Clinical Impact of Rapid Bacterial Microbiological Identification with the MALDI-TOF MS
Abstract
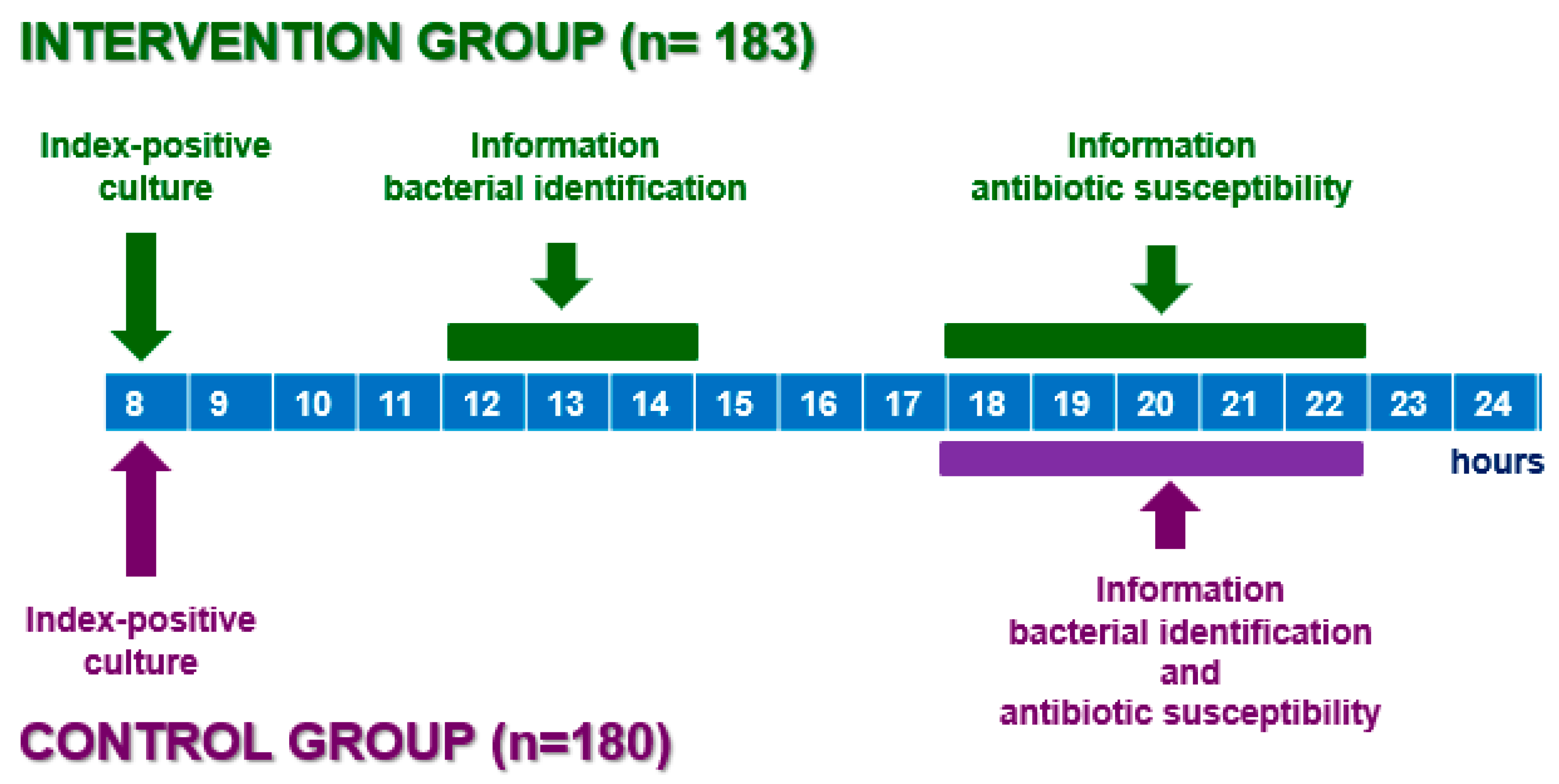
:1. Introduction
2. Results
2.1. Demographics and Clinical Basal Characteristics of Patients
2.2. Samples and Infectious Syndrome
2.3. Outcomes in Antibiotic Therapy of Early Microbiological Information
2.4. Hospital Impact and Mortality Outcomes
3. Discussion
4. Materials and Methods
4.1. Setting
4.2. Power Analysis and Sample Size Calculation
4.3. Statistical Analysis
4.4. Variables Recorded Included
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clerc, O.; Prod’hom, G.; Vogne, C.; Bizzini, A.; Calandra, T.; Greub, G. Impact of Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry on the Clinical Management of Patients with Gram-Negative Bacteremia: A Prospective Observational Study. Clin. Infect. Dis. 2013, 56, 1101–1107. [Google Scholar] [CrossRef] [PubMed]
- Timbrook, T.T.; Morton, J.B.; McConeghy, K.W.; Caffrey, A.R.; Mylonakis, E.; LaPlante, K.L. The Effect of Molecular Rapid Diagnostic Testing on Clinical Outcomes in Bloodstream Infections: A Systematic Review and Meta-Analysis. Clin. Infect. Dis. 2017, 64, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Galar, A.; Leiva, J.; Espinosa, M.; Guillén-Grima, F.; Hernáez, S.; Yuste, J.R. Clinical and Economic Evaluation of the Impact of Rapid Microbiological Diagnostic Testing. J. Infect. 2012, 65, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, G.; Midiri, A.; Gerace, E.; Biondo, C. Bacterial Antibiotic Resistance: The Most Critical Pathogens. Pathogens 2021, 10, 1310. [Google Scholar] [CrossRef]
- Galar, A.; Yuste, J.R.; Espinosa, M.; Guillén-Grima, F.; Hernáez-Crespo, S.; Leiva, J. Clinical and Economic Impact of Rapid Reporting of Bacterial Identification and Antimicrobial Susceptibility Results of the Most Frequently Processed Specimen Types. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 2445–2452. [Google Scholar] [CrossRef]
- Nadjm, B.; Dat, V.Q.; Campbell, J.I.; Dung, V.T.V.; Torre, A.; Tu, N.T.C.; Van, N.T.T.; Trinh, D.T.; Lan, N.P.H.; Trung, N.V.; et al. A Randomised Controlled Trial of Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDITOF-MS) versus Conventional Microbiological Methods for Identifying Pathogens: Impact on Optimal Antimicrobial Therapy of Invasive Bacterial and Fungal Infections in Vietnam. J. Infect. 2019, 78, 454–460. [Google Scholar] [CrossRef]
- Roux-Dalvai, F.; Gotti, C.; Leclercq, M.; Helie, M.; Boissinot, M.; Arrey, T.; Dauly, C.; Fournier, F.; Kelly, I.; Marcoux, J.; et al. Fast and Accurate Bacterial Species Identification in Urine Specimens Using LC-MS/MS Mass Spectrometry and Machine Learning. Mol. Cell. Proteom. 2019, 18, 2492–2505. [Google Scholar] [CrossRef]
- De la Pedrosa, E.G.G.; Gimeno, C.; Soriano, A.; Cantón, R. Studies of the cost effectiveness of MALDI-TOF and clinical impact. Enferm. Infecc. Microbiol. Clin. 2016, 34 (Suppl. 2), 47–52. [Google Scholar] [CrossRef]
- Fenselau, C.; Demirev, P.A. Characterization of Intact Microorganisms by MALDI Mass Spectrometry. Mass. Spectrom. Rev. 2001, 20, 157–171. [Google Scholar] [CrossRef]
- Tsuchida, S.; Umemura, H.; Nakayama, T. Current Status of Matrix-Assisted Laser Desorption/Ionization-Time-of-Flight Mass Spectrometry (MALDI-TOF MS) in Clinical Diagnostic Microbiology. Molecules 2020, 25, 4775. [Google Scholar] [CrossRef] [PubMed]
- Yo, C.-H.; Shen, Y.-H.; Hsu, W.-T.; Mekary, R.A.; Chen, Z.R.; Lee, W.-T.J.; Chen, S.-C.; Lee, C.-C. MALDI-TOF Mass Spectrometry Rapid Pathogen Identification and Outcomes of Patients with Bloodstream Infection: A Systematic Review and Meta-Analysis. Microb. Biotechnol. 2022, 15, 2667–2682. [Google Scholar] [CrossRef]
- Oros, D.; Ceprnja, M.; Zucko, J.; Cindric, M.; Hozic, A.; Skrlin, J.; Barisic, K.; Melvan, E.; Uroic, K.; Kos, B.; et al. Identification of Pathogens from Native Urine Samples by MALDI-TOF/TOF Tandem Mass Spectrometry. Clin. Proteom. 2020, 17, 25. [Google Scholar] [CrossRef]
- Muñoz Bellido, J.L.; González Buitrago, J.M. MALDI-TOF mass spectrometry in clinical microbiology. Current situation and future perspectives. Enferm. Infecc. Microbiol. Clin. 2015, 33, 369–371. [Google Scholar] [CrossRef] [PubMed]
- Samaranayake, W.A.M.P.; Dempsey, S.; Howard-Jones, A.R.; Outhred, A.C.; Kesson, A.M. Rapid Direct Identification of Positive Paediatric Blood Cultures by MALDI-TOF MS Technology and Its Clinical Impact in the Paediatric Hospital Setting. BMC Res. Notes 2020, 13, 12. [Google Scholar] [CrossRef]
- Legarraga, P.; Moraga, M.; Lam, M.; Geoffroy, E.; Zumarán, C.; García, P. [Impact of mass spectrometry by MALDI-TOF MS for the rapid identification of aerobic and anaerobic bacteria of clinical importance]. Rev. Chilena Infectol. 2013, 30, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Astrup, L.B.; Pedersen, K.; Farre, M. Microbiological Diagnoses on Clinical Mastitis—Comparison between Diagnoses Made in Veterinary Clinics versus in Laboratory Applying MALDI-TOF MS. Antibiotics 2022, 11, 271. [Google Scholar] [CrossRef] [PubMed]
- Pawlak, Z.; Andrusiów, S.; Pajączkowska, M.; Janczura, A. Identification of Fungi Isolated from Oral Cavity of Patients with HIV Using MALDI–TOF MS. J. Clin. Med. 2021, 10, 1570. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.D.; Seong, H.; Kim, D.; Ahn, M.Y.; Jung, I.Y.; Jeong, S.J.; Choi, J.Y.; Song, Y.G.; Yong, D.; Lee, K.; et al. Impact of Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometric Evaluation on the Clinical Outcomes of Patients with Bacteremia and Fungemia in Clinical Settings Lacking an Antimicrobial Stewardship Program: A Pre-Post Quasi Experimental Study. BMC Infect. Dis. 2018, 18, 385. [Google Scholar] [CrossRef]
- Rindi, L.; Puglisi, V.; Franconi, I.; Fais, R.; Lupetti, A. Rapid and Accurate Identification of Nontuberculous Mycobacteria Directly from Positive Primary MGIT Cultures by MALDI-TOF MS. Microorganisms 2022, 10, 1447. [Google Scholar] [CrossRef] [PubMed]
- Yoon, E.-J.; Jeong, S.H. MALDI-TOF Mass Spectrometry Technology as a Tool for the Rapid Diagnosis of Antimicrobial Resistance in Bacteria. Antibiotics 2021, 10, 982. [Google Scholar] [CrossRef]
- Huang, A.M.; Newton, D.; Kunapuli, A.; Gandhi, T.N.; Washer, L.L.; Isip, J.; Collins, C.D.; Nagel, J.L. Impact of Rapid Organism Identification via Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Combined with Antimicrobial Stewardship Team Intervention in Adult Patients with Bacteremia and Candidemia. Clin. Infect. Dis. 2013, 57, 1237–1245. [Google Scholar] [CrossRef] [PubMed]
- Niwa, T.; Yonetamari, J.; Hayama, N.; Fujibayashi, A.; Ito-Takeichi, S.; Suzuki, K.; Ohta, H.; Niwa, A.; Tsuchiya, M.; Yamamoto, M.; et al. Clinical Impact of Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry Combined with Antimicrobial Stewardship Interventions in Patients with Bloodstream Infections in a Japanese Tertiary Hospital. Int. J. Clin. Pract. 2019, 73, e13332. [Google Scholar] [CrossRef] [PubMed]
- Kyaw, B.M.; Tudor Car, L.; van Galen, L.S.; van Agtmael, M.A.; Costelloe, C.E.; Ajuebor, O.; Campbell, J.; Car, J. Health Professions Digital Education on Antibiotic Management: Systematic Review and Meta-Analysis by the Digital Health Education Collaboration. J. Med. Internet Res. 2019, 21, e14984. [Google Scholar] [CrossRef]
- Yu, V.L.; Stoehr, G.P.; Starling, R.C.; Shogan, J.E. Empiric Antibiotic Selection by Physicians: Evaluation of Reasoning Strategies. Am. J. Med. Sci. 1991, 301, 165–172. [Google Scholar] [CrossRef]
- Faron, M.L.; Buchan, B.W.; Ledeboer, N.A. Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry for Use with Positive Blood Cultures: Methodology, Performance, and Optimization. J. Clin. Microbiol. 2017, 55, 3328–3338. [Google Scholar] [CrossRef] [PubMed]
- Patel, R. MALDI-TOF MS for the Diagnosis of Infectious Diseases. Clin. Chem. 2015, 61, 100–111. [Google Scholar] [CrossRef]
- Segawa, S.; Sawai, S.; Murata, S.; Nishimura, M.; Beppu, M.; Sogawa, K.; Watanabe, M.; Satoh, M.; Matsutani, T.; Kobayashi, M.; et al. Direct Application of MALDI-TOF Mass Spectrometry to Cerebrospinal Fluid for Rap-id Pathogen Identification in a Patient with Bacterial Meningitis. Clin Chim Acta 2014, 435, 59–61. [Google Scholar] [CrossRef]
- Lotte, R.; Courdurié, A.; Gaudart, A.; Emery, A.; Chevalier, A.; Tran, A.; Payen, M.; Ruimy, R. Spontaneous Bacterial Peritonitis: The Incremental Value of a Fast and Direct Bacterial Identification from Ascitic Fluids Inoculated in Blood Culture Bottles by MALDI-TOF MS for a Better Management of Patients. Microorganisms 2022, 10, 1188. [Google Scholar] [CrossRef]
- Guo, L.; Ye, L.; Zhao, Q.; Ma, Y.; Yang, J.; Luo, Y. Comparative Study of MALDI-TOF MS and VITEK 2 in Bacteria Identification. J. Thorac. Dis. 2014, 6, 534–538. [Google Scholar] [CrossRef]
- Torres, I.; Pinto, C.; Oltra, R.; Pascual, T.; Carbonell, N.; Colomina, J.; Tormo, M.; Albert, E.; Aguilar, G.; Solano, C.; et al. Early Adjustment of Empirical Antibiotic Therapy of Bloodstream Infections on the Basis of Direct Identification of Bacteria by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry and Gram Staining Results. J. Infect. Chemother. 2020, 26, 963–969. [Google Scholar] [CrossRef]
- Patel, T.S.; Kaakeh, R.; Nagel, J.L.; Newton, D.W.; Stevenson, J.G. Cost Analysis of Implementing Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry Plus Real-Time Antimicrobial Stewardship Intervention for Bloodstream Infections. J. Clin. Microbiol. 2017, 55, 60–67. [Google Scholar] [CrossRef]
- Paul, M.; Shani, V.; Muchtar, E.; Kariv, G.; Robenshtok, E.; Leibovici, L. Systematic Review and Meta-Analysis of the Efficacy of Appropriate Empiric Antibiotic Therapy for Sepsis. Antimicrob. Agents Chemother. 2010, 54, 4851–4863. [Google Scholar] [CrossRef]
- Mertz, D.; Koller, M.; Haller, P.; Lampert, M.L.; Plagge, H.; Hug, B.; Koch, G.; Battegay, M.; Flückiger, U.; Bassetti, S. Outcomes of Early Switching from Intravenous to Oral Antibiotics on Medical Wards. J. Antimicrob. Chemother. 2009, 64, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Pérez-López, A.; Elamin, N.; Nabor, R.; Dumindin, S.; Roscoe, D.; Hasan, M.R.; Suleiman, M.; Tang, P. Performance and Impact on Initial Antibiotic Choice of Direct Identification of Pathogens from Pediatric Blood Culture Bottles Using an In-House MALDI-TOF MS Protocol. Microbiol. Spectr. 2021, 9, e0190521. [Google Scholar] [CrossRef] [PubMed]
- Osthoff, M.; Gürtler, N.; Bassetti, S.; Balestra, G.; Marsch, S.; Pargger, H.; Weisser, M.; Egli, A. Impact of MALDI-TOF-MS-Based Identification Directly from Positive Blood Cultures on Patient Management: A Controlled Clinical Trial. Clin. Microbiol. Infect. 2017, 23, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Porreca Kratz, A.M.; Sullivan, K.V.; Gallagher, J.C. Clinical Impact of Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry for the Management of Inpatient Pneumonia without Additional Antimicrobial Stewardship Support. Infect. Control Hosp. Epidemiol. 2019, 40, 1053–1055. [Google Scholar] [CrossRef]
- Florio, W.; Tavanti, A.; Barnini, S.; Ghelardi, E.; Lupetti, A. Recent Advances and Ongoing Challenges in the Diagnosis of Microbial Infections by MALDI-TOF Mass Spectrometry. Front. Microbiol. 2018, 9, 1097. [Google Scholar] [CrossRef]
- Angeletti, S. Matrix Assisted Laser Desorption Time of Flight Mass Spectrometry (MALDI-TOF MS) in Clinical Microbiology. J. Microbiol. Methods 2017, 138, 20–29. [Google Scholar] [CrossRef]
- Bai, A.D.; Lo, C.K.L.; Komorowski, A.S.; Suresh, M.; Guo, K.; Garg, A.; Tandon, P.; Senecal, J.; Del Corpo, O.; Stefanova, I.; et al. Staphylococcus Aureus Bacteraemia Mortality: A Systematic Review and Meta-Analysis. Clin. Microbiol. Infect. 2022, 28, 1076–1084. [Google Scholar] [CrossRef]
- Lewis, P.O.; Heil, E.L.; Covert, K.L.; Cluck, D.B. Treatment Strategies for Persistent Methicillin-Resistant Staphylococcus Aureus Bacteraemia. J. Clin. Pharm. Ther. 2018, 43, 614–625. [Google Scholar] [CrossRef]
- Sánchez-Romero, M.I.; Moya, J.G.G.-L.; López, J.J.G.; Mira, N.O. Collection, Transport and General Processing of Clinical Specimens in Microbiology Laboratory. Enfermedades Infecc. Microbiol. Clin. (Engl. Ed.) 2019, 37, 127–134. [Google Scholar] [CrossRef] [PubMed]
- McCABE, W.R. Gram-Negative Bacteremia: I. Etiology and Ecology. Arch. Intern. Med. 1962, 110, 847. [Google Scholar] [CrossRef]
- Charlson, M.E.; Pompei, P.; Ales, K.L.; MacKenzie, C.R. A New Method of Classifying Prognostic Comorbidity in Longitudinal Studies: Development and Validation. J. Chronic Dis. 1987, 40, 373–383. [Google Scholar] [CrossRef] [PubMed]
| Control Group n = 183 | Intervention Group n = 180 | Total n = 363 | p-Value | |
|---|---|---|---|---|
| Mean age ± SD, years | 65.9 ± 16.5 | 65.5 ± 18.1 | 65.7 ± 17.3 | 0.85 |
| (range) | (5–96) | (0–97) | (0–97) | |
| Male Gender, no. (%) | 105 (57.4) | 118 (65.6) | 223 (61.4) | 0.11 |
| McCabe–Jackson, no. (%) | ||||
| Non-fatal | 53 (29) | 69 (38) | 122 (34) | 0.06 |
| Rapidly fatal | 25 (14) | 22 (12) | 47 (13) | 0.68 |
| Ultimately fatal | 105 (57) | 89 (49) | 194 (53) | 0.13 |
| Charlson index, median (Q1–Q3) | 4.2 (2–6) | 3.8 (1.25–6) | 4.0 (2–6) | 0.54 |
| Severity of Infection, no. (%) | ||||
| No Sepsis | 162 (88.5) | 167 (92.8) | 329 (90.6) | 0.16 |
| Sepsis | 12 (6.5) | 9 (5) | 21 (5.8) | 0.52 |
| Septic Shock | 9 (5) | 4 (2.2) | 13 (3.6) | 0.16 |
| Department in charge, no. (%) | ||||
| Medical | 139 (76.0) | 121 (67.2) | 260 (71.7) | 0.06 |
| Surgical | 22 (12.0) | 37 (20.6) | 59 (16.2) | 0.01 |
| Medical-surgical | 22 (12.0) | 22 (12.2) | 44 (12.1) | 0.95 |
| Place of acquisition, no. (%) | ||||
| Community | 57 (31.1) | 60 (33.3) | 117 (32.2) | 0.69 |
| Nosocomial | 75 (41) | 73 (40.6) | 148 (40.8) | 0.92 |
| Healthcare related | 51 (27.9) | 47 (26.1) | 98 (27) | 0.69 |
| Index positive culture, no. (%) | ||||
| Biological fluids * | 12 (6.6) | 9 (5) | 21 (5.8) | 0.55 |
| Biopsies | 6 (3.3) | 5 (2.8) | 11 (3) | 0.76 |
| Blood cultures | 28 (15.3) | 39 (21.7) | 67 (18.5) | 0.11 |
| Urine | 59 (32.2) | 56 (31.1) | 115 (31.7) | 0.84 |
| Respiratory | 48 (26.2) | 32 (17.7) | 80 (22) | 0.06 |
| Prosthesis | 2 (1.1) | 0 (0) | 2 (0.5) | 0.16 |
| Wounds and abscesses | 28 (15.3) | 39 (21.7) | 67 (18.5) | 0.11 |
| Infectious syndrome, no. (%) | ||||
| Endovascular infections | ||||
| Catheter site infection | 2 (1.1) | 1 (0.6) | 3 (0.8) | 0.57 |
| Endocarditis vascular | 2(1.1) | 0 (0) | 2 (0.6) | 0.16 |
| Abdominal | ||||
| Intraabdominal | 18 (9.8) | 13 (7.2) | 31 (8.5) | 0.37 |
| Biliary | 6 (3.3) | 4 (2.2) | 10 (2.8) | 0.55 |
| Respiratory | ||||
| Pneumonia | 23 (12.6) | 8 (4.4) | 31 (8.5) | 0.01 |
| Upper respiratory tract | 0 (0) | 2 (1.1) | 2 (0.6) | 0.15 |
| Lower respiratory tract (not pneumonia) | 29 (15.8) | 38 (21.1) | 67 (18.4) | 0.20 |
| Bone and joint | 2 (1.1) | 7 (3.9) | 9 (2.5) | 0.09 |
| Skin and soft tissue | 25 (13.7) | 40 (22.2) | 65 (17.9) | 0.03 |
| Central nervous system | 1 (0.5) | 1 (0.6) | 2 (0.6) | 0.76 |
| Urinary | 74 (40.4) | 63 (35.0) | 137 (37.7) | 0.28 |
| No focus | 1 (0.6) | 3 (1.7) | 4 (1.1) | 0.31 |
| Bacterial Isolate | Control Group n = 183 | Intervention Group n = 180 | p-Value | |
|---|---|---|---|---|
| Gram-positive | Staphylococcus aureus | 10 (4.5) | 26 (11.9) | 0.01 |
| CNS | 16 (7.2) | 7 (3.2) | 0.06 | |
| Enterococcus spp. | 40 (17.9) | 36 (16.4) | 0.67 | |
| Streptococcus spp. | 5 (2.2) | 6 (2.7) | 0.73 | |
| Other Gram-positive | 2 (0.9) | 3 (1.4) | 0.64 | |
| Anaerobes | 4 (1.8) | 6 (2.7) | 0.50 | |
| Gram-negative | Enterobacteriaceae | 104 (46.6) | 97 (44.3) | 0.62 |
| Non fermenter | 34 (15.2) | 34 (15.5) | 0.94 | |
| Other Gram-negative | 8 (3.6) | 4 (1.8) | 0.25 | |
| Total isolates | 223 | 220 |
| 0–10 h | 10–24 h | 24–36 h | 36–48 h | 48–96 h | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. (%) | CG | IG | p | CG | IG | p | CG | IG | p | CG | IG | p | CG | IG | p |
| Substitution | 31 | 29 | 0.83 | 21 | 22 | 0.82 | 5 | 6 | 0.74 | 12 | 9 | 0.52 | 5 | 11 | 0.11 |
| (16.9) | (16.1) | (11.5) | (12.2) | (2.7) | (3.3) | (6.6) | (5.0) | (2.8) | (6.1) | ||||||
| Instauration | 20 | 25 | 0.39 | 1 | 6 | 0.05 | 1 | 1 | 0.99 | 2 | 0 | 0.16 | 1 | 2 | 0.55 |
| (10.9) | (13.9) | (0.5) | (3.3) | (0.5) | (0.6) | (1.1) | (0.5) | (1.1) | |||||||
| Elimination | 12 | 14 | 0.65 | 5 | 6 | 0.74 | 4 | 0 | 0.05 | 2 | 1 | 0.57 | 4 | 5 | 0.42 |
| (6.6) | (7.8) | (2.7) | (3.3) | (2.2) | (1.1) | (0.6) | (2.2) | (2.8) | |||||||
| Addition | 18 | 6 | 0.01 | 7 | 2 | 0.09 | 1 | 0 | 0.32 | 5 | 2 | 0.26 | 3 | 1 | 0.32 |
| (9.8) | (3.3) | (3.8) | (1.1) | (0.5) | (2.7) | (1.1) | (1.6) | (0.6) | |||||||
| Total changes | 81 | 74 | 0.54 | 34 | 36 | 0.73 | 11 | 7 | 0.35 | 21 | 12 | 0.11 | 13 | 19 | 0.24 |
| (44.3) | (41.1) | (18.6) | (20.0) | (6.0) | (3.9) | (11.5) | (6.7) | (7.1) | (10.6) | ||||||
| No changes | 102 | 106 | 0.54 | 149 | 144 | 0.73 | 172 | 173 | 0.35 | 162 | 168 | 0.11 | 170 | 161 | 0.24 |
| (55.7) | (58.9) | (81.4) | (80.0) | (94.0) | (96.1) | (88.5) | (93.3) | (92.9) | (89.4) | ||||||
| Conversion to oral route | 3 | 7 | 0.19 | 4 | 2 | 0.42 | 1 | 1 | 0.99 | 1 | 2 | 0.55 | 1 | 3 | 0.31 |
| (1.6) | (3.8) | (2.2) | (1.1) | (0.5) | (0.5) | (0.5) | (1.1) | (0.5) | (3.8) | ||||||
| Control Group | Intervention Group | p-Value | |
|---|---|---|---|
| Patients in total say no. (%) | 183 (100) | 180 (100) | 1 |
| Length of the total, say, median ± SD (days) | 10.72 ± 16.69 | 11.14 ± 48.5 | 0.91 |
| Patients in CHU, no. (%) | 178 (97.3) | 172 (95.5) | 0.39 |
| Length of stay in CHU, median ± SD (days) | 9.75 ± 16.33 | 9.87 ± 48.24 | 0.97 |
| Patients in special hospitalization area, no. (%) | 16 (8.7) | 18 (10) | 0.68 |
| Length of stay in special hospitalization area, median ± SD (days) | 3.95 ± 5.49 | 6.21 ± 11.42 | 0.47 |
| Patients in ICU, no. (%) | 40 (21.8) | 26 (14.4) | 0.06 |
| Length of stay in ICU, median ± SD (days) | 2.81 ± 8.13 | 5.96 ± 13.47 | 0.29 |
| Patients with endotracheal intubation from IPC, no. (%) | 12 (6.5) | 12 (6.6) | 0.96 |
| Endotracheal intubation from IPC, median ± SD (days) | 3 ± 8.16 | 4.5 ± 6.34 | 0.62 |
| Patients with mechanical ventilation from IPC, no. (%) | 15 (8.1) | 15 (8.3) | 0.96 |
| Mechanical ventilation from IPC, median ± SD (days) | 27 ± 91.37 | 4 ± 40.25 | 0.38 |
| Inpatient mortality, no. (%) | 31 (17.2) | 21 (11.5) | 0.15 |
| Infection-related | 14 (7.8) | 8 (4.3) | 0.21 |
| 30-day mortality, no. (%) | 0 (0) | 3 (1.6) | 0.08 |
| Infection-related | 0 (0) | 2 (1.1) | 0.15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uzuriaga, M.; Leiva, J.; Guillén-Grima, F.; Rua, M.; Yuste, J.R. Clinical Impact of Rapid Bacterial Microbiological Identification with the MALDI-TOF MS. Antibiotics 2023, 12, 1660. https://doi.org/10.3390/antibiotics12121660
Uzuriaga M, Leiva J, Guillén-Grima F, Rua M, Yuste JR. Clinical Impact of Rapid Bacterial Microbiological Identification with the MALDI-TOF MS. Antibiotics. 2023; 12(12):1660. https://doi.org/10.3390/antibiotics12121660
Chicago/Turabian StyleUzuriaga, Miriam, José Leiva, Francisco Guillén-Grima, Marta Rua, and José R. Yuste. 2023. "Clinical Impact of Rapid Bacterial Microbiological Identification with the MALDI-TOF MS" Antibiotics 12, no. 12: 1660. https://doi.org/10.3390/antibiotics12121660






