.A.; Wu, X.-Y.; Barchia, I.; Bettelheim, K.A.; Driesen, S.; Trott, D.; Wilson, M.; Chin, J.J.-C. Comparison of Virulence Gene Profiles of Escherichia coli Strains Isolated from Healthy and Diarrheic Swine. Appl. Environ. Microbiol. 2006, 72, 4782–4795. [Google Scholar] [CrossRef]
- Pandit, R.; Awal, B.; Shrestha, S.S.; Joshi, G.; Rijal, B.P.; Parajuli, N.P. Extended-Spectrum β-Lactamase (ESBL) Genotypes among Multidrug-Resistant Uropathogenic Escherichia coli Clinical Isolates from a Teaching Hospital of Nepal. Interdiscip. Perspect. Infect. Dis. 2020, 2020, 6525826. [Google Scholar] [CrossRef]


| ESBL-Producing E. coli Genes | No of Strains | Virulence Genes | No. of Strains |
|---|---|---|---|
| TEM, CTX-M, SHV | 1 | pai, hly, afa, sfa | 2 |
| TEM, CTX-M | 58 | pai, hly, afa, | 18 |
| TEM | 4 | pai, hly, sfa, cnf-1 | 1 |
| CTX-M | 1 | pai, hly | 21 |
| pai, afa | 5 | ||
| hly, afa | 2 | ||
| pai, hly, sfa | 5 | ||
| pai, hly, cnf-1 | 3 | ||
| pai, sfa | 1 | ||
| pai | 5 | ||
| hly | 1 | ||
| afa | 1 |
| Types | Genes | Oligonucleotide Sequence (5′–3′) Forward and Reverse | Amplicon Size (bp) | References |
|---|---|---|---|---|
| Species-specific gene | uidA | F:5′-CATTACGGCAAAGTGTGGGTCAAT-3′ | 658 bp | [81] |
| R:5′-CCATCAGCACGTTATCGAATCCTT-3′ | ||||
| Virulence genes | pai | F: 5′-GGACATCCTGGTACAGCGCGCA-3′ | 930 bp | [82] |
| R: 5′-TCGCCACCAATCACAGCCGAAC-3′ | ||||
| afa | F: 5′-GCTGGGCAGCAAACTGATAACTCTC-3′ | 750 bp | [83] | |
| R: 5′-CATCAAGCTGTTTGTTCGTCCGCCG-3′ | ||||
| hlyA | F:5′-AGATTCTTGGGCATGTATCCT-3′ | 565 bp | [84] | |
| R:5′-TTGCTTTGCAGACTGTAGTGT-3′ | ||||
| cnf-1 | F: 5′-AAGATGGAGTTTCCTATGCAGGAG-3′ | 498 bp | [81,85] | |
| R: 5′-CATTCAGAGTCCTGCCCTCATTATT-3′ | ||||
| sfa | F: 5′-GTGGATACGACGATTACTGTG- 3′ | 240 bp | [85] | |
| R: 5′-CCGCCAGCATTCCCTGTATTC-3′ | ||||
| ESBL genes | CTX-M | F: 5′-GAAGGTCATCAAGAAGGTGCG-3′ | 560 bp | [86] |
| R: 5′-GCATTGCCACGCTTTTCATAG-3′ | ||||
| TEM | F:5′-GAGACAATAACCCTGGTAAAT-3′ | 459 bp | ||
| R-5′-AGAAGTAAGTTGGCAGCAGTG-3′ | ||||
| SHV | F: 5′-GTCAGCGAAAAACACCTTGCC3′ | 383 bp | ||
| R: 5′-GTCTTATCGGCGATAAACCAG3′ |
| Genes | Initial Denaturation | Denaturation | Annealing | Extension | Final Extension | References |
|---|---|---|---|---|---|---|
| uidA | 94 °C | 92 °C | 58 °C | 72 °C | 72 °C | [81] |
| 10 min | 1 min | I min | 30 s | 5 min | ||
| 1 cycle | 35 cycles | 1 cycle | ||||
| afa | 94 °C | 94 °C | 63 °C | 68 °C | 72 °C | [83] |
| 5 min | 1 min | 1 min | 3 min | 7 min | ||
| 1 cycle | 30 cycles | 1 cycle | ||||
| sfa | 95 °C | 94 °C | 63 °C | 68 °C | 72 °C | [85] |
| 3 min | 30 s | 30 s | 4 min | 10 min | ||
| 1 cycle | 30 cycles | 1 cycle | ||||
| hly | 94 °C | 94 °C | 55 °C | 72 °C | 72 °C | [84] |
| 4 min | 30 s | 30 s | 1 min | 5 min | ||
| 1 cycle | 30 cycles | 1 cycle | ||||
| cnf-1 | 95 °C | 94 °C | 68 °C | 68 °C | 72 °C | [81,85] |
| 3 min | 30 sec | 30 s | 4 min | 10 min | ||
| 1 cycle | 25 cycles | 1 cycle | ||||
| pai | 94 °C | 94 °C | 63 °C | 72 °C | 72 °C | [82,85] |
| 1 min | 1 min | 30 s | 1.30 min | 5 min | ||
| 1 cycle | 30 cycles | 1 cycle | ||||
| TEM | 94 °C | 94 °C | 55 °C | 72 °C | 72 °C | [86] |
| 3 min | 45 s | 30 sec | 3 min | 2 min | ||
| 1 cycle | 35 cycles | 1 cycle | ||||
| SHV and CTX | 94 °C | 94 °C | 60 °C | 72 °C | 72 °C | [86] |
| 3 min | 45 s | 30 s | 3 min | 2 min | ||
| 1 cycle | 35 cycles | 1 cycle | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hasan, S.M.; Ibrahim, K.S. Molecular Characterization of Extended Spectrum β-Lactamase (ESBL) and Virulence Gene-Factors in Uropathogenic Escherichia coli (UPEC) in Children in Duhok City, Kurdistan Region, Iraq. Antibiotics 2022, 11, 1246. https://doi.org/10.3390/antibiotics11091246
Hasan SM, Ibrahim KS. Molecular Characterization of Extended Spectrum β-Lactamase (ESBL) and Virulence Gene-Factors in Uropathogenic Escherichia coli (UPEC) in Children in Duhok City, Kurdistan Region, Iraq. Antibiotics. 2022; 11(9):1246. https://doi.org/10.3390/antibiotics11091246
Chicago/Turabian StyleHasan, Salwa Muhsin, and Khalid S. Ibrahim. 2022. "Molecular Characterization of Extended Spectrum β-Lactamase (ESBL) and Virulence Gene-Factors in Uropathogenic Escherichia coli (UPEC) in Children in Duhok City, Kurdistan Region, Iraq" Antibiotics 11, no. 9: 1246. https://doi.org/10.3390/antibiotics11091246
APA StyleHasan, S. M., & Ibrahim, K. S. (2022). Molecular Characterization of Extended Spectrum β-Lactamase (ESBL) and Virulence Gene-Factors in Uropathogenic Escherichia coli (UPEC) in Children in Duhok City, Kurdistan Region, Iraq. Antibiotics, 11(9), 1246. https://doi.org/10.3390/antibiotics11091246







