The Identification of a Novel Spider Toxin Peptide, Lycotoxin-Pa2a, with Antibacterial and Anti-Inflammatory Activities
Abstract
:1. Introduction
2. Results
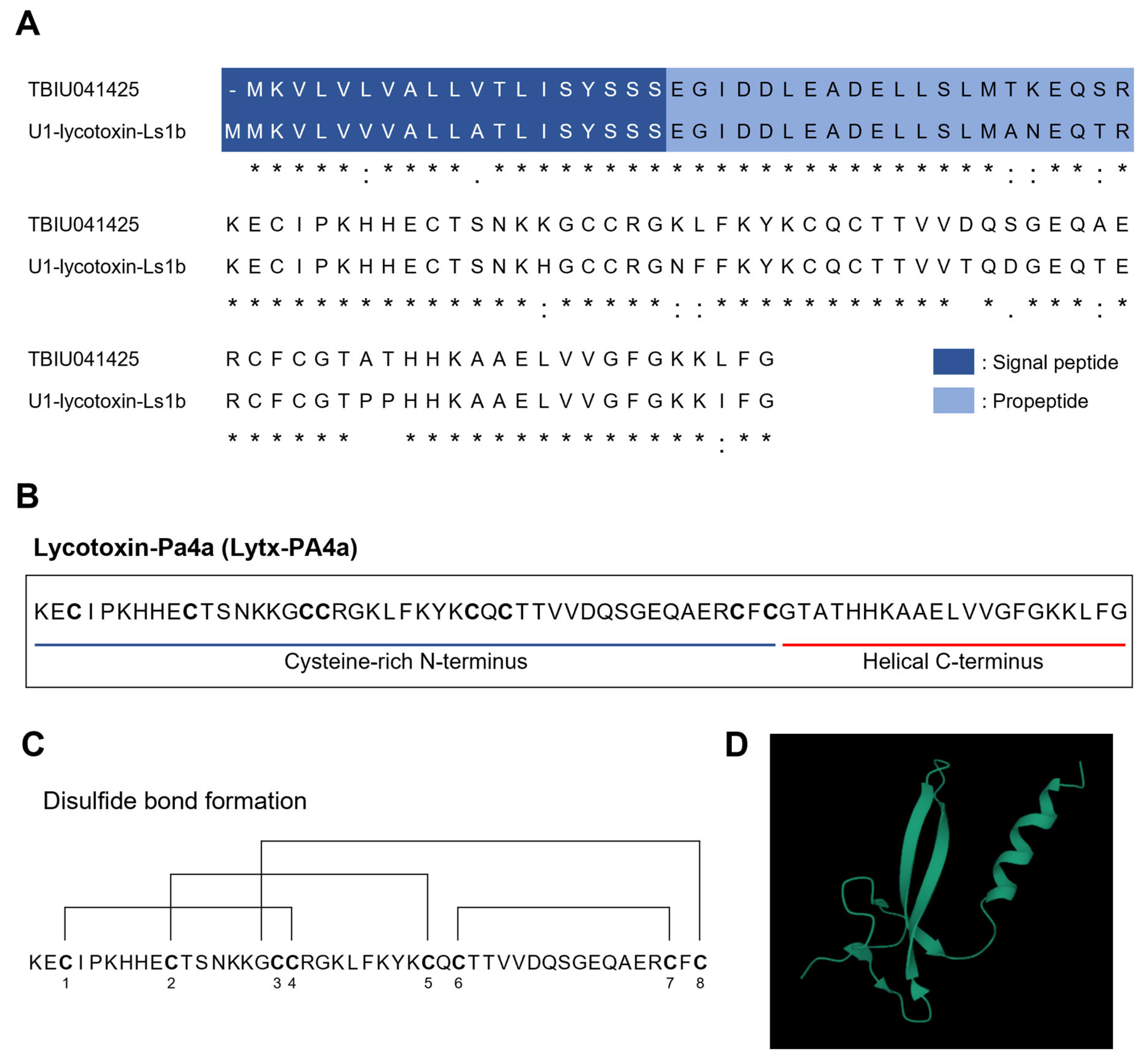
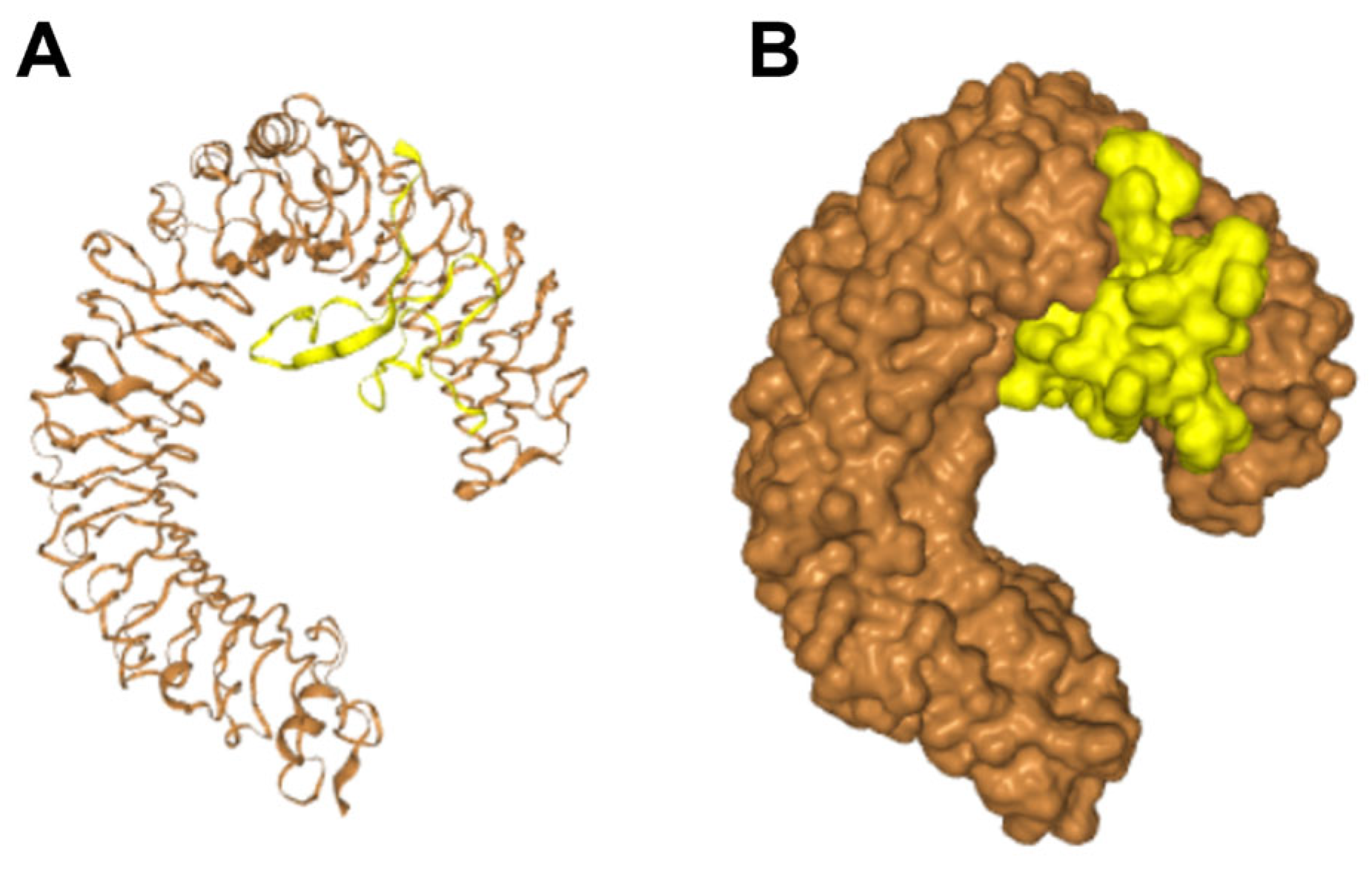
2.1. Identification of a Novel Toxin Peptide from the Transccriptome of Spider Pardosa astrigera
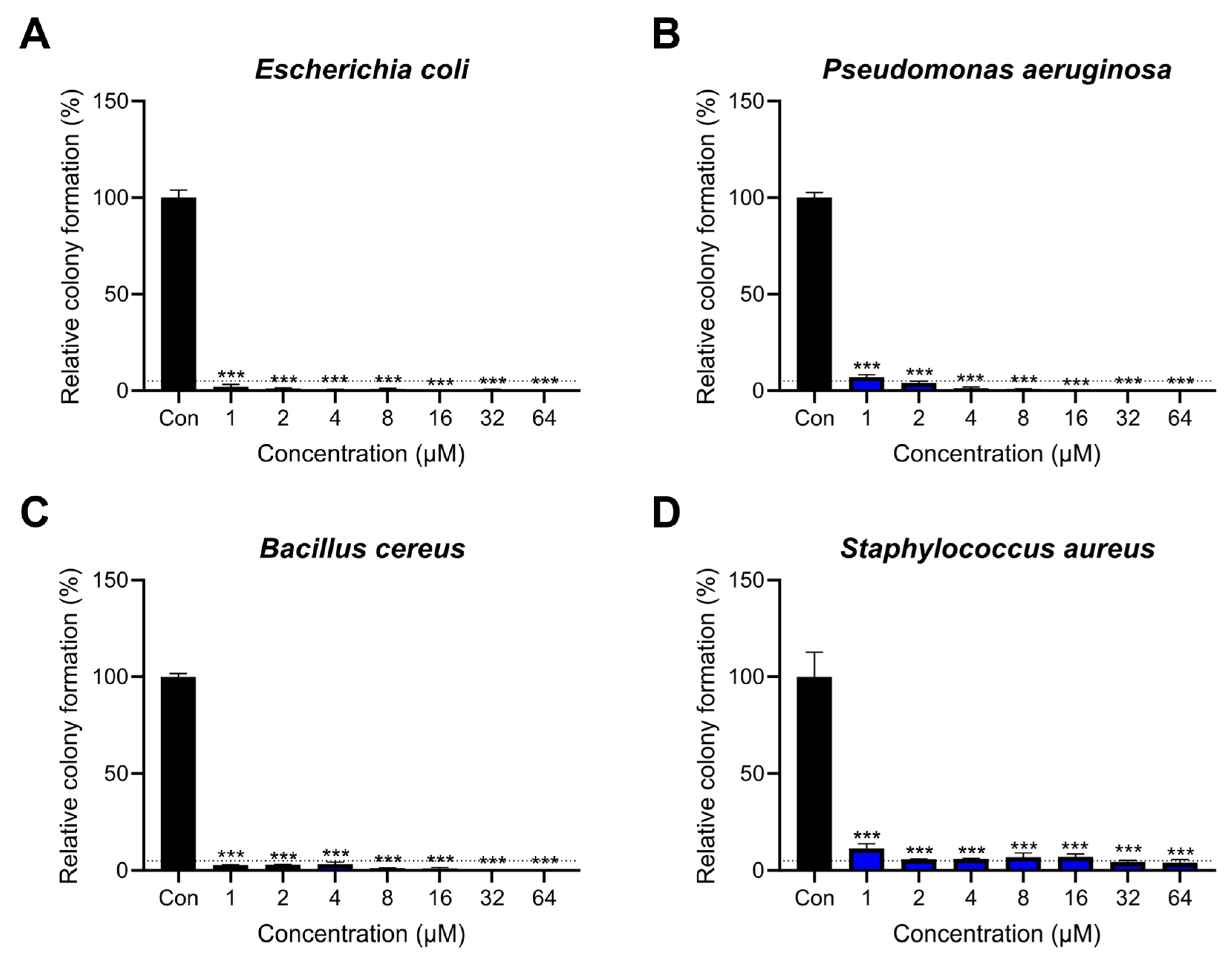
2.2. Antibacterial Activity of Lytx-Pa2a against Pathogenic Bacteria
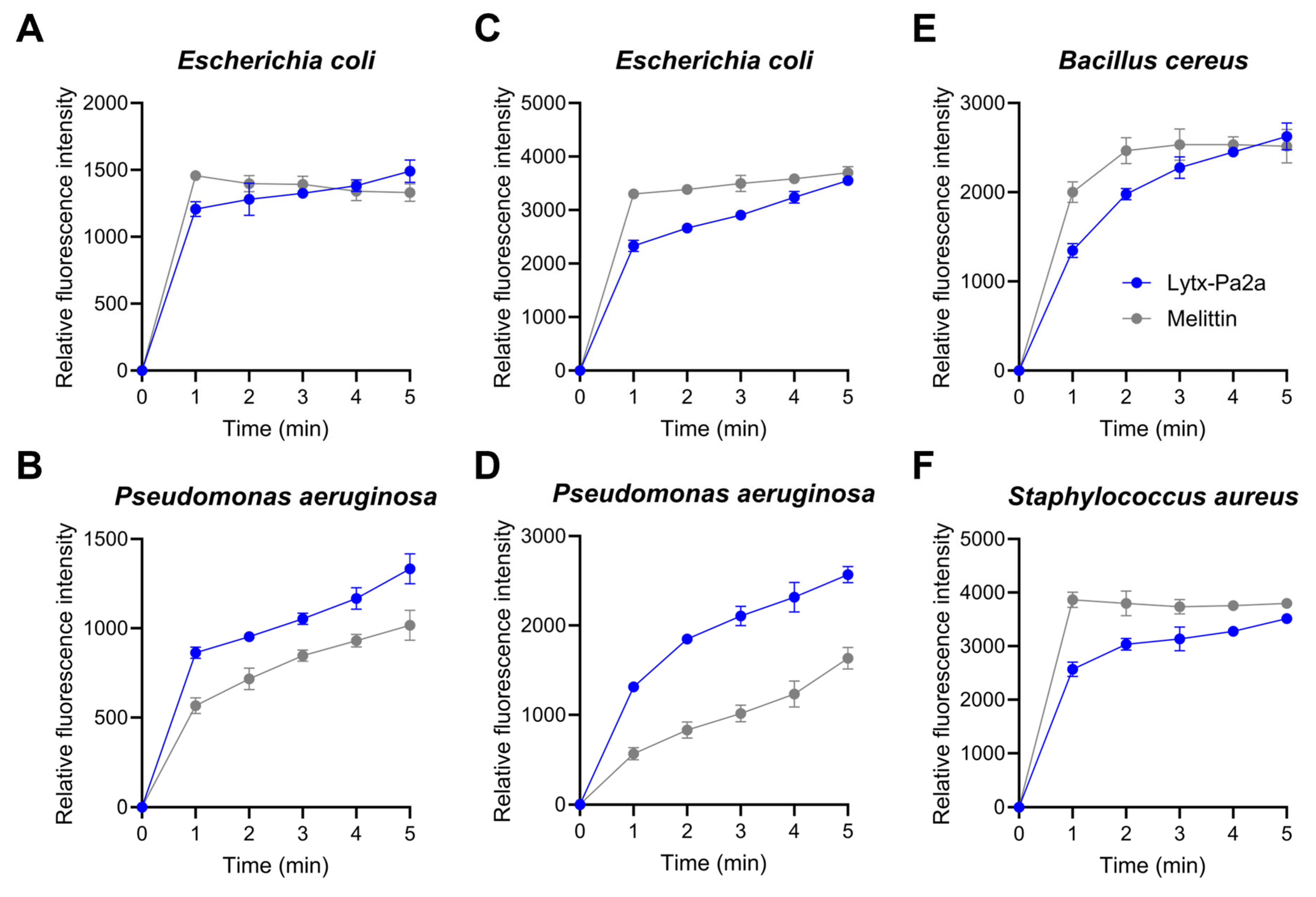
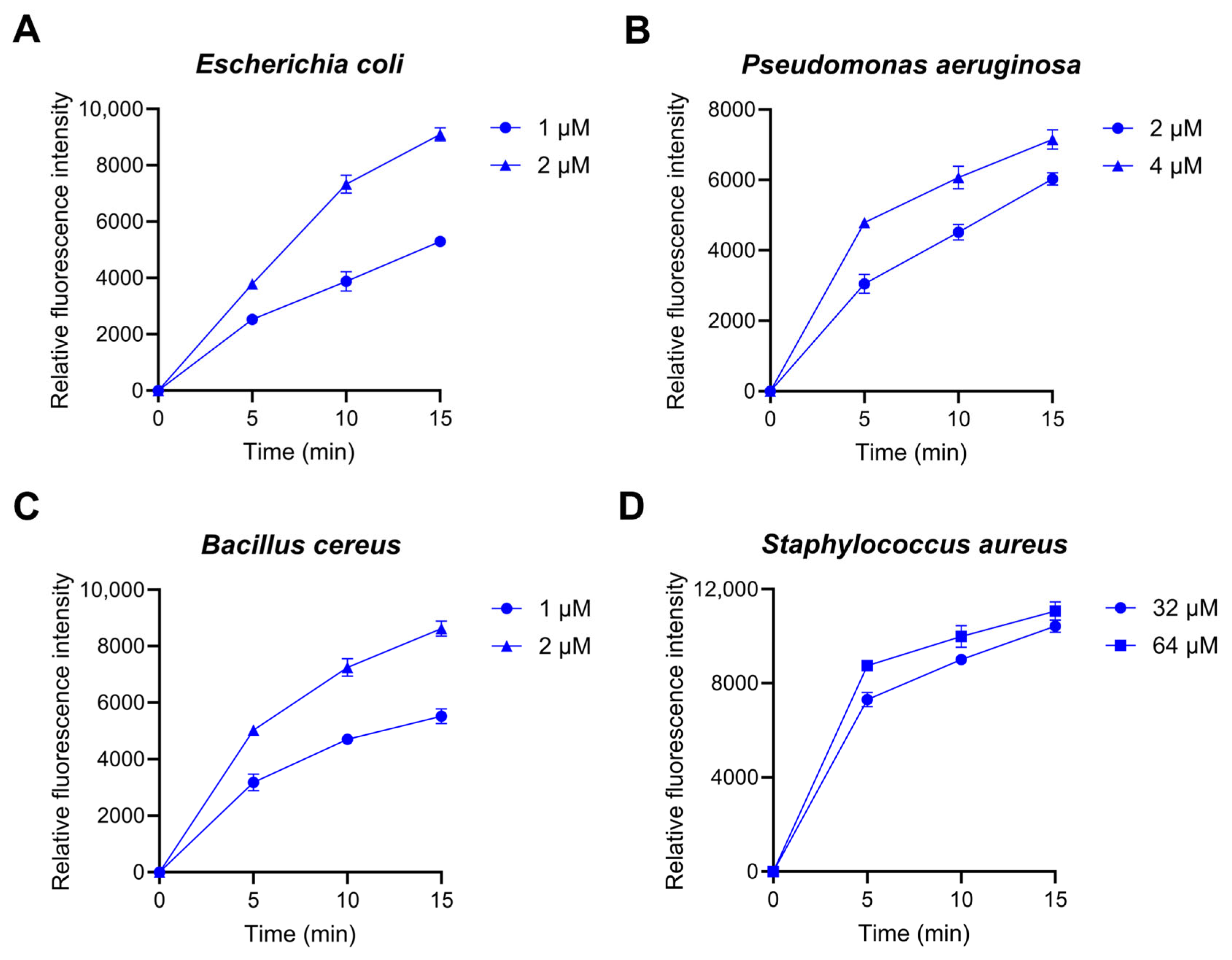
2.3. Bacterial Membrane Permeation and ROS Production by Lytx-Pa2a
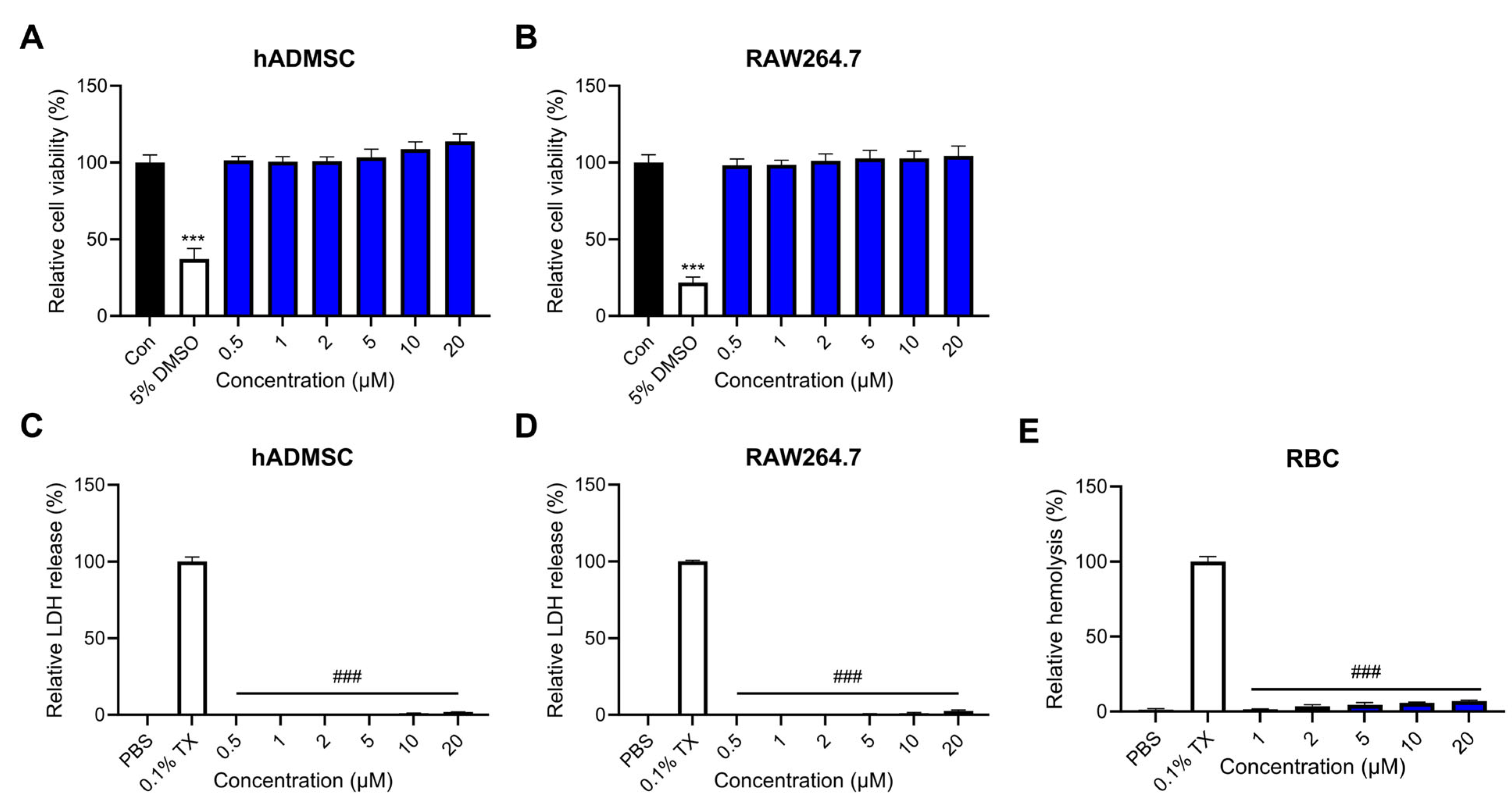
2.4. Effects of Lytx-Pa2a on Mammalian Cells
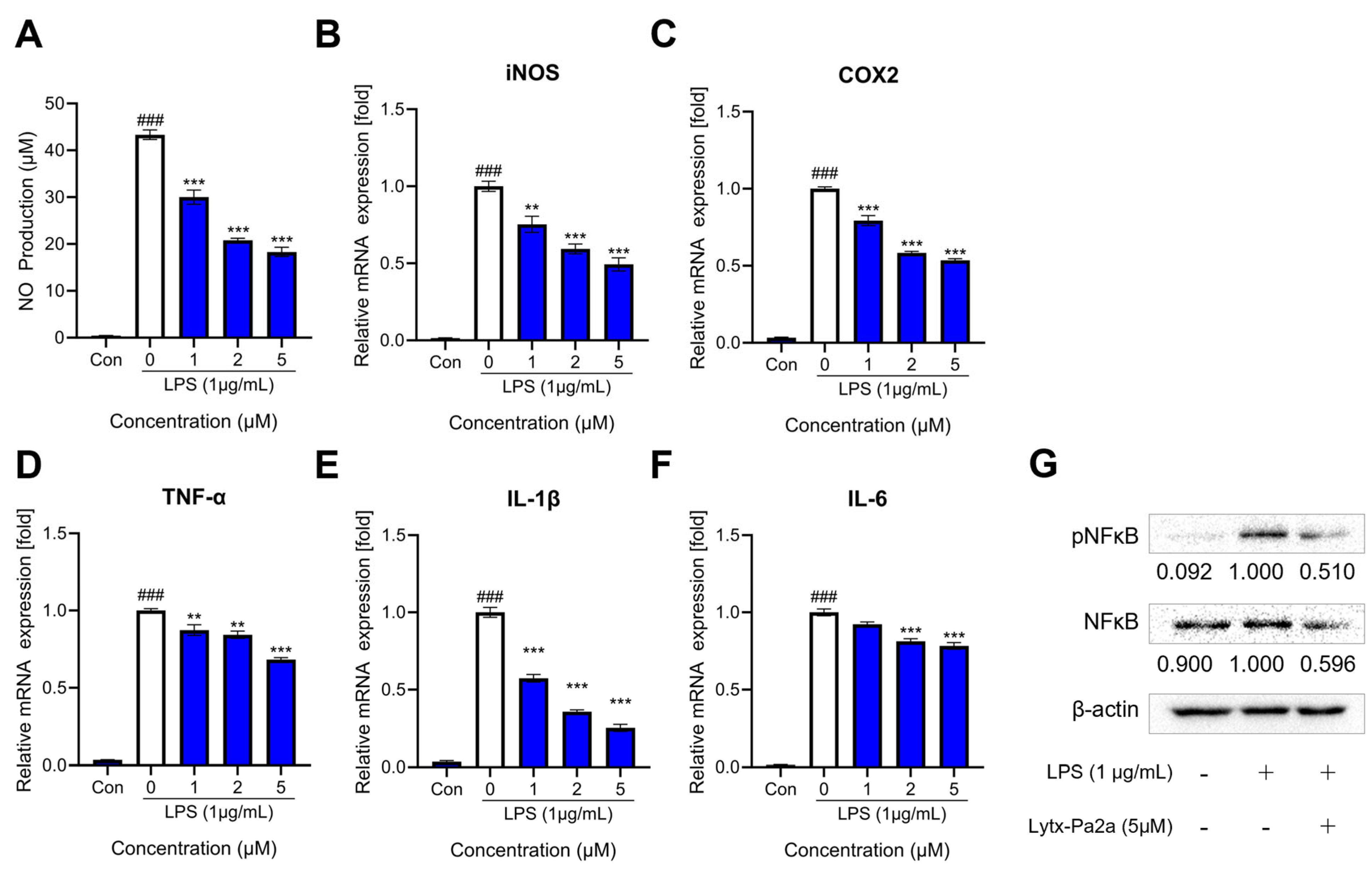
2.5. Inhibition of LPS-Induced Inflammation by Lytx-Pa2a
3. Discussion
4. Materials and Methods
4.1. In Silico Analysis of Peptide Sequence
4.2. Bacterial Strains and Cell Lines
4.3. Antimicrobial Activity Assay
4.4. Intrabacterial ROS Detection
4.5. Cell Viablility Assay and LDH-Release Assay
4.6. Hemolytic Activity Measurement
4.7. NO Quatification
4.8. RT-qPCR
4.9. Western Blot Analysis
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bowler, P.G. Antibiotic resistance and biofilm tolerance: A combined threat in the treatment of chronic infections. J. Wound Care 2018, 27, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Arnold, B.J.; Huang, I.T.; Hanage, W.P. Horizontal gene transfer and adaptive evolution in bacteria. Nat. Rev. Microbiol. 2022, 20, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and Barriers to, Horizontal Gene Transfer between Bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Adermann, K.; John, H.; Ständker, L.; Forssmann, W.-G. Exploiting natural peptide diversity: Novel research tools and drug leads. Curr. Opin. Biotechnol. 2004, 15, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Dutescu, I.A.; Hillier, S.A. Encouraging the Development of New Antibiotics: Are Financial Incentives the Right Way Forward? A Systematic Review and Case Study. Infect. Drug Resist. 2021, 14, 415–434. [Google Scholar] [CrossRef] [PubMed]
- Gould, I.M.; Bal, A.M. New antibiotic agents in the pipeline and how they can help overcome microbial resistance. Virulence 2013, 4, 185–191. [Google Scholar] [CrossRef]
- Pacios, O.; Blasco, L.; Bleriot, I.; Fernandez-Garcia, L.; González Bardanca, M.; Ambroa, A.; López, M.; Bou, G.; Tomás, M. Strategies to Combat Multidrug-Resistant and Persistent Infectious Diseases. Antibiotics 2020, 9, 65. [Google Scholar] [CrossRef] [PubMed]
- Utkin, Y.N. Animal venom studies: Current benefits and future developments. World J. Biol. Chem. 2015, 6, 28–33. [Google Scholar] [CrossRef]
- Zhang, Y. Why do we study animal toxins? Dongwuxue Yanjiu 2015, 36, 183–222. [Google Scholar] [CrossRef] [PubMed]
- Perumal Samy, R.; Stiles, B.G.; Franco, O.L.; Sethi, G.; Lim, L.H.K. Animal venoms as antimicrobial agents. Biochem. Pharmacol. 2017, 134, 127–138. [Google Scholar] [CrossRef]
- Schendel, V.; Rash, L.D.; Jenner, R.A.; Undheim, E.A.B. The Diversity of Venom: The Importance of Behavior and Venom System Morphology in Understanding Its Ecology and Evolution. Toxins 2019, 11, 666. [Google Scholar] [CrossRef]
- Zhang, Y.-Y.; Huang, Y.; He, Q.-Z.; Luo, J.; Zhu, L.; Lu, S.-S.; Liu, J.-Y.; Huang, P.-F.; Zeng, X.-Z.; Liang, S.-P. Structural and Functional Diversity of Peptide Toxins from Tarantula Haplopelma hainanum (Ornithoctonus hainana) Venom Revealed by Transcriptomic, Peptidomic, and Patch Clamp Approaches. J. Biol. Chem. 2015, 290, 14192–14207. [Google Scholar] [CrossRef] [PubMed]
- Dias, N.B.; de Souza, B.M.; Gomes, P.C.; Palma, M.S. Peptide diversity in the venom of the social wasp Polybia paulista (Hymenoptera): A comparison of the intra- and inter-colony compositions. Peptides 2014, 51, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Badr, G.; Hozzein, W.N.; Badr, B.M.; Al Ghamdi, A.; Saad Eldien, H.M.; Garraud, O. Bee Venom Accelerates Wound Healing in Diabetic Mice by Suppressing Activating Transcription Factor-3 (ATF-3) and Inducible Nitric Oxide Synthase (iNOS)-Mediated Oxidative Stress and Recruiting Bone Marrow-Derived Endothelial Progenitor Cells. J. Cell Physiol. 2016, 231, 2159–2171. [Google Scholar] [CrossRef] [PubMed]
- Guilhelmelli, F.; Vilela, N.; Smidt, K.S.; de Oliveira, M.A.; da Cunha Morales Álvares, A.; Rigonatto, M.C.L.; da Silva Costa, P.H.; Tavares, A.H.; Freitas, S.M.D.; Nicola, A.M.; et al. Activity of Scorpion Venom-Derived Antifungal Peptides against Planktonic Cells of Candida spp. and Cryptococcus neoformans and Candida albicans Biofilms. Front. Microbiol. 2016, 7, 1844. [Google Scholar] [CrossRef]
- Sousa, S.R.; McArthur, J.R.; Brust, A.; Bhola, R.F.; Rosengren, K.J.; Ragnarsson, L.; Dutertre, S.; Alewood, P.F.; Christie, M.J.; Adams, D.J.; et al. Novel analgesic ω-conotoxins from the vermivorous cone snail Conus moncuri provide new insights into the evolution of conopeptides. Sci. Rep. 2018, 8, 13397. [Google Scholar] [CrossRef]
- Minutti-Zanella, C.; Gil-Leyva, E.J.; Vergara, I. Immunomodulatory properties of molecules from animal venoms. Toxicon 2021, 191, 54–68. [Google Scholar] [CrossRef] [PubMed]
- Mohamed Abd El-Aziz, T.; Garcia Soares, A.; Stockand, J.D. Snake Venoms in Drug Discovery: Valuable Therapeutic Tools for Life Saving. Toxins 2019, 11, 564. [Google Scholar] [CrossRef]
- Messadi, E. Snake Venom Components as Therapeutic Drugs in Ischemic Heart Disease. Biomolecules 2023, 13, 1539. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.T.; Craik, D.J.; Kaas, Q. Bibliometric Review of the Literature on Cone Snail Peptide Toxins from 2000 to 2022. Mar. Drugs 2023, 21, 154. [Google Scholar] [CrossRef] [PubMed]
- Pennington, M.W.; Czerwinski, A.; Norton, R.S. Peptide therapeutics from venom: Current status and potential. Bioorg. Med. Chem. 2018, 26, 2738–2758. [Google Scholar] [CrossRef] [PubMed]
- King, G.F. Venoms as a platform for human drugs: Translating toxins into therapeutics. Expert Opin. Biol. Ther. 2011, 11, 1469–1484. [Google Scholar] [CrossRef]
- Baig, M.H.; Ahmad, K.; Saeed, M.; Alharbi, A.M.; Barreto, G.E.; Ashraf, G.M.; Choi, I. Peptide based therapeutics and their use for the treatment of neurodegenerative and other diseases. Biomed. Pharmacother. 2018, 103, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Benson, N.; Cucurull-Sanchez, L.; Demin, O.; Smirnov, S.; van der Graaf, P. Reducing Systems Biology to Practice in Pharmaceutical Company Research; Selected Case Studies. In Proceedings of the Advances in Systems Biology, 2012th ed.; Springer: New York, NY, USA, 2012; pp. 607–615. [Google Scholar]
- Chang, R.Y.K.; Nang, S.C.; Chan, H.-K.; Li, J. Novel antimicrobial agents for combating antibiotic-resistant bacteria. Adv. Drug Deliv. Rev. 2022, 187, 114378. [Google Scholar] [CrossRef] [PubMed]
- Hof, W.V.T.; Veerman, E.C.I.; Helmerhorst, E.J.; Amerongen, A.V.N. Antimicrobial Peptides: Properties and Applicability; Walter de Gruyter: Berlin, Germnay, 2001; Volume 382, pp. 597–619. [Google Scholar] [CrossRef]
- Vamathevan, J.; Clark, D.; Czodrowski, P.; Dunham, I.; Ferran, E.; Lee, G.; Li, B.; Madabhushi, A.; Shah, P.; Spitzer, M.; et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 2019, 18, 463–477. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Srivastava, D.; Sahu, M.; Tiwari, S.; Ambasta, R.K.; Kumar, P. Artificial intelligence to deep learning: Machine intelligence approach for drug discovery. Mol. Divers. 2021, 25, 1315–1360. [Google Scholar] [CrossRef] [PubMed]
- Mutz, K.-O.; Heilkenbrinker, A.; Lönne, M.; Walter, J.-G.; Stahl, F. Transcriptome analysis using next-generation sequencing. Curr. Opin. Biotechnol. 2013, 24, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Bordon, K.C.F.; Cologna, C.T.; Fornari-Baldo, E.C.; Pinheiro-Júnior, E.L.; Cerni, F.A.; Amorim, F.G.; Anjolette, F.A.P.; Cordeiro, F.A.; Wiezel, G.A.; Cardoso, I.A.; et al. From Animal Poisons and Venoms to Medicines: Achievements, Challenges and Perspectives in Drug Discovery. Front. Pharmacol. 2020, 11, 1132. [Google Scholar] [CrossRef] [PubMed]
- Nayfach, S.; Pollard, K.S. Toward Accurate and Quantitative Comparative Metagenomics. Cell 2016, 166, 1103–1116. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.K.; Lee, B.; Kim, S.T.; Yoo, J.S.; Sung, J.S. Designing a Novel Functional Peptide with Dual Antimicrobial and Anti-inflammatory Activities via in Silico Methods. Front. Immunol. 2022, 13, 821070. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, N.; Yapp, E.K.Y.; Le, N.Q.K.; Kamaraj, B.; Al-Subaie, A.M.; Yeh, H.-Y. Application of Computational Biology and Artificial Intelligence Technologies in Cancer Precision Drug Discovery. BioMed Res. Int. 2019, 2019, 8427042. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.K.; Hwang, I.-W.; Kim, Y.; Kim, S.T.; Jang, W.; Lee, S.; Bang, W.Y.; Bae, C.-H.; Sung, J.-S. Antibacterial and Anti-Inflammatory Effects of Novel Peptide Toxin from the Spider Pardosa astrigera. Antibiotics 2020, 9, 422. [Google Scholar] [CrossRef] [PubMed]
- King, G.F.; Gentz, M.C.; Escoubas, P.; Nicholson, G.M. A rational nomenclature for naming peptide toxins from spiders and other venomous animals. Toxicon 2008, 52, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Jiang, C. Antimicrobial peptides: Structure, mechanism, and modification. Eur. J. Med. Chem. 2023, 255, 115377. [Google Scholar] [CrossRef]
- Memariani, H.; Memariani, M.; Shahidi-Dadras, M.; Nasiri, S.; Akhavan, M.M.; Moravvej, H. Melittin: From honeybees to superbugs. Appl. Microbiol. Biotechnol. 2019, 103, 3265–3276. [Google Scholar] [CrossRef] [PubMed]
- Askari, P.; Namaei, M.H.; Ghazvini, K.; Hosseini, M. In vitro and in vivo toxicity and antibacterial efficacy of melittin against clinical extensively drug-resistant bacteria. BMC Pharmacol. Toxicol. 2021, 22, 42. [Google Scholar] [CrossRef]
- Benfield, A.H.; Henriques, S.T. Mode-of-Action of Antimicrobial Peptides: Membrane Disruption vs. Intracellular Mechanisms. Front. Med. Technol. 2020, 2, 610997. [Google Scholar] [CrossRef]
- Kang, H.K.; Seo, C.H.; Luchian, T.; Park, Y. Pse-T2, an Antimicrobial Peptide with High-Level, Broad-Spectrum Antimicrobial Potency and Skin Biocompatibility against Multidrug-Resistant Pseudomonas aeruginosa Infection. Antimicrob. Agents Chemother. 2018, 62, e01493-18. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Zhang, F.; Yao, J.; Zhu, Y.; Zhu, N.; Zhang, J.; Ouyang, X.; Zhang, T.; Li, B.; Xie, J.; et al. New Antimicrobial Peptides with Repeating Unit against Multidrug-Resistant Bacteria. ACS Infect. Dis. 2021, 7, 1619–1637. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Kawasaki, T.; Kawai, T. Toll-Like Receptor Signaling Pathways. Front. Immunol. 2014, 5, 461. [Google Scholar] [CrossRef] [PubMed]
- Ain, Q.U.; Batool, M.; Choi, S. TLR4-Targeting Therapeutics: Structural Basis and Computer-Aided Drug Discovery Approaches. Molecules 2020, 25, 627. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, N.; Zhang, W.; Cheng, X.; Yan, Z.; Shao, G.; Wang, X.; Wang, R.; Fu, C. Therapeutic peptides: Current applications and future directions. Signal Transduct. Target. Ther. 2022, 7, 48. [Google Scholar] [CrossRef] [PubMed]
- Yacoub, T.; Rima, M.; Karam, M.; Sabatier, J.-M.; Fajloun, Z. Antimicrobials from Venomous Animals: An Overview. Molecules 2020, 25, 2402. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.C.-L.; Harris, J.L.; Khanna, K.K.; Hong, J.-H. A Comprehensive Review on Current Advances in Peptide Drug Development and Design. Int. J. Mol. Sci. 2019, 20, 2383. [Google Scholar] [CrossRef]
- Lüddecke, T.; Herzig, V.; von Reumont, B.M.; Vilcinskas, A. The biology and evolution of spider venoms. Biol. Rev. 2022, 97, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Saez, N.J.; Senff, S.; Jensen, J.E.; Er, S.Y.; Herzig, V.; Rash, L.D.; King, G.F. Spider-venom peptides as therapeutics. Toxins 2010, 2, 2851–2871. [Google Scholar] [CrossRef]
- Agwa, A.J.; Tran, P.; Mueller, A.; Tran, H.N.T.; Deuis, J.R.; Israel, M.R.; McMahon, K.L.; Craik, D.J.; Vetter, I.; Schroeder, C.I. Manipulation of a spider peptide toxin alters its affinity for lipid bilayers and potency and selectivity for voltage-gated sodium channel subtype 1.7. J. Biol. Chem. 2020, 295, 5067–5080. [Google Scholar] [CrossRef] [PubMed]
- Kuhn-Nentwig, L.; Müller, J.; Schaller, J.; Walz, A.; Dathe, M.; Nentwig, W. Cupiennin 1, a New Family of Highly Basic Antimicrobial Peptides in the Venom of the Spider Cupiennius salei(Ctenidae)*. J. Biol. Chem. 2002, 277, 11208–11216. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Adams, M.E. Lycotoxins, antimicrobial peptides from venom of the wolf spider Lycosa carolinensis. J. Biol. Chem. 1998, 273, 2059–2066. [Google Scholar] [CrossRef]
- Zhang, Q.-Y.; Yan, Z.-B.; Meng, Y.-M.; Hong, X.-Y.; Shao, G.; Ma, J.-J.; Cheng, X.-R.; Liu, J.; Kang, J.; Fu, C.-Y. Antimicrobial peptides: Mechanism of action, activity and clinical potential. Mil. Med. Res. 2021, 8, 48. [Google Scholar] [CrossRef]
- Laverty, G.; Gorman, S.P.; Gilmore, B.F. The Potential of Antimicrobial Peptides as Biocides. Int. J. Mol. Sci. 2011, 12, 6566–6596. [Google Scholar] [CrossRef]
- Seixas, A.F.; Quendera, A.P.; Sousa, J.P.; Silva, A.F.Q.; Arraiano, C.M.; Andrade, J.M. Bacterial Response to Oxidative Stress and RNA Oxidation. Front. Genet. 2022, 12, 821535. [Google Scholar] [CrossRef] [PubMed]
- Diamond, G.; Beckloff, N.; Weinberg, A.; Kisich, K. The Roles of Antimicrobial Peptides in Innate Host Defense. Curr. Pharm. Des. 2009, 15, 2377–2392. [Google Scholar] [CrossRef] [PubMed]
- Buccini, D.F.; Roriz, B.C.; Rodrigues, J.M.; Franco, O.L. Antimicrobial peptides could antagonize uncontrolled inflammation via Toll-like 4 receptor. Front. Bioeng. Biotechnol. 2022, 10, 1037147. [Google Scholar] [CrossRef] [PubMed]
- Robles-Loaiza, A.A.; Pinos-Tamayo, E.A.; Mendes, B.; Ortega-Pila, J.A.; Proaño-Bolaños, C.; Plisson, F.; Teixeira, C.; Gomes, P.; Almeida, J.R. Traditional and Computational Screening of Non-Toxic Peptides and Approaches to Improving Selectivity. Pharmaceuticals 2022, 15, 323. [Google Scholar] [CrossRef]
- Mhlongo, J.T.; Waddad, A.Y.; Albericio, F.; de la Torre, B.G. Antimicrobial Peptide Synergies for Fighting Infectious Diseases. Adv. Sci. 2023, 10, 2300472. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Pineda, S.S.; Chaumeil, P.-A.; Kunert, A.; Kaas, Q.; Thang, M.W.C.; Le, L.; Nuhn, M.; Herzig, V.; Saez, N.J.; Cristofori-Armstrong, B.; et al. ArachnoServer 3.0: An online resource for automated discovery, analysis and annotation of spider toxins. Bioinformatics 2017, 34, 1074–1076. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.-Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Lee, H.-T.; Lee, C.-C.; Yang, J.-R.; Lai, J.Z.C.; Chang, K.Y. A Large-Scale Structural Classification of Antimicrobial Peptides. BioMed Res. Int. 2015, 2015, 475062. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Lv, H.; Guo, Y.; Peng, W.; Liu, B. sAMPpred-GAT: Prediction of antimicrobial peptide by graph attention network and predicted peptide structure. Bioinformatics 2022, 39, btac715. [Google Scholar] [CrossRef] [PubMed]
- Burdukiewicz, M.; Sidorczuk, K.; Rafacz, D.; Pietluch, F.; Chilimoniuk, J.; Rödiger, S.; Gagat, P. Proteomic Screening for Prediction and Design of Antimicrobial Peptides with AmpGram. Int. J. Mol. Sci. 2020, 21, 4310. [Google Scholar] [CrossRef] [PubMed]
- Khatun, M.S.; Hasan, M.M.; Kurata, H. PreAIP: Computational Prediction of Anti-inflammatory Peptides by Integrating Multiple Complementary Features. Front. Genet. 2019, 10, 129. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Lv, H.; Wen, J.; Guo, Y.; Xu, Y.; Liu, B. PreTP-Stack: Prediction of Therapeutic Peptides Based on the Stacked Ensemble Learing. IEEE/ACM Trans. Comput. Biol. Bioinform. 2023, 20, 1337–1344. [Google Scholar] [CrossRef] [PubMed]
- Pirtskhalava, M.; Amstrong, A.A.; Grigolava, M.; Chubinidze, M.; Alimbarashvili, E.; Vishnepolsky, B.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M. DBAASP v3: Database of antimicrobial/cytotoxic activity and structure of peptides as a resource for development of new therapeutics. Nucleic Acids Res. 2020, 49, D288–D297. [Google Scholar] [CrossRef]
- Swedan, S.; Shubair, Z.; Almaaytah, A. Synergism of cationic antimicrobial peptide WLBU2 with antibacterial agents against biofilms of multi-drug resistant Acinetobacter baumannii and Klebsiella pneumoniae. Infect. Drug Resist. 2019, 12, 2019–2030. [Google Scholar] [CrossRef]
- Dwivedi, S.; Wahab, R.; Khan, F.; Mishra, Y.K.; Musarrat, J.; Al-Khedhairy, A.A. Reactive Oxygen Species Mediated Bacterial Biofilm Inhibition via Zinc Oxide Nanoparticles and Their Statistical Determination. PLoS ONE 2014, 9, e111289. [Google Scholar] [CrossRef] [PubMed]
- Jang, B.-Y.; Shin, M.K.; Han, D.-H.; Sung, J.-S. Curcumin Disrupts a Positive Feedback Loop between ADMSCs and Cancer Cells in the Breast Tumor Microenvironment via the CXCL12/CXCR4 Axis. Pharmaceutics 2023, 15, 2627. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.; Ma, Q.; Shan, A.; Lv, Y.; Hu, W.; Gu, Y.; Li, Y. Strand Length-Dependent Antimicrobial Activity and Membrane-Active Mechanism of Arginine- and Valine-Rich β-Hairpin-Like Antimicrobial Peptides. Antimicrob. Agents Chemother. 2012, 56, 2994–3003. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.-H.; Park, T.; Kang, M.-G.; Park, D. Anti-Inflammatory Activity and ROS Regulation Effect of Sinapaldehyde in LPS-Stimulated RAW 264.7 Macrophages. Molecules 2020, 25, 4089. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Jang, H.-J.; Baek, S.-Y.; Choi, K.-J.; Han, D.-H.; Sung, J.-S. Regulation of base excision repair during adipogenesis and osteogenesis of bone marrow-derived mesenchymal stem cells. Sci. Rep. 2023, 13, 16384. [Google Scholar] [CrossRef] [PubMed]
| Antimicrobial Activity | Anti-Inflammatory Activity | Hemolytic Activity | |||
|---|---|---|---|---|---|
| ADAM | sAMPred-GAT | AmpGram | PreAIP | PreTP-Stack | DBAASP 1 |
| 0.7 (AMP) | AMP | 0.9894 | 0.589 (AIP) | AIP | Not active |
| MIC (μM) | E. coli | P. aeruginosa | B. cereus | S. aureus |
|---|---|---|---|---|
| Lytx-Pa2a | 1 | 1 | 1 | 8 |
| Melittin | 1 | 8 | 1 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, M.K.; Hwang, I.-W.; Jang, B.-Y.; Bu, K.-B.; Han, D.-H.; Lee, S.-H.; Oh, J.W.; Yoo, J.S.; Sung, J.-S. The Identification of a Novel Spider Toxin Peptide, Lycotoxin-Pa2a, with Antibacterial and Anti-Inflammatory Activities. Antibiotics 2023, 12, 1708. https://doi.org/10.3390/antibiotics12121708
Shin MK, Hwang I-W, Jang B-Y, Bu K-B, Han D-H, Lee S-H, Oh JW, Yoo JS, Sung J-S. The Identification of a Novel Spider Toxin Peptide, Lycotoxin-Pa2a, with Antibacterial and Anti-Inflammatory Activities. Antibiotics. 2023; 12(12):1708. https://doi.org/10.3390/antibiotics12121708
Chicago/Turabian StyleShin, Min Kyoung, In-Wook Hwang, Bo-Young Jang, Kyung-Bin Bu, Dong-Hee Han, Seung-Ho Lee, Jin Wook Oh, Jung Sun Yoo, and Jung-Suk Sung. 2023. "The Identification of a Novel Spider Toxin Peptide, Lycotoxin-Pa2a, with Antibacterial and Anti-Inflammatory Activities" Antibiotics 12, no. 12: 1708. https://doi.org/10.3390/antibiotics12121708





