Genomic Insights into Methicillin-Resistant Staphylococci and Mammaliicocci from Bulk Tank Milk of Dairy Farms in Serbia
Abstract
:1. Introduction
2. Results
2.1. The Occurrence of MRS/MRM in BTM Samples
2.2. Species Identification of MRS and MRM
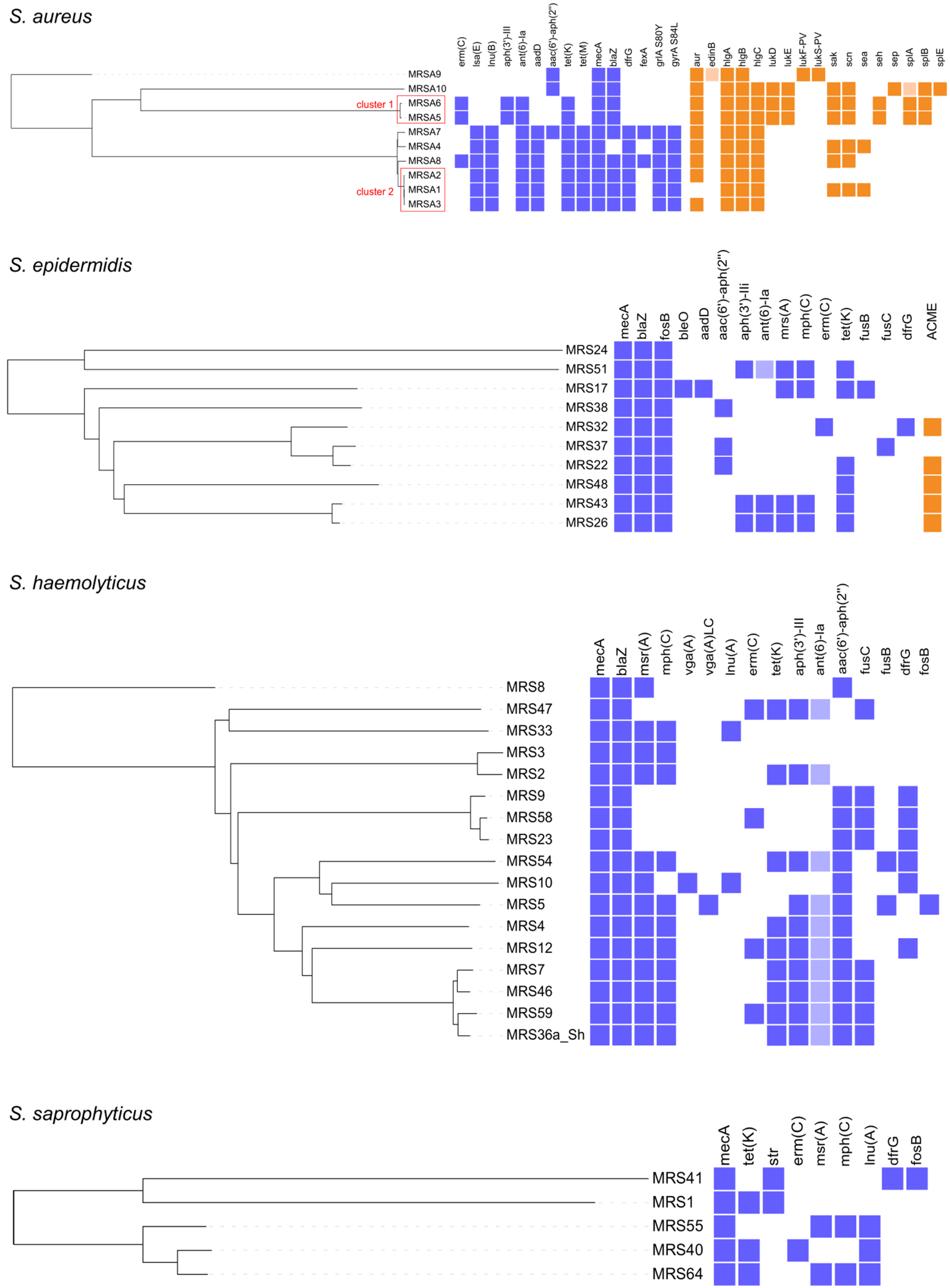
2.3. WGS Characterization of MRS and MRM
2.4. Risk Factors for the Occurrence of MRS/MRM in Dairy Farms
3. Discussion
4. Materials and Methods
4.1. Study Design and Sample Collection
4.2. Isolation and Confirmation of Presumptive MRS and MRM
4.3. Species Identification of MRS and MRM Using VITEK 2 and MALDI-TOF VITEK MS Systems
4.4. WGS and Bioinformatics Analyses
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Howden, B.P.; Giulieri, S.G.; Wong Fok Lung, T.; Baines, S.L.; Sharkey, L.K.; Lee, J.Y.H.; Hachani, A.; Monk, I.R.; Stinear, T.P. Staphylococcus aureus host interactions and adaptation. Nat. Rev. Microbiol. 2023, 21, 380–395. [Google Scholar] [CrossRef]
- Campos, B.; Pickering, A.C.; Souza Rocha, L.; Pereira Aguilar, A.; Fabres-Klein, M.H.; de Oliveira Mendes, T.A.; Ross Fitzgerald, J.; de Oliveira Barros Ribon, A. Diversity and pathogenesis of Staphylococcus aureus from bovine mastitis: Current understanding and future perspectives. BMC Vet. Res. 2022, 18, 115. [Google Scholar] [CrossRef]
- Fetsch, A.; Johler, S. Staphylococcus aureus as a foodborne pathogen. Curr. Clin. Microbiol. Rep. 2018, 5, 88–96. [Google Scholar] [CrossRef]
- De Buck, J.; Ha, V.; Naushad, S.; Nobrega, D.B.; Luby, C.; Middleton, J.R.; De Vliegher, S.; Barkema, H.W. Non-aureus staphylococci and bovine udder health: Current understanding and knowledge gaps. Front. Vet. Sci. 2021, 8, 658031. [Google Scholar] [CrossRef]
- Schnitt, A.; Tenhagen, B.A. Risk factors for the occurrence of methicillin-resistant Staphylococcus aureus in dairy herds: An update. Foodborne Pathog. Dis. 2020, 17, 585–596. [Google Scholar] [CrossRef]
- Khanal, S.; Boonyayatra, S.; Awaiwanont, N. Prevalence of methicillin-resistant Staphylococcus aureus in dairy farms: A systematic review and meta-analysis. Front. Vet. Sci. 2022, 9, 947154. [Google Scholar] [CrossRef]
- Zaatout, N.; Hezil, D. A meta-analysis of the global prevalence of methicillin-resistant Staphylococcus aureus (MRSA) isolated from clinical and subclinical bovine mastitis. J. Appl. Microbiol. 2022, 132, 140–154. [Google Scholar] [CrossRef]
- Vanderhaeghen, W.; Vandendriessche, S.; Crombé, F.; Nemeghaire, S.; Dispas, M.; Denis, O.; Hermans, K.; Haesebrouck, F.; Butaye, P. Characterization of methicillin-resistant non-Staphylococcus aureus staphylococci carriage isolates from different bovine populations. J. Antimicrob. Chemother. 2013, 68, 300–307. [Google Scholar] [CrossRef]
- Argudin, M.A.; Vanderhaeghen, W.; Butaye, P. Diversity of antimicrobial resistance and virulence genes in methicillin-resistant non-Staphylococcus aureus staphylococci from veal calves. Res. Vet. Sci. 2015, 99, 10–16. [Google Scholar] [CrossRef]
- Schnitt, A.; Lienen, T.; Wichmann-Schauer, H.; Tenhagen, B.A. The occurrence of methicillin-resistant non-aureus staphylococci in samples from cows, young stock, and the environment on German dairy farms. J. Dairy Sci. 2021, 104, 4604–4614. [Google Scholar] [CrossRef]
- Saraiva, M.M.S.; de Leon, C.; Silva, N.; Raso, T.F.; Serafini, P.P.; Givisiez, P.E.N.; Gebreyes, W.A.; Oliveira, C.J.B. Staphylococcus sciuri as a reservoir of mecA to Staphylococcus aureus in non-migratory seabirds from a remote oceanic island. Microb. Drug Resist. 2021, 27, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Lienen, T.; Schnitt, A.; Hammerl, J.A.; Maurischat, S.; Tenhagen, B.-A. Mammaliicoccus spp. from German dairy farms exhibit a wide range of antimicrobial resistance genes and non-wildtype phenotypes to several antibiotic classes. Biology 2022, 11, 152. [Google Scholar] [CrossRef] [PubMed]
- Madhaiyan, M.; Wirth, J.S.; Saravanan, V.S. Phylogenomic analyses of the Staphylococcaceae family suggest the reclassification of five species within the genus Staphylococcus as heterotypic synonyms, the promotion of five subspecies to novel species, the taxonomic reassignment of five Staphylococcus species to Mammaliicoccus gen. nov., and the formal assignment of Nosocomiicoccus to the family Staphylococcaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 5926–5936. [Google Scholar] [CrossRef] [PubMed]
- Nemeghaire, S.; Argudín, M.A.; Feßler, A.T.; Hauschild, T.; Schwarz, S.; Butaye, P. The ecological importance of the Staphylococcus sciuri species group as a reservoir for resistance and virulence genes. Vet. Microbiol. 2014, 171, 342–356. [Google Scholar] [CrossRef]
- Harrison, E.M.; Paterson, G.K.; Holden, M.T.G.; Ba, X.; Rolo, J.; Morgan, F.J.E.; Pichon, B.; Kearns, A.; Zadoks, R.N.; Peacock, S.J.; et al. A novel hybrid SCCmec-mecC region in Staphylococcus sciuri. J. Antimicrob. Chemother. 2013, 69, 911–918. [Google Scholar] [CrossRef]
- Paterson, G.K. Genomic epidemiology of methicillin-resistant Staphylococcus sciuri carrying a SCCmec-mecC hybrid element. Infect. Genet. Evol. 2020, 79, 104148. [Google Scholar] [CrossRef]
- SORS (Statistical Office of the Republic of Serbia). Statistical Yearbook of the Republic of Serbia: Agriculture; Publication of SORS: Belgrade, Serbia, 2022; pp. 211–250. Available online: https://publikacije.stat.gov.rs/G2022/PdfE/G20222055.pdf (accessed on 27 February 2023).
- OGSR (Official Gazette of the Republic of Serbia). OGRS No. 23/2023: Regulation on Animal Health Program for the Year 2023. in Serbian. Available online: http://demo.paragraf.rs/demo/combined/Old/t/t2023_03/SG_023_2023_008.htm (accessed on 29 September 2023).
- Vidović, J.; Stojanović, D.; Cagnardi, P.; Kladar, N.; Horvat, O.; Ćirković, I.; Bijelić, K.; Stojanac, N.; Kovačević, Z. Farm animal veterinarians’ knowledge and attitudes toward antimicrobial resistance and antimicrobial use in the Republic of Serbia. Antibiotics 2022, 11, 64. [Google Scholar] [CrossRef]
- Ćirković, I.; Sørum, M.; Radenković, D.; Švabić Vlahović, M.; Larsen, A.R. National surveillance reveals findings of Panton-Valentine leukocidin positive methicillin-resistant Staphylococcus aureus in Serbia. J. Med. Microbiol. 2013, 62, 342–344. [Google Scholar] [CrossRef]
- Ćirković, I.; Đukić, S.; Carević, B.; Mazić, N.; Mioljević, V.; Stepanović, S. Methicillin-resistant Staphylococcus aureus nasal carriage among hospitalized patients and health care workers in medical center of Serbia. Arch. Biol. Sci. Belgrade 2014, 66, 87–92. [Google Scholar] [CrossRef]
- Cirkovic, I.; Stepanovic, S.; Skov, R.; Trajkovic, J.; Grgurevic, A.; Larsen, A.R. Carriage and genetic diversity of methicillin-resistant Staphylococcus aureus among patients and healthcare workers in a Serbian university hospital. PLoS ONE 2015, 10, e0127347. [Google Scholar] [CrossRef]
- Rakonjac, B.; Lepšanović, Z.; Šuljagić, V.; Jovčić, B.; Kojić, M.; Larsen, A.R.; Urić, M.; Ćirković, I. Predominance of t355/ST152/SCCmec V clonal type among PVL-positive MRSA isolates in a tertiary care hospital in Belgrade, Serbia. PLoS ONE 2022, 17, e0273474. [Google Scholar] [CrossRef] [PubMed]
- Velebit, B.; Fetsch, A.; Mirilovic, M.; Teodorovic, V.; Jovanovic, M. MRSA in pigs in Serbia. Vet. Rec. 2010, 167, 183–184. [Google Scholar] [CrossRef]
- Zutic, M.; Cirkovic, I.; Pavlovic, L.; Asanin, J.; Jovanovic, S.; Zutic, J.; Asanin, R. First isolation of methicillin-resistant Staphylococcus aureus from pigs’ clinical samples in Serbia. Acta Vet. Brno 2012, 81, 225–227. [Google Scholar] [CrossRef]
- Zutic, M.; Cirkovic, I.; Pavlovic, L.; Zutic, J.; Asanin, J.; Radanovic, O.; Pavlovic, N. Occurrence of methicillin-resistant Staphylococcus aureus in milk samples from Serbian cows with subclinical mastitis. Afr. J. Microbiol. Res. 2012, 6, 5887–5889. [Google Scholar] [CrossRef]
- Asanin, J.; Misic, D.; Aksentijevic, K.; Tambur, Z.; Rakonjac, B.; Kovacevic, I.; Spergser, J.; Loncaric, I. Genetic profiling and comparison of human and animal methicillin-resistant Staphylococcus aureus (MRSA) isolates from Serbia. Antibiotics 2019, 8, 26. [Google Scholar] [CrossRef] [PubMed]
- Pajić, M.J.; Rašić, Z.B.; Velebit, B.M.; Boboš, S.F.; Mihajlović-Ukropina, M.M.; Radinović, M.Ž.; Galfi, A.L.; Petković, J.M.; Trojačanec, S.I. The Prevalence of Methicillin Resistance and Panton-Valentine leukocidin Synthesis Genes in Staphylococcus aureus Isolates of Bovine and Human Origin. Vet. Arhiv 2014, 84, 205–214. Available online: https://hrcak.srce.hr/121942 (accessed on 23 March 2023).
- Bulajic, S.; Colovic, S.; Misic, D.; Djordjevic, J.; Savic-Radovanovic, R.; Asanin, J.; Ledina, T. Enterotoxin production and antimicrobial susceptibility in Staphylococci isolated from traditional raw milk cheeses in Serbia. J. Environ. Sci. Health B 2017, 52, 864–870. [Google Scholar] [CrossRef]
- Kreausukon, K.; Fetsch, A.; Kraushaar, B.; Alt, K.; Müller, K.; Krömker, V.; Zessin, K.-H.; Käsbohrer, A.; Tenhagen, B.-A. Prevalence, antimicrobial resistance, and molecular characterization of methicillin-resistant Staphylococcus aureus from bulk tank milk of dairy herds. J. Dairy Sci. 2012, 95, 4382–4388. [Google Scholar] [CrossRef]
- Paterson, G.K.; Morgan, F.J.E.; Harrison, E.M.; Peacock, S.J.; Parkhill, J.; Zadoks, R.N.; Holmes, M.A. Prevalence and properties of mecC methicillin-resistant Staphylococcus aureus (MRSA) in bovine bulk tank milk in Great Britain. J. Antimicrob. Chemother. 2014, 69, 598–602. [Google Scholar] [CrossRef]
- Cortimiglia, C.; Luini, M.; Bianchini, V.; Marzagalli, L.; Vezzoli, F.; Avisani, D.; Bertoletti, M.; Ianzano, A.; Franco, A.; Battisti, A. Prevalence of Staphylococcus aureus and of methicillin-resistant S. aureus clonal complexes in bulk tank milk from dairy cattle herds in Lombardy Region (Northern Italy). Epidemiol. Infect. 2016, 144, 3046–3051. [Google Scholar] [CrossRef]
- Locatelli, C.; Cremonesi, P.; Bertocchi, L.; Zanoni, M.G.; Barberio, A.; Drigo, I.; Varisco, G.; Castiglioni, B.; Bronzo, V.; Moroni, P. Short communication: Methicillin-resistant Staphylococcus aureus in bulk tank milk of dairy cows and effect of swine population density. J. Dairy Sci. 2016, 99, 2151–2156. [Google Scholar] [CrossRef]
- Tenhagen, B.-A.; Alt, K.; Pfefferkorn, B.; Wiehle, L.; Käsbohrer, A.; Fetsch, A. Short communication: Methicillin-resistant Staphylococcus aureus in conventional and organic dairy herds in Germany. J. Dairy Sci. 2018, 101, 3380–3386. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Ba, X.; Holmes, M.A. Prevalence and characterization of mecC MRSA in bovine bulk tank milk in Great Britain, 2017–2018. JAC Antimicrob. Resist. 2021, 3, dlaa125. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- MAEP-RS (Ministry of Agriculture and Environmental Protection Republic of Serbia). Republic of Serbia IPARD Programme for 2014–2020. 2019. Available online: https://uap.gov.rs/wp-content/uploads/2017/11/IPARDII-final-III-modification-ENG27062019.pdf (accessed on 12 September 2023).
- SORS (Statistical Office of the Republic of Serbia). Area, Livestock (LSU), Labour Force and Standard Output (SO) by Legal Status of Holding and Agricultural Size of Farm (UAA). STAT Database (Agriculture, Forestry and Fishery, Census of Agriculture, Farm Structure 2018, Key Farm Variables); SORS: Belgrade, Serbia, 2018. Available online: https://data.stat.gov.rs/Home/Result/1300020101?languageCode=en-US&displayMode=table (accessed on 27 February 2023).
- Ostojić Andrić, D.; Hristov, S.; Petrović, M.M.; Pantelić, V.; Bojkovski, J.; Novaković, Ž.; Lazarević, M.; Nikšić, D. Housing Conditions and Welfare of Dairy Cows in Serbia. In Proceedings of the 4th International Congress ‘New Perspectives and Challenges of Sustainable Livestock Production’, Belgrade, Serbia, 7–9 October 2015; pp. 62–73. Available online: https://r.istocar.bg.ac.rs/bitstream/handle/123456789/591/Proceedings-2015-Dusica-Ostojic-Andric.pdf?sequence=1&isAllowed=y (accessed on 12 September 2023).
- Relic, R.; Lakic, N.; Jankovic, L.; Davidovic, V.; Staric, J.; Jezek, J. Factors affecting rearing practices and health of calves on family farms. Span. J. Agric. Res. 2021, 19, e0501. [Google Scholar] [CrossRef]
- Virgin, J.E.; Van Slyke, T.M.; Lombard, J.E.; Zadoks, R.N. Short Communication: Methicillin-resistant Staphylococcus aureus detection in US bulk tank milk. J. Dairy Sci. 2009, 92, 4988–4991. [Google Scholar] [CrossRef]
- Spohr, M.; Rau, J.; Friedrich, A.; Klittich, G.; Fetsch, A.; Guerra, B.; Hammerl, J.A.; Tenhagen, B.-A. Methicillin-resistant Staphylococcus aureus (MRSA) in three dairy herds in southwest Germany. Zoonoses Public Health 2011, 58, 252–261. [Google Scholar] [CrossRef]
- Haran, K.P.; Godden, S.M.; Boxrud, D.; Jawahir, S.; Bender, J.B.; Sreevatsan, S. Prevalence and characterization of Staphylococcus aureus, including methicillin-resistant Staphylococcus aureus, isolated from bulk tank milk from Minnesota dairy farms. J. Clin. Microbiol. 2012, 50, 688–695. [Google Scholar] [CrossRef]
- Parisi, A.; Caruso, M.; Normanno, G.; Latorre, L.; Sottili, R.; Miccolupo, A.; Fraccalvieri, R.; Santagada, G. Prevalence, antimicrobial susceptibility and molecular typing of methicillin-resistant Staphylococcus aureus (MRSA) in bulk tank milk from southern Italy. Food Microbiol. 2016, 58, 36–42. [Google Scholar] [CrossRef]
- Locatelli, C.; Cremonesi, P.; Caprioli, A.; Carfora, V.; Ianzano, A.; Barberio, A.; Morandi, S.; Casula, A.; Castiglioni, B.; Bronzo, V.; et al. Occurrence of methicillin-resistant Staphylococcus aureus in dairy cattle herds, related swine farms, and humans in contact with herds. J. Dairy Sci. 2017, 100, 608–619. [Google Scholar] [CrossRef]
- Papadopoulos, P.; Papadopoulos, T.; Angelidis, A.S.; Boukouvala, E.; Zdragas, A.; Papa, A.; Hadjichristodoulou, C.; Sergelidis, D. Prevalence of Staphylococcus aureus and of methicillin-resistant S. aureus (MRSA) along the production chain of dairy products in north-western Greece. Food Microbiol. 2018, 69, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Selim, A.; Kelis, K.; AlKahtani, M.D.F.; Albohairy, F.M.; Attia, K.A. Prevalence, antimicrobial susceptibilities and risk factors of Methicillin resistant Staphylococcus aureus (MRSA) in dairy bovines. BMC Vet. Res. 2022, 18, 293. [Google Scholar] [CrossRef] [PubMed]
- Tenhagen, B.A.; Alt, K.; Grobbel, M.; Maurischat, S. MRSA in bulk tank milk of dairy herds in Germany—Changes over time. Tierarztl. Prax. Ausg. G Grosstiere Nutztiere. 2023, 51, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Antoci, E.; Pinzone, M.R.; Nunnari, G.; Stefani, S.; Cacopardo, B. Prevalence and Molecular Characteristics of Methicillin-Resistant Staphylococcus aureus (MRSA) among Subjects Working on Bovine Dairy Farms. Infez. Med. 2013, 21, 125–129. Available online: https://www.infezmed.it/media/journal/Vol_21_2_2013_5.pdf (accessed on 12 September 2023). [PubMed]
- Schnitt, A.; Lienen, T.; Wichmann-Schauer, H.; Cuny, C.; Tenhagen, B.A. The occurrence and distribution of livestock-associated methicillin-resistant Staphylococcus aureus ST398 on German dairy farms. J. Dairy Sci. 2020, 103, 11806–11819. [Google Scholar] [CrossRef] [PubMed]
- Huber, H.; Ziegler, D.; Pflüger, V.; Vogel, G.; Zweifel, C.R.; Stephan, R. Prevalence and characteristics of methicillin-resistant coagulase-negative staphylococci from livestock, chicken carcasses, bulk tank milk, minced meat, and contact persons. BMC Vet. Res. 2011, 7, 6. [Google Scholar] [CrossRef]
- Frey, Y.; Rodriguez, J.P.; Thomann, A.; Schwendener, S.; Perreten, V. Genetic characterization of antimicrobial resistance in coagulase-negative staphylococci from bovine mastitis milk. J. Dairy Sci. 2013, 96, 2247–2257. [Google Scholar] [CrossRef]
- Cicconi-Hogan, K.M.; Belomestnykh, N.; Gamroth, M.; Ruegg, P.L.; Tikofsky, L.; Schukken, Y.H. Short communication: Prevalence of methicillin resistance in coagulase-negative staphylococci and Staphylococcus aureus isolated from bulk milk on organic and conventional dairy farms in the United States. J. Dairy Sci. 2014, 97, 2959–2964. [Google Scholar] [CrossRef]
- Fisher, E.A.; Paterson, G.K. Prevalence and characterisation of methicillin-resistant staphylococci from bovine bulk tank milk in England and Wales. J. Glob. Antimicrob. Resist. 2020, 22, 139–144. [Google Scholar] [CrossRef]
- Rolo, J.; Worning, P.; Boye Nielsen, J.; Sobral, R.; Bowden, R.; Bouchami, O.; Damborg, P.; Guardabassi, L.; Perreten, V.; Westh, H.; et al. Evidence for the evolutionary steps leading to mecA-mediated β-lactam resistance in staphylococci. PLoS Genet. 2017, 13, e1006674. [Google Scholar] [CrossRef]
- Traversari, J.; van den Borne, B.H.P.; Dolder, C.; Thomann, A.; Perreten, V.; Bodmer, M. Non-aureus staphylococci species in the teat canal and milk in four commercial Swiss dairy herds. Front. Vet. Sci. 2019, 6, 186. [Google Scholar] [CrossRef] [PubMed]
- Rosa, N.M.; Penati, M.; Fusar-Poli, S.; Addis, M.F.; Tola, S. Species identification by MALDI-TOF MS and gap PCR-RFLP of non-aureus Staphylococcus, Mammaliicoccus, and Streptococcus spp. associated with sheep and goat mastitis. Vet. Res. 2022, 53, 84. [Google Scholar] [CrossRef]
- Nobrega, D.B.; Naushad, S.; Naqvi, S.A.; Condas, L.A.Z.; Saini, V.; Kastelic, J.P.; Luby, C.; De Buck, J.; Barkema, H.W. Prevalence and genetic basis of antimicrobial resistance in non-aureus staphylococci isolated from Canadian dairy herds. Front. Microbiol. 2018, 9, 256. [Google Scholar] [CrossRef] [PubMed]
- Martin, N.H.; Evanowski, R.L.; Wiedmann, M. Invited review: Redefining raw milk quality—Evaluation of raw milk microbiological parameters to ensure high-quality processed dairy products. J. Dairy Sci. 2023, 106, 1502–1517. [Google Scholar] [CrossRef]
- Zigo, F.; Vasil’, M.; Ondrašovicová, S.; Výrostková, J.; Bujok, J.; Pecka-Kielb, E. Maintaining optimal mammary gland health and prevention of mastitis. Front. Vet. Sci. 2021, 8, 607311. [Google Scholar] [CrossRef] [PubMed]
- Latorre, A.A.; Pachá, P.A.; González-Rocha, G.; San Martín, I.; Quezada-Aguiluz, M.; Aguayo-Reyes, A.; Bello-Toledo, H.; Oliva, R.; Estay, A.; Pugin, J.; et al. On-farm surfaces in contact with milk: The role of Staphylococcus aureus-containing biofilms for udder health and milk quality. Foodborne Pathog. Dis. 2020, 17, 44–51. [Google Scholar] [CrossRef]
- Pacha, P.A.; Munoz, M.A.; González-Rocha, G.; San Martín, I.; Quezada-Aguiluz, M.; Aguayo-Reyes, A.; Bello-Toledo, H.; Latorre, A.A. Molecular diversity of Staphylococcus aureus and the role of milking equipment adherences or biofilm as a source for bulk tank milk contamination. J. Dairy Sci. 2021, 104, 3522–3531. [Google Scholar] [CrossRef]
- Smistad, M.; Bakka, H.C.; Sølverød, L.; Jørgensen, H.J.; Wolff, C. Prevalence of udder pathogens in milk samples from Norwegian dairy cows recorded in a national database in 2019 and 2020. Acta Vet. Scand. 2023, 65, 19. [Google Scholar] [CrossRef]
- Nonnemann, B.; Lyhs, U.; Svennesen, L.; Kristensen, K.A.; Klaas, I.C.; Pedersen, K. Bovine mastitis bacteria resolved by MALDI-TOF mass spectrometry. J. Dairy Sci. 2019, 102, 2515–2524. [Google Scholar] [CrossRef]
- Conesa, A.; Dieser, S.; Barberis, C.; Bonetto, C.; Lasagno, M.; Vay, C.; Odierno, L.; Porporatto, C.; Raspanti, C. Differentiation of non-aureus staphylococci species isolated from bovine mastitis by PCR-RFLP of groEL and gap genes in comparison to MALDI-TOF mass spectrometry. Microb. Pathog. 2020, 149, 104489. [Google Scholar] [CrossRef]
- Hamel, J.; Zhang, Y.; Went, N.; Krömker, V. Non-S. aureus staphylococci (NAS) in milk samples: Infection or contamination? Vet. Microbiol. 2020, 242, 108594. [Google Scholar] [CrossRef] [PubMed]
- Bergeron, M.; Dauwalder, O.; Gouy, M.; Freydiere, A.M.; Bes, M.; Meugnier, H.; Benito, Y.; Etienne, J.; Lina, G.; Vandenesch, F.; et al. Species identification of staphylococci by amplification and sequencing of the tuf gene compared to the gap gene and by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Mahmmod, Y.S.; Nonnemann, B.; Svennesen, L.; Pedersen, K.; Klaas, I.C. Typeability of MALDI-TOF assay for identification of non-aureus staphylococci associated with bovine intramammary infections and teat apex colonization. J. Dairy Sci. 2018, 101, 9430–9438. [Google Scholar] [CrossRef] [PubMed]
- Titouche, Y.; Akkou, M.; Houali, K.; Auvray, F.; Hennekinne, J.-A. Role of milk and milk products in the spread of methicillin-resistant Staphylococcus aureus in the dairy production chain. J. Food Sci. 2022, 87, 3699–3723. [Google Scholar] [CrossRef]
- Tegegne, H.A.; Florianová, M.; Gelbíčová, T.; Karpíšková, R.; Koláčková, I. Detection and molecular characterization of methicillin-resistant Staphylococcus aureus isolated from bulk tank milk of cows, sheep, and goats. Foodborne Pathog. Dis. 2018, 16, 68–73. [Google Scholar] [CrossRef]
- Hansen, J.E.; Ronco, T.; Stegger, M.; Sieber, R.N.; Fertner, M.E.; Martin, H.L.; Farre, M.; Toft, N.; Larsen, A.R.; Pedersen, K. LA-MRSA CC398 in dairy cattle and veal calf farms indicates spillover from pig production. Front. Microbiol. 2019, 10, 2733. [Google Scholar] [CrossRef]
- Kadlec, K.; Entorf, M.; Peters, T. Occurrence and characteristics of livestock-associated methicillin-resistant Staphylococcus aureus in quarter milk samples from dairy cows in Germany. Front. Microbiol. 2019, 10, 1295. [Google Scholar] [CrossRef]
- Lienen, T.; Schnitt, A.; Cuny, C.; Maurischat, S.; Tenhagen, B.-A. Phylogenetic tracking of LA-MRSA ST398 intra-farm transmission among animals, humans and the environment on German dairy farms. Microorganisms 2021, 9, 1119. [Google Scholar] [CrossRef]
- Alba, P.; Feltrin, F.; Cordaro, G.; Porrero, M.C.; Kraushaar, B.; Argudín, M.A.; Nykäsenoja, S.; Monaco, M.; Stegger, M.; Aarestrup, F.M.; et al. Livestock-associated methicillin resistant and methicillin susceptible Staphylococcus aureus sequence type (CC)1 in European farmed animals: High genetic relatedness of isolates from Italian cattle herds and humans. PLoS ONE 2015, 10, e0137143. [Google Scholar] [CrossRef]
- Monaco, M.; Pedroni, P.; Sanchini, A.; Bonomini, A.; Indelicato, A.; Pantosti, A. Livestock-associated methicillin-resistant Staphylococcus aureus responsible for human colonization and infection in an area of Italy with high density of pig farming. BMC Infect. Dis. 2013, 13, 258. [Google Scholar] [CrossRef]
- Huber, H.; Koller, S.; Giezendanner, N.; Stephan, R.; Zweifel, C. Prevalence and characteristics of meticillin-resistant Staphylococcus aureus in humans in contact with farm animals, in livestock, and in food of animal origin, Switzerland, 2009. Euro Surveill. 2010, 15, 19542. [Google Scholar] [CrossRef] [PubMed]
- Pilla, R.; Castiglioni, V.; Gelain, M.E.; Scanziani, E.; Lorenzi, V.; Anjum, M.; Piccinini, R. Long-term study of MRSA ST1, t127 mastitis in a dairy cow. Vet. Rec. 2012, 170, 312. [Google Scholar] [CrossRef]
- Juhász-Kaszanyitzky, E.; Jánosi, S.; Somogyi, P.; Dán, A.; van der Graaf-van Bloois, L.; van Duijkeren, E.; Wagenaar, J.A. MRSA transmission between cows and humans. Emerg. Infect. Dis. 2007, 13, 630–632. [Google Scholar] [CrossRef] [PubMed]
- Baig, S.; Rhod Larsen, A.; Martins Simões, P.; Laurent, F.; Johannesen, T.B.; Lilje, B.; Tristan, A.; Schaumburg, F.; Egyir, B.; Cirkovic, I.; et al. Evolution and population dynamics of clonal complex 152 community-associated methicillin-resistant Staphylococcus aureus. mSphere 2020, 5, e00226-20. [Google Scholar] [CrossRef] [PubMed]
- Basanisi, M.G.; La Bella, G.; Nobili, G.; Franconieri, I.; La Salandra, G. Genotyping of methicillin-resistant Staphylococcus aureus (MRSA) isolated from milk and dairy products in South Italy. Food Microbiol. 2017, 62, 141–146. [Google Scholar] [CrossRef]
- Crespo-Piazuelo, D.; Lawlor, P.G. Livestock associated methicillin resistant Staphylococcus aureus (LA MRSA) prevalence in humans in close contact with animals and measures to reduce on farm colonization. Ir. Vet. J. 2021, 74, 21. [Google Scholar] [CrossRef]
- Patel, K.; Godden, S.M.; Royster, E.E.; Crooker, B.A.; Johnson, T.J.; Smith, E.A.; Sreevatsan, S. Prevalence, antibiotic resistance, virulence and genetic diversity of Staphylococcus aureus isolated from bulk tank milk samples of U.S. dairy herds. BMC Genom. 2021, 22, 367. [Google Scholar] [CrossRef]
- Sabat, A.; Kosowska, K.; Poulsen, K.; Kasprowicz, A.; Sekowska, A.; van Den Burg, B.; Travis, J.; Potempa, J. Two allelic forms of the aureolysin gene (aur) within Staphylococcus aureus. Infect. Immun. 2000, 68, 973–976. [Google Scholar] [CrossRef]
- Cui, M.; Li, J.; Ali, T.; Kalim, K.; Wang, H.; Song, L.; Li, Z.; Ren, X.; Ma, F.; Zou, M.; et al. Emergence of livestock-associated MRSA ST398 from bulk tank milk, China. J. Antimicrob. Chemother. 2020, 75, 3471–3474. [Google Scholar] [CrossRef]
- Sieber, R.N.; Urth, T.R.; Petersen, A.; Moller, C.H.; Price, L.B.; Skov, R.L.; Larsen, A.R.; Stegger, M.; Larsen, J. Phage-mediated immune evasion and transmission of livestock-associated methicillin-resistant Staphylococcus aureus in humans. Emerg. Infect. Dis. 2020, 26, 2578–2585. [Google Scholar] [CrossRef]
- Chaguza, C.; Smith, J.T.; Bruce, S.A.; Gibson, R.; Martin, I.W.; Andam, C.P. Prophage-encoded immune evasion factors are critical for Staphylococcus aureus host infection, switching, and adaptation. Cell Genom. 2022, 2, 100194. [Google Scholar] [CrossRef] [PubMed]
- Monecke, S.; Müller, E.; Dorneanu, O.S.; Vremerǎ, T.; Ehricht, R. Molecular typing of MRSA and of clinical Staphylococcus aureus isolates from Iaşi, Romania. PLoS ONE 2014, 9, e97833. [Google Scholar] [CrossRef] [PubMed]
- Benito, D.; Lozano, C.; Rezusta, A.; Ferrer, I.; Vasquez, M.A.; Ceballos, S.; Zarazaga, M.; Revillo, M.J.; Torres, C. Characterization of tetracycline and methicillin resistant Staphylococcus aureus strains in a Spanish hospital: Is livestock-contact a risk factor in infections caused by MRSA CC398? Int. J. Med. Microbiol. 2014, 304, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Hummerjohann, J.; Naskova, J.; Baumgartner, A.; Graber, H.U. Enterotoxin-producing Staphylococcus aureus genotype B as a major contaminant in Swiss raw milk cheese. J. Dairy Sci. 2014, 97, 1305–1312. [Google Scholar] [CrossRef]
- Earls, M.R.; Shore, A.C.; Brennan, G.I.; Simbeck, A.; Schneider-Brachert, W.; Vremerǎ, T.; Dorneanu, O.S.; Slickers, P.; Ehricht, R.; Monecke, S.; et al. A novel multidrug-resistant PVL-negative CC1-MRSA-IV clone emerging in Ireland and Germany likely originated in South-Eastern Europe. Infect. Genet. Evol. 2019, 69, 117–126. [Google Scholar] [CrossRef]
- Ikeda, T.; Tamate, N.; Yamaguchi, K.; Makino, S. Mass outbreak of food poisoning disease caused by small amounts of staphylococcal enterotoxins A and H. Appl. Environ. Microbiol. 2005, 71, 2793–2795. [Google Scholar] [CrossRef] [PubMed]
- Kadariya, J.; Smith, T.C.; Thapaliya, D. Staphylococcus aureus and staphylococcal food-borne disease: An ongoing challenge in public health. Biomed. Res. Int. 2014, 2014, 827965. [Google Scholar] [CrossRef]
- Bastos, C.P.; Bassani, M.T.; Mata, M.M.; Lopes, G.V.; da Silva, W.P. Prevalence and expression of staphylococcal enterotoxin genes in Staphylococcus aureus isolated from food poisoning outbreaks. Can. J. Microbiol. 2017, 63, 834–840. [Google Scholar] [CrossRef]
- Linage, B.; Rodríguez-Calleja, J.M.; Otero, A.; García-López, M.L.; Santos, J.A. Characterization of coagulase-positive staphylococci isolated from tank and silo ewe milk. J. Dairy Sci. 2012, 95, 1639–1644. [Google Scholar] [CrossRef]
- Vitale, M.; Gaglio, S.; Galluzzo, P.; Cascone, G.; Piraino, C.; Di Marco Lo Presti, V.; Alduina, R. Antibiotic resistance profiling, analysis of virulence aspects and molecular genotyping of Staphylococcus aureus isolated in Sicily, Italy. Foodborne Pathog. Dis. 2018, 15, 177–185. [Google Scholar] [CrossRef]
- Fang, R.; Cui, J.; Cui, T.; Guo, H.; Ono, H.K.; Park, C.-H.; Okamura, M.; Nakane, A.; Hu, D.-L. Staphylococcal enterotoxin C is an important virulence factor for mastitis. Toxins 2019, 11, 141. [Google Scholar] [CrossRef] [PubMed]
- Cuny, C.; Wieler, L.H.; Witte, W. Livestock-associated MRSA: The impact on humans. Antibiotics 2015, 4, 521–543. [Google Scholar] [CrossRef] [PubMed]
- van Wamel, W.J.B.; Rooijakkers, S.H.M.; Ruyken, M.; van Kessel, K.P.M.; van Strijp, J.A.G. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J. Bacteriol. 2006, 188, 1310–1315. [Google Scholar] [CrossRef] [PubMed]
- Naushad, S.; Naqvi, S.A.; Nobrega, D.; Luby, C.; Kastelic, J.P.; Barkema, H.W.; De Buck, J. Comprehensive virulence gene profiling of bovine non-aureus staphylococci based on whole-genome sequencing data. mSystems 2019, 4, e00098-18. [Google Scholar] [CrossRef]
- Belhout, C.; Boyen, F.; Vereecke, N.; Theuns, S.; Taibi, N.; Stegger, M.; de la Fé-Rodríguez, P.Y.; Bouayad, L.; Elgroud, R.; Butaye, P. Prevalence and molecular characterization of methicillin-resistant staphylococci (MRS) and mammaliicocci (MRM) in dromedary camels from Algeria: First detection of SCCmec-mecC hybrid in methicillin-resistant Mammaliicoccus lentus. Antibiotics 2023, 12, 674. [Google Scholar] [CrossRef]
- Diep, B.A.; Stone, G.G.; Basuino, L.; Graber, C.J.; Miller, A.; des Etages, S.A.; Jones, A.; Palazzolo-Ballance, A.M.; Perdreau-Remington, F.; Sensabaugh, G.F.; et al. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: Convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 2008, 197, 1523–1530. [Google Scholar] [CrossRef]
- Tong, C.; Wu, Z.; Zhao, X.; Xue, H. Arginine catabolic mobile elements in livestock-associated methicillin-resistant staphylococcal isolates from bovine mastitic milk in China. Front. Microbiol. 2018, 9, 1031. [Google Scholar] [CrossRef]
- ALIMS (Medicines and Medical Devices Agency of Serbia). Promet Veterinarskih Lekova 2019–2020; ALIMS (Agencija za Lekova I Medicinska Sredstva Srbije): Belgrade, Serbia, 2021. Available online: https://www.alims.gov.rs/wp-content/uploads/2022/03/PPL-VET-2020.pdf (accessed on 4 May 2023). (In Serbian)
- Schwarz, S.; Feßler, A.T.; Loncaric, I.; Wu, C.; Kadlec, K.; Wang, Y.; Shen, J. Antimicrobial resistance among staphylococci of animal origin. Microbiol. Spectr. 2018, 6, ARBA-0010-2017. [Google Scholar] [CrossRef]
- Monecke, S.; Kuhnert, P.; Hotzel, H.; Slickers, P.; Ehricht, R. Microarray based study on virulence-associated genes and resistance determinants of Staphylococcus aureus isolates from cattle. Vet. Microbiol. 2007, 125, 128–140. [Google Scholar] [CrossRef]
- Chua, K.; Laurent, F.; Coombs, G.; Grayson, M.L.; Howden, B.P. Not community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA)! A clinician’s guide to community MRSA—Its evolving antimicrobial resistance and implications for therapy. Clin. Infect. Dis. 2011, 52, 99–114. [Google Scholar] [CrossRef]
- Said-Salim, B.; Dunman, P.M.; McAleese, F.M.; Macapagal, D.; Murphy, E.; McNamara, P.J.; Arvidson, S.; Foster, T.J.; Projan, S.J.; Kreiswirth, B.N. Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 2003, 185, 610–619. [Google Scholar] [CrossRef] [PubMed]
- Shoaib, M.; Xu, J.; Meng, X.; Wu, Z.; Hou, X.; He, Z.; Shang, R.; Zhang, H.; Pu, W. Molecular epidemiology and characterization of antimicrobial-resistant Staphylococcus haemolyticus strains isolated from dairy cattle milk in Northwest, China. Front. Cell. Infect. Microbiol. 2023, 13, 1183390. [Google Scholar] [CrossRef] [PubMed]
- Fergestad, M.E.; Touzain, F.; De Vliegher, S.; De Visscher, A.; Thiry, D.; Tchamba, C.N.; Mainil, J.G.; L’Abee-Lund, T.; Blanchard, Y.; Wasteson, Y. Whole genome sequencing of staphylococci isolated from bovine milk samples. Front. Microbiol. 2021, 12, 715851. [Google Scholar] [CrossRef]
- Osada, M.; Aung, M.S.; Urushibara, N.; Kawaguchiya, M.; Ohashi, N.; Hirose, M.; Kobayashi, N. Prevalence and antimicrobial resistance of Staphylococcus aureus and coagulase-negative Staphylococcus/Mammaliicoccus from retail ground meat: Identification of broad genetic diversity in fosfomycin resistance gene fosB. Pathogens 2022, 11, 469. [Google Scholar] [CrossRef] [PubMed]
- Lüthje, P.; Schwarz, S. Antimicrobial resistance of coagulase-negative staphylococci from bovine subclinical mastitis with particular reference to macrolide-lincosamide resistance phenotypes and genotypes. J. Antimicrob. Chemother. 2006, 57, 966–969. [Google Scholar] [CrossRef]
- Schauer, B.; Szostak, M.P.; Ehricht, R.; Monecke, S.; Feßler, A.T.; Schwarz, S.; Spergser, J.; Krametter-Frötscher, R.; Loncaric, I. Diversity of methicillin-resistant coagulase-negative Staphylococcus spp. and methicillin-resistant Mammaliicoccus spp. isolated from ruminants and New World camelids. Vet. Microbiol. 2021, 254, 109005. [Google Scholar] [CrossRef]
- EUCAST (European Committee on Antimicrobial Susceptibility Testing). Breakpoint Tables for Interpretation of MICs and Zone Diameters; Version 11.0; EUCAST: Växjö, Sweden, 2021; Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_11.0_Breakpoint_Tables.pdf (accessed on 4 May 2023).
- Schouls, L.M.; Spalburg, E.C.; van Luit, M.; Huijsdens, X.W.; Pluister, G.N.; van Santen-Verheuvel, M.G.; van der Heide, H.G.J.; Grundmann, H.; Heck, M.E.O.C.; de Neeling, A.J. Multiple-locus variable number tandem repeat analysis of Staphylococcus aureus: Comparison with pulsed-field gel electrophoresis and spa-typing. PLoS ONE 2009, 4, e5082. [Google Scholar] [CrossRef]
- Stegger, M.; Andersen, P.S.; Kearns, A.; Pichon, B.; Holmes, M.A.; Edwards, G.; Laurent, F.; Teale, C.; Skov, R.; Larsen, A.R. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecALGA251. Clin. Microbiol. Infect. 2012, 18, 395–400. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomics analysis with Kraken 2. Genome Biol. 2019, 20, e257. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R.; Glöckner, F.O.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, e19126-31. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.D.; Zhou, S.Y.; Chen, L.H.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef]
- Bartels, M.D.; Petersen, A.; Worning, P.; Nielsen, J.B.; Larner-Svensson, H.; Johansen, H.K.; Andersen, L.P.; Jarløv, J.O.; Boye, K.; Larsen, A.R.; et al. Comparing whole-genome sequencing with Sanger sequencing for spa typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2014, 52, 4305–4308. [Google Scholar] [CrossRef]
- Jolley, K.A.; Maiden, M.C. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 2010, 11, e595. [Google Scholar] [CrossRef]
- Kaya, H.; Hasman, H.; Larsen, J.; Stegger, M.; Johannesen, T.B.; Allesøe, R.L.; Lemvigh, C.K.; Aarestrup, F.M.; Lund, O.; Larsen, A.R. SCCmecFinder, a web-based tool for typing of Staphylococcal Cassette Chromosome mec in Staphylococcus aureus using whole-genome sequence data. mSphere 2018, 3, e00612-17. [Google Scholar] [CrossRef]
- Silva, M.; Machado, M.P.; Silva, D.N.; Rossi, M.; Moran-Gilad, J.; Santos, S.; Ramirez, M.; Carriço, J.A. chewBBACA: A complete suite for gene-by-gene schema creation and strain identification. Microb. Genom. 2018, 4, e000166. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.F.; Sergeant, M.J.; Luhmann, N.; Vaz, C.; Francisco, A.P.; Carriço, J.A.; Achtman, M. GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018, 28, 1395–1404. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Schürch, A.C.; Arredondo-Alonso, S.; Willems, R.J.L.; Goering, R.V. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene-based approaches. Clin. Microbiol. Infect. 2018, 24, 350–354. [Google Scholar] [CrossRef] [PubMed]
- EU (European Union). Corrigendum to Regulation (EC) No 853/2004 of the European Parliament and of the Council of 29 April 2004 Laying down Specific Hygiene Rules for Food of Animal Origin. Off. J. Eur. Union 2004, L 226, 22–82. Available online: https://eur-lex.europa.eu/eli/reg/2004/853/corrigendum/2004-06-25/oj (accessed on 11 September 2023).
- SORS (Statistical Office of the Republic of Serbia). Farms and Heads of Animals by Livestock Units (LSU). STAT Database (Agriculture, Census of Agriculture, Census 2012, Livestock and Bees); SORS: Belgrade, Serbia, 2012. Available online: https://data.stat.gov.rs/Home/Result/1300010204?languageCode=en-US (accessed on 6 June 2023).

| Variable | n | MRS/MRM- Positive Farms (%) | OR | 95% CI | p |
|---|---|---|---|---|---|
| Milking system | |||||
| Bucket | 125 | 16.8 | 1.00 | 0.008 | |
| Pipeline | 137 | 32.8 | 2.42 | 1.34–4.37 | |
| Robot | 3 | 33.3 | 2.48 | 0.26–28.57 | |
| Milking parlor | 18 | 5.6 | 0.29 | 0.04–2.31 | |
| Housing system | |||||
| Tie-stall | 263 | 25.1 | 1.00 | 0.146 | |
| Free-stall | 20 | 10.0 | 0.33 | 0.07–1.47 | |
| Herd size (no. of dairy cows) | |||||
| >10 | 194 | 24.2 | 1.00 | 0.908 | |
| <10 | 89 | 23.6 | 0.97 | 0.54–1.74 | |
| Breed | |||||
| Simmental | 157 | 28.0 | 1.00 | 0.216 | |
| Holstein Friesian | 83 | 19.3 | 0.61 | 0.32–1.17 | |
| Mix | 43 | 18.6 | 0.59 | 0.25–1.36 | |
| TBC (CFU/mL) | |||||
| <100,000 | 193 | 17.6 | 1.00 | 0.001 | |
| >100,000 | 90 | 37.8 | 2.84 | 1.61–4.99 | |
| SCC per mL | |||||
| <400,000 | 227 | 22.5 | 1.00 | 0.218 | |
| >400,000 | 56 | 30.4 | 1.50 | 0.79–2.88 | |
| Total | 283 | 24.0 |
| Variable | Adjusted OR | 95% CI | p |
|---|---|---|---|
| Milking system | |||
| Bucket | 1.00 | ||
| Pipeline | 2.51 | 1.37–4.59 | 0.003 |
| Robot | 2.57 | 0.21–31.79 | 0.461 |
| Milking parlor | 0.37 | 0.05–3.00 | 0.353 |
| TBC (CFU/mL) | |||
| <100,000 | 1.00 | ||
| >100,000 | 2.78 | 1.55–4.97 | 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kos, A.; Papić, B.; Golob, M.; Avberšek, J.; Kušar, D.; Ledina, T.; Đorđević, J.; Bulajić, S. Genomic Insights into Methicillin-Resistant Staphylococci and Mammaliicocci from Bulk Tank Milk of Dairy Farms in Serbia. Antibiotics 2023, 12, 1529. https://doi.org/10.3390/antibiotics12101529
Kos A, Papić B, Golob M, Avberšek J, Kušar D, Ledina T, Đorđević J, Bulajić S. Genomic Insights into Methicillin-Resistant Staphylococci and Mammaliicocci from Bulk Tank Milk of Dairy Farms in Serbia. Antibiotics. 2023; 12(10):1529. https://doi.org/10.3390/antibiotics12101529
Chicago/Turabian StyleKos, Andrea, Bojan Papić, Majda Golob, Jana Avberšek, Darja Kušar, Tijana Ledina, Jasna Đorđević, and Snežana Bulajić. 2023. "Genomic Insights into Methicillin-Resistant Staphylococci and Mammaliicocci from Bulk Tank Milk of Dairy Farms in Serbia" Antibiotics 12, no. 10: 1529. https://doi.org/10.3390/antibiotics12101529
APA StyleKos, A., Papić, B., Golob, M., Avberšek, J., Kušar, D., Ledina, T., Đorđević, J., & Bulajić, S. (2023). Genomic Insights into Methicillin-Resistant Staphylococci and Mammaliicocci from Bulk Tank Milk of Dairy Farms in Serbia. Antibiotics, 12(10), 1529. https://doi.org/10.3390/antibiotics12101529





