Comparative Meropenem Pharmacodynamics and Emergence of Resistance against Carbapenem-Susceptible Non-Carbapenemase-Producing and Carbapenemase-Producing Enterobacterales: A Pharmacodynamic Study in a Hollow-Fiber Infection Model
Abstract
:1. Introduction
2. Results
2.1. Transconjugant and Mutant K. pneumoniae and E. coli Strains, Meropenem Susceptibility at Standard and High Inocula
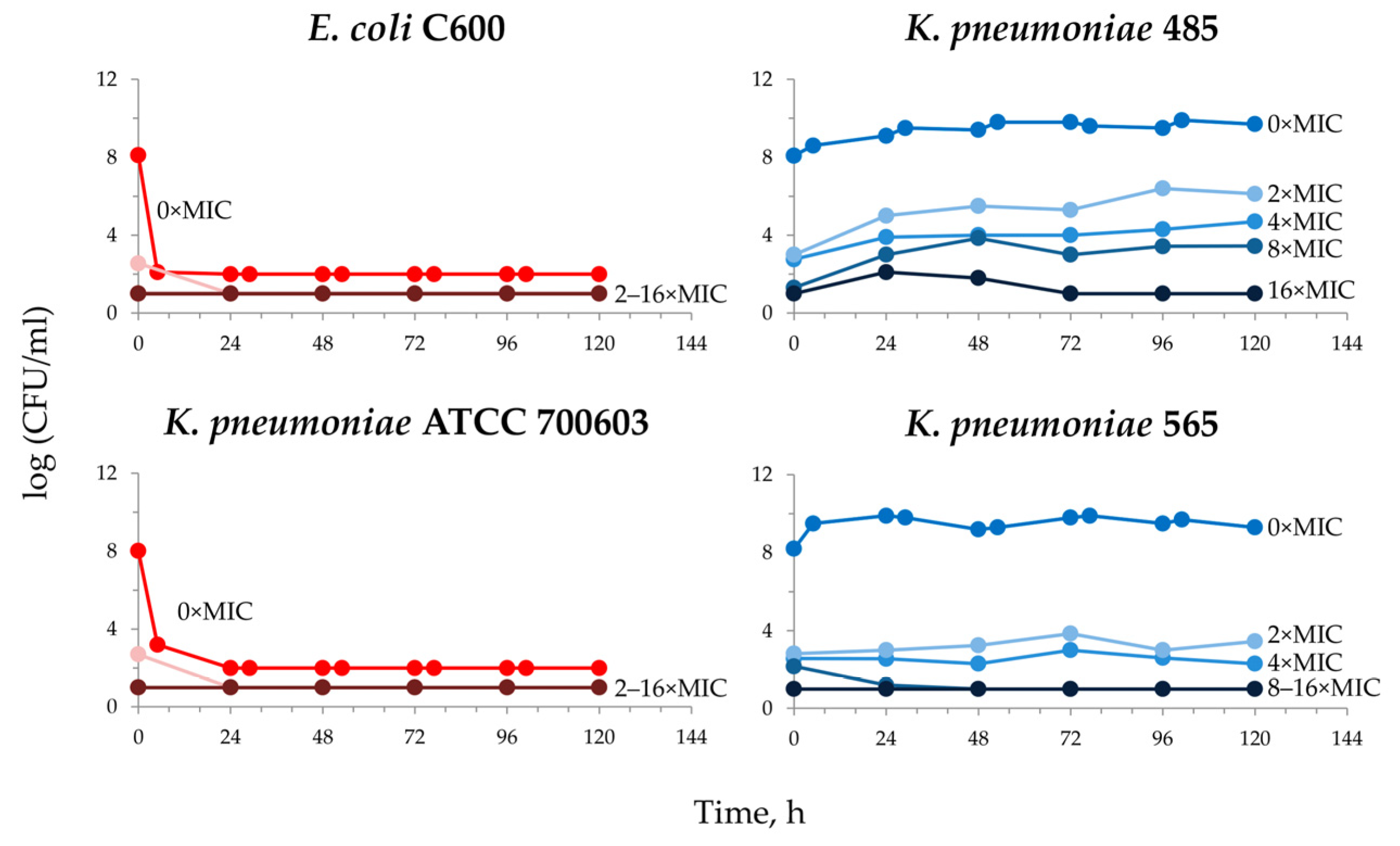
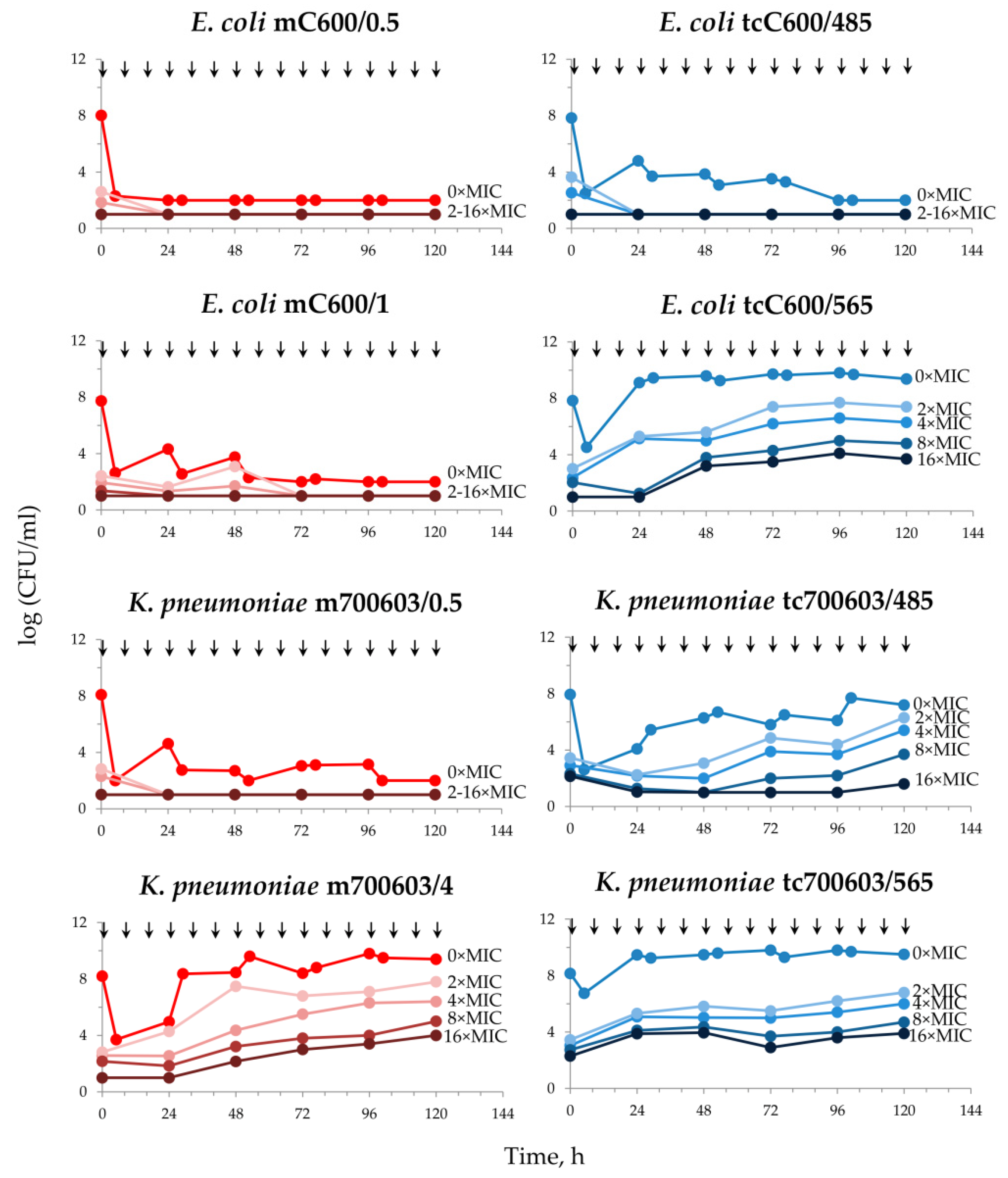
2.2. Meropenem Pharmacodynamics with E. coli and K. pneumoniae Total and Resistant Subpopulations
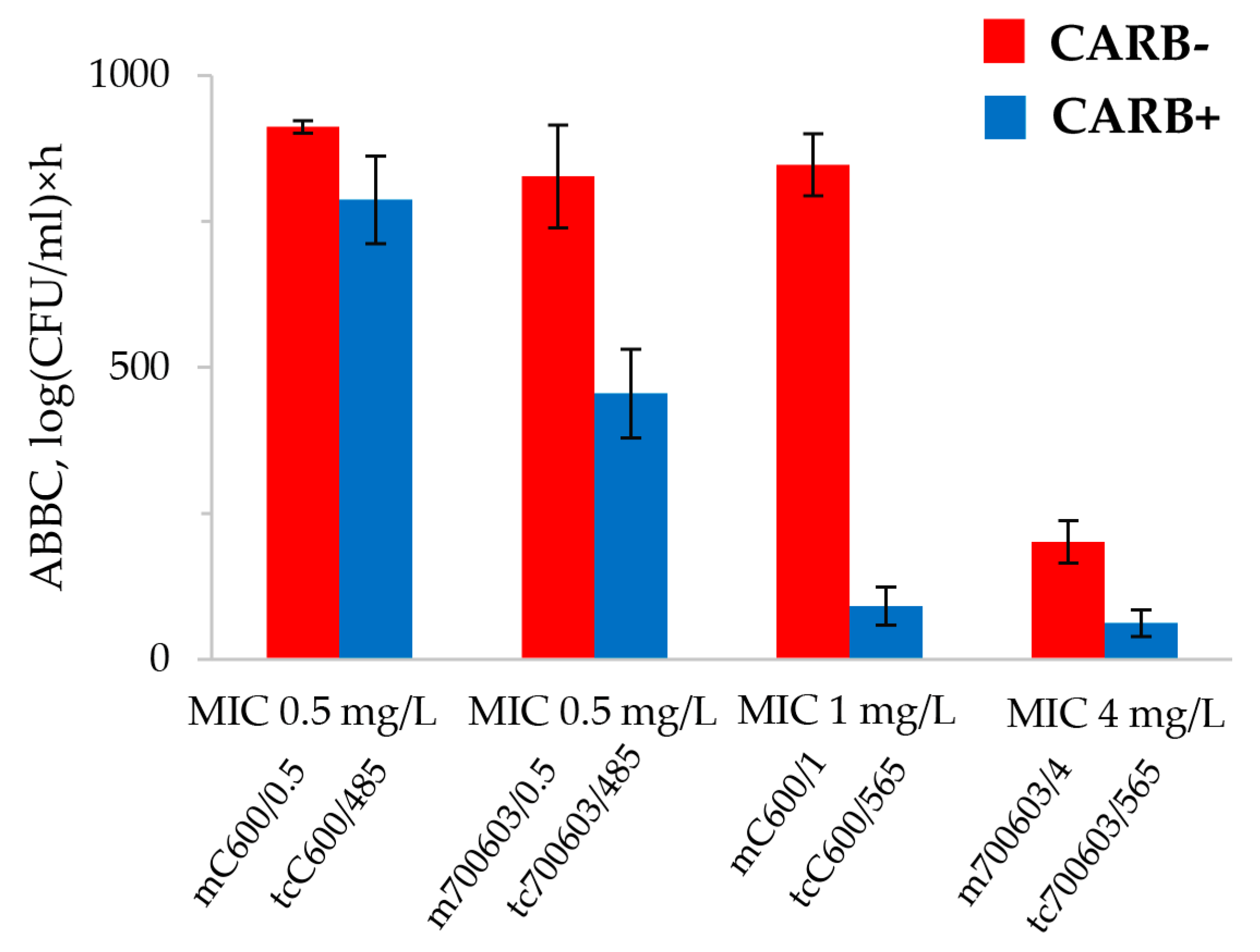
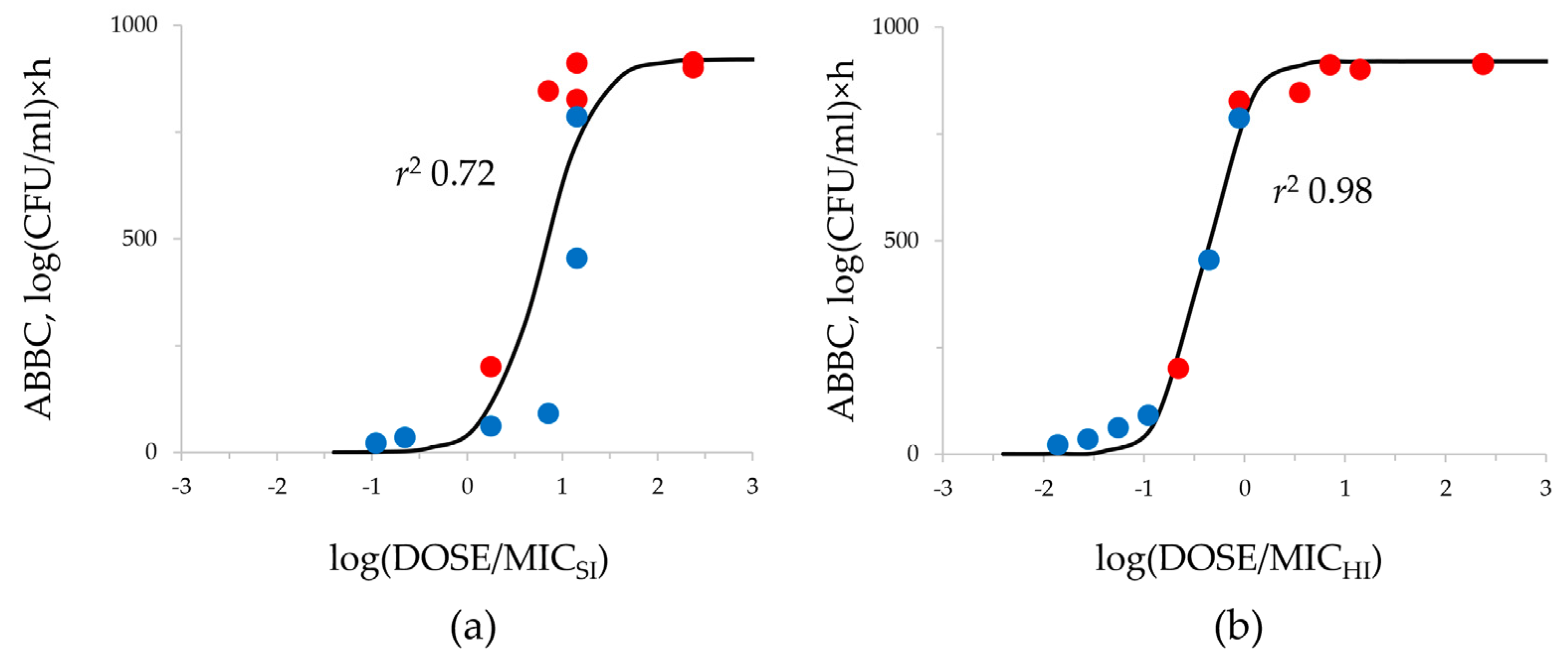
2.3. DOSE/MIC Relationships with the Meropenem Effect and Emergence of Resistance
3. Discussion
4. Materials and Methods
4.1. Antimicrobial Agent, Bacterial Isolates, and Susceptibility Testing
4.2. Mating Experiments
4.3. Resistance Selection Studies under Static Conditions
4.4. In Vitro Dynamic Model and Operational Procedure Used in the Pharmacodynamic Experiments
4.5. Antibiotic Dosing Regimens and Simulated Pharmacokinetic Profiles
4.6. Quantitation of the Antimicrobial Effect
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lai, C.-C.; Yu, W.-L. Klebsiella pneumoniae Harboring Carbapenemase Genes in Taiwan: Its Evolution over 20 Years, 1998–2019. Int. J. Antimicrob. Agents 2021, 58, 106354. [Google Scholar] [CrossRef] [PubMed]
- Ragheb, S.M.; Tawfick, M.M.; El-Kholy, A.A.; Abdulall, A.K. Phenotypic and Genotypic Features of Klebsiella pneumoniae Harboring Carbapenemases in Egypt: OXA-48-Like Carbapenemases as an Investigated Model. Antibiotics 2020, 9, 852. [Google Scholar] [CrossRef]
- Raro, O.H.F.; da Silva, R.M.C.; Filho, E.M.R.; Sukiennik, T.C.T.; Stadnik, C.; Dias, C.A.G.; Iglesias, J.O.; Pérez-Vázquez, M. Carbapenemase-Producing Klebsiella pneumoniae From Transplanted Patients in Brazil: Phylogeny, Resistome, Virulome and Mobile Genetic Elements Harboring blaKPC–2 or blaNDM–1. Front. Microbiol. 2020, 11, 1563. [Google Scholar] [CrossRef] [PubMed]
- Michaelis, C.; Grohmann, E. Horizontal Gene Transfer of Antibiotic Resistance Genes in Biofilms. Antibiotics 2023, 12, 328. [Google Scholar] [CrossRef] [PubMed]
- Dell’annunziata, F.; Folliero, V.; Giugliano, R.; De Filippis, A.; Santarcangelo, C.; Izzo, V.; Daglia, M.; Galdiero, M.; Arciola, C.R.; Franci, G. Gene Transfer Potential of Outer Membrane Vesicles of Gram-Negative Bacteria. Int. J. Mol. Sci. 2021, 22, 5985. [Google Scholar] [CrossRef]
- Emamalipour, M.; Seidi, K.; Vahed, S.Z.; Jahanban-Esfahlan, A.; Jaymand, M.; Majdi, H.; Amoozgar, Z.; Chitkushev, L.T.; Javaheri, T.; Jahanban-Esfahlan, R.; et al. Horizontal Gene Transfer: From Evolutionary Flexibility to Disease Progression. Front. Cell Dev. Biol. 2020, 8, 229. [Google Scholar] [CrossRef] [PubMed]
- Forero-Hurtado, D.; Corredor-Rozo, Z.L.; Ruiz-Castellanos, J.S.; Márquez-Ortiz, R.A.; Abril, D.; Vanegas, N.; Lafaurie, G.I.; Chambrone, L.; Escobar-Pérez, J. Worldwide Dissemination of blaKPC Gene by Novel Mobilization Platforms in Pseudomonas aeruginosa: A Systematic Review. Antibiotics 2023, 12, 658. [Google Scholar] [CrossRef]
- Meng, L.; Liu, Z.; Liu, C.; Li, C.; Shen, H.; Cao, X. The distribution characteristics of global blaOXA-carrying Klebsiella pneumoniae. BMC Infect. Dis. 2023, 23, 1–9. [Google Scholar] [CrossRef]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 13.1. 2023. Available online: http://www.eucast.org (accessed on 1 December 2023).
- CLSI document M100; Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 33rd ed. Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2023.
- Alieva, K.N.; Golikova, M.V.; Dovzhenko, S.A.; Kobrin, M.B.; Strukova, E.N.; Ageevets, V.A.; Avdeeva, A.A.; Sulian, O.S.; Sidorenko, S.V.; Zinner, S.H. Testing the mutant selection window hypothesis with meropenem: In vitro model study with OXA-48-producing Klebsiella pneumoniae. PLoS ONE 2023, 18, e0288660. [Google Scholar] [CrossRef]
- Blaser, J.; Zinner, S.H. In vitro models for the study of antibiotic activities. Prog Drug Res. 1987, 31, 349–381. [Google Scholar] [CrossRef]
- Sadouki, Z.; McHugh, T.D.; Aarnoutse, R.; Canseco, J.O.; Darlow, C.; Hope, W.; van Ingen, J.; Longshaw, C.; Manissero, D.; Mead, A.; et al. Application of the hollow fibre infection model (HFIM) in antimicrobial development: A systematic review and recommendations of reporting. J. Antimicrob. Chemother. 2021, 76, 2252–2259. [Google Scholar] [CrossRef] [PubMed]
- Lenhard, J.R.; Bulman, Z.P. Inoculum effect of β-lactam antibiotics. J. Antimicrob. Chemother. 2019, 74, 2825–2843. [Google Scholar] [CrossRef] [PubMed]
- Veeraraghavan, B.; Bakthavatchalam, Y.D.; Anandan, S. Laboratory detection and clinical implication of oxacillinase-48 like carbapenemase: The hidden threat. J. Glob. Infect. Dis. 2016, 8, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Hleba, L.; Hlebová, M.; Kováčik, A.; Čuboň, J.; Medo, J. Carbapenemase Producing Klebsiella pneumoniae (KPC): What Is the Best MALDI-TOF MS Detection Method. Antibiotics 2021, 10, 1549. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Wang, Q.; Wang, Y.; Wen, X.; Peng, H.; Peng, R.; Shi, Q.; Xie, X.; Li, L. Outer Membrane Porins Contribute to Antimicrobial Resistance in Gram-Negative Bacteria. Microorganisms 2023, 11, 1690. [Google Scholar] [CrossRef] [PubMed]
- Padilla, E.; Llobet, E.; Doménech-Sánchez, A.; Martínez-Martínez, L.; Bengoechea, J.A.; Albertí, S. Klebsiella pneumoniae AcrAB Efflux Pump Contributes to Antimicrobial Resistance and Virulence. Antimicrob. Agents Chemother. 2010, 54, 177–183. [Google Scholar] [CrossRef]
- Bodmann, K.-F.; Der Infektliga, U.D.E. Komplizierte intraabdominelle Infektionen: Erreger, Resistenzen. Der Chir. 2009, 81, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Golikova, M.V.; Strukova, E.N.; Alieva, K.N.; Ageevets, V.A.; Avdeeva, A.A.; Sulian, O.S.; Zinner, S.H. Meropenem MICs at Standard and High Inocula and Mutant Prevention Concentration Inter-Relations: Comparative Study with Non-Carbapenemase-Producing and OXA-48-, KPC- and NDM-Producing Klebsiella pneumoniae. Antibiotics 2023, 12, 872. [Google Scholar] [CrossRef]
- Su, C.-F.; Chuang, C.; Lin, Y.-T.; Chan, Y.-J.; Lin, J.-C.; Lu, P.-L.; Wang, J.-T.; Siu, L.K.; Fung, C.-P. Treatment outcome of non-carbapenemase-producing carbapenem-resistant Klebsiella pneumoniae infections: A multicenter study in Taiwan. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 37, 651–659. [Google Scholar] [CrossRef]
- Lim, F.K.; Liew, Y.X.; Cai, Y.; Lee, W.; Teo, J.Q.M.; Lay, W.Q.; Chung, J.; Kwa, A.L.H. Treatment and Outcomes of Infections Caused by Diverse Carbapenemase-Producing Carbapenem-Resistant Enterobacterales. Front. Cell. Infect. Microbiol. 2020, 10, 579462. [Google Scholar] [CrossRef]
- Debnath, A.; Pillinger, K.E.; Martin, A.J.; Dobrzynski, D.; Cameron, A.; Shulder, S. Clinical Outcomes and Treatment Strategies in Patients with Non-Carbapenemase-producing Carbapenem-Resistant Versus Carbapenem-Susceptible Enterobacterales Infections. Ann. Pharmacother. 2022, 57, 803–812. [Google Scholar] [CrossRef] [PubMed]
- Oka, K.; Matsumoto, A.; Tetsuka, N.; Morioka, H.; Iguchi, M.; Ishiguro, N.; Nagamori, T.; Takahashi, S.; Saito, N.; Tokuda, K.; et al. Clinical characteristics and treatment outcomes of carbapenem-resistant Enterobacterales infections in Japan. J. Glob. Antimicrob. Resist. 2022, 29, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.-Y.; Tsai, C.-S.; Syue, L.-S.; Chen, P.-L.; Li, C.-W.; Li, M.-C.; Ko, W.-C. Treatment Outcome of Bacteremia Due to Non–Carbapenemase-producing Carbapenem-Resistant Klebsiella pneumoniae Bacteremia: Role of Carbapenem Combination Therapy. Clin. Ther. 2020, 42, e33–e44. [Google Scholar] [CrossRef] [PubMed]
- Tamma, P.D.; Huang, Y.; Opene, B.N.A.; Simner, P.J. Determining the Optimal Carbapenem MIC That Distinguishes Carbapenemase-Producing and Non-Carbapenemase-Producing Carbapenem-Resistant Enterobacteriaceae. Antimicrob. Agents Chemother. 2016, 60, 6425–6429. [Google Scholar] [CrossRef] [PubMed]
- Soga, Y.; Ohge, H.; Ikawa, K.; Morikawa, N.; Ikeda, K.; Sueda, T. Peritoneal Pharmacokinetics and Pharmacodynamic Target Attainment of Meropenem in Patients Undergoing Abdominal Surgery. J. Chemother. 2010, 22, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Ageevets, V.; Sopova, J.; Lazareva, I.; Malakhova, M.; Ilina, E.; Kostryukova, E.; Babenko, V.; Carattoli, A.; Lobzin, Y.; Uskov, A.; et al. Genetic Environment of the blaKPC-2 Gene in a Klebsiella pneumoniae Isolate That May Have Been Imported to Russia from Southeast Asia. Antimicrob. Agents Chemother. 2017, 61, e01856-16. [Google Scholar] [CrossRef]
- Van Der Zwaluw, K.; De Haan, A.; Pluister, G.N.; Bootsma, H.J.; De Neeling, A.J.; Schouls, L.M. The Carbapenem Inactivation Method (CIM), a Simple and Low-Cost Alternative for the Carba NP Test to Assess Phenotypic Carbapenemase Activity in Gram-Negative Rods. PLoS ONE 2015, 10, e0123690. [Google Scholar] [CrossRef]
- ISO 20776-1:2019; Susceptibility Testing of Infectious Agents and Evaluation of Performance of Antimicrobial Susceptibility Test Devices—Part 1: Broth Micro-Dilution Reference Method for Testing the in vitro Activity of Antimicrobial Agents against Rapidly Growing Aerobic Bacteria Involved in Infectious Diseases. ISO: Geneva, Switzerland, 2019.
- Canene-Adams, K. General PCR. Lab. Methods Enzymol. DNA 2013, 529, 291–298. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, 3rd ed.; Cold Spring Harbor: New York, NY, USA, 2001. [Google Scholar]
- Firsov, A.A.; Alieva, K.N.; Strukova, E.N.; Golikova, M.V.; Portnoy, Y.A.; Dovzhenko, S.A.; Kobrin, M.B.; Romanov, A.V.; Edelstein, M.V.; Zinner, S.H. Testing the mutant selection window hypothesis with Staphylococcus aureus exposed to linezolid in an in vitro dynamic model. J. Antimicrob. Chemother. 2017, 72, 3100–3107. [Google Scholar] [CrossRef]
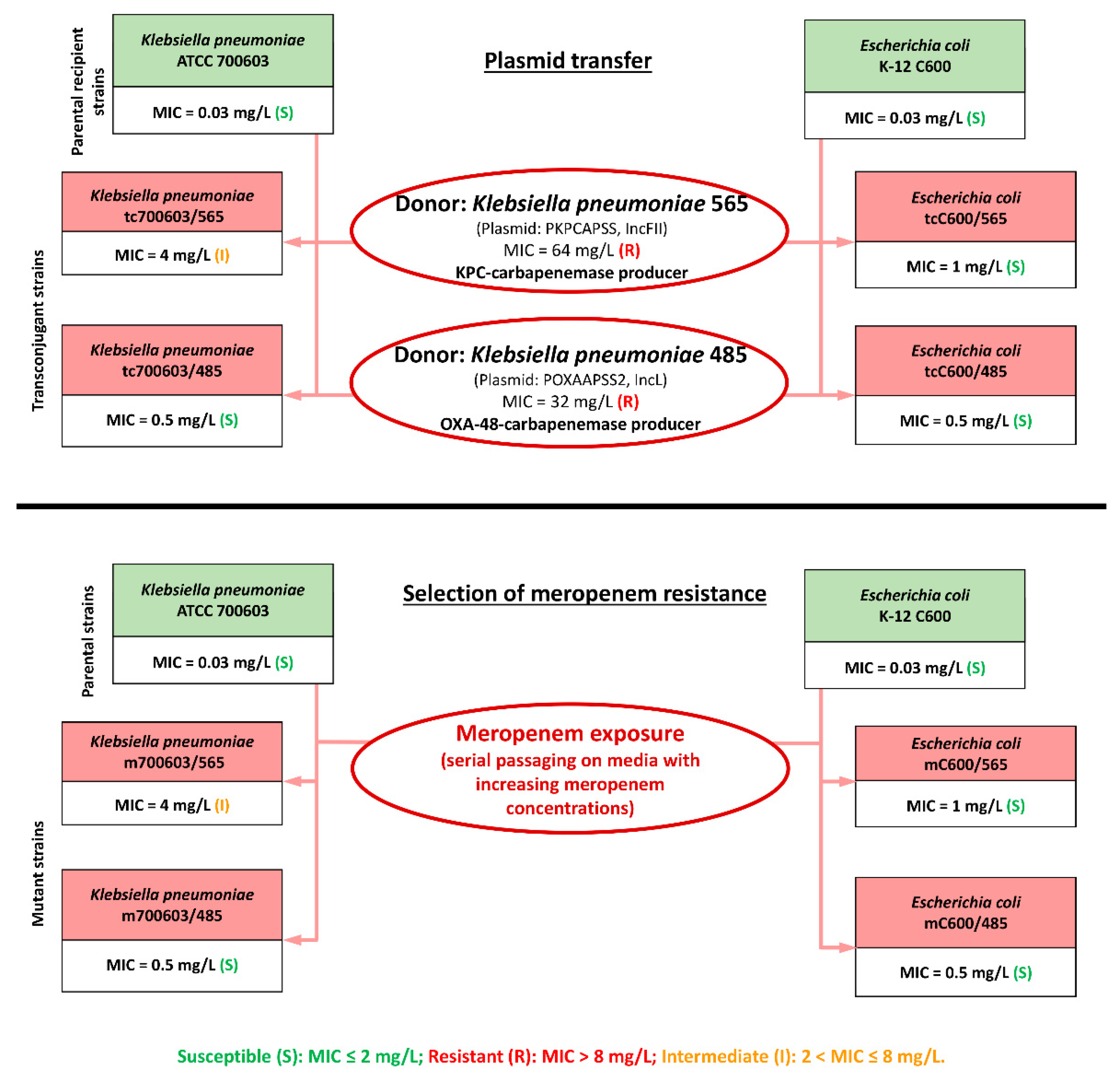

| Bacterial Strain | Carbapenemase Producer | Meropenem MIC at SI, mg/L | Meropenem MIC at HI, mg/L | Fold MIC Increase | IE |
|---|---|---|---|---|---|
| C600 | No | 0.03 | 0.03 | 1 | − |
| ATCC 700603 | No | 0.03 | 0.5 | 16 * | + |
| 485 | Yes | 32 | 256 | 8 * | + |
| 565 | Yes | 64 | 512 | 8 * | + |
| mC600/0.5 | No | 0.5 | 2 | 4 | − |
| mC600/1 | No | 1 | 4 | 4 | − |
| tcC600/485 | Yes | 0.5 | 8 | 16 * | + |
| tcC600/565 | Yes | 1 | 64 | 64 * | + |
| m700603/0.5 | No | 0.5 | 4 | 8 * | + |
| m700603/4 | No | 4 | 32 | 8 * | + |
| tc700603/485 | Yes | 0.5 | 8 | 16 * | + |
| tc700603/565 | Yes | 4 | 128 | 32 * | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Golikova, M.V.; Alieva, K.N.; Strukova, E.N.; Kondratieva, D.A.; Petrova, N.F.; Petrova, M.A.; Zinner, S.H. Comparative Meropenem Pharmacodynamics and Emergence of Resistance against Carbapenem-Susceptible Non-Carbapenemase-Producing and Carbapenemase-Producing Enterobacterales: A Pharmacodynamic Study in a Hollow-Fiber Infection Model. Antibiotics 2023, 12, 1717. https://doi.org/10.3390/antibiotics12121717
Golikova MV, Alieva KN, Strukova EN, Kondratieva DA, Petrova NF, Petrova MA, Zinner SH. Comparative Meropenem Pharmacodynamics and Emergence of Resistance against Carbapenem-Susceptible Non-Carbapenemase-Producing and Carbapenemase-Producing Enterobacterales: A Pharmacodynamic Study in a Hollow-Fiber Infection Model. Antibiotics. 2023; 12(12):1717. https://doi.org/10.3390/antibiotics12121717
Chicago/Turabian StyleGolikova, Maria V., Kamilla N. Alieva, Elena N. Strukova, Daria A. Kondratieva, Nika F. Petrova, Mayya A. Petrova, and Stephen H. Zinner. 2023. "Comparative Meropenem Pharmacodynamics and Emergence of Resistance against Carbapenem-Susceptible Non-Carbapenemase-Producing and Carbapenemase-Producing Enterobacterales: A Pharmacodynamic Study in a Hollow-Fiber Infection Model" Antibiotics 12, no. 12: 1717. https://doi.org/10.3390/antibiotics12121717





