Characterization of Two Novel AmpC Beta-Lactamases from the Emerging Opportunistic Pathogen, Cedecea neteri
Abstract
:1. Introduction
2. Results
2.1. Sequence Analysis of AmpC Proteins
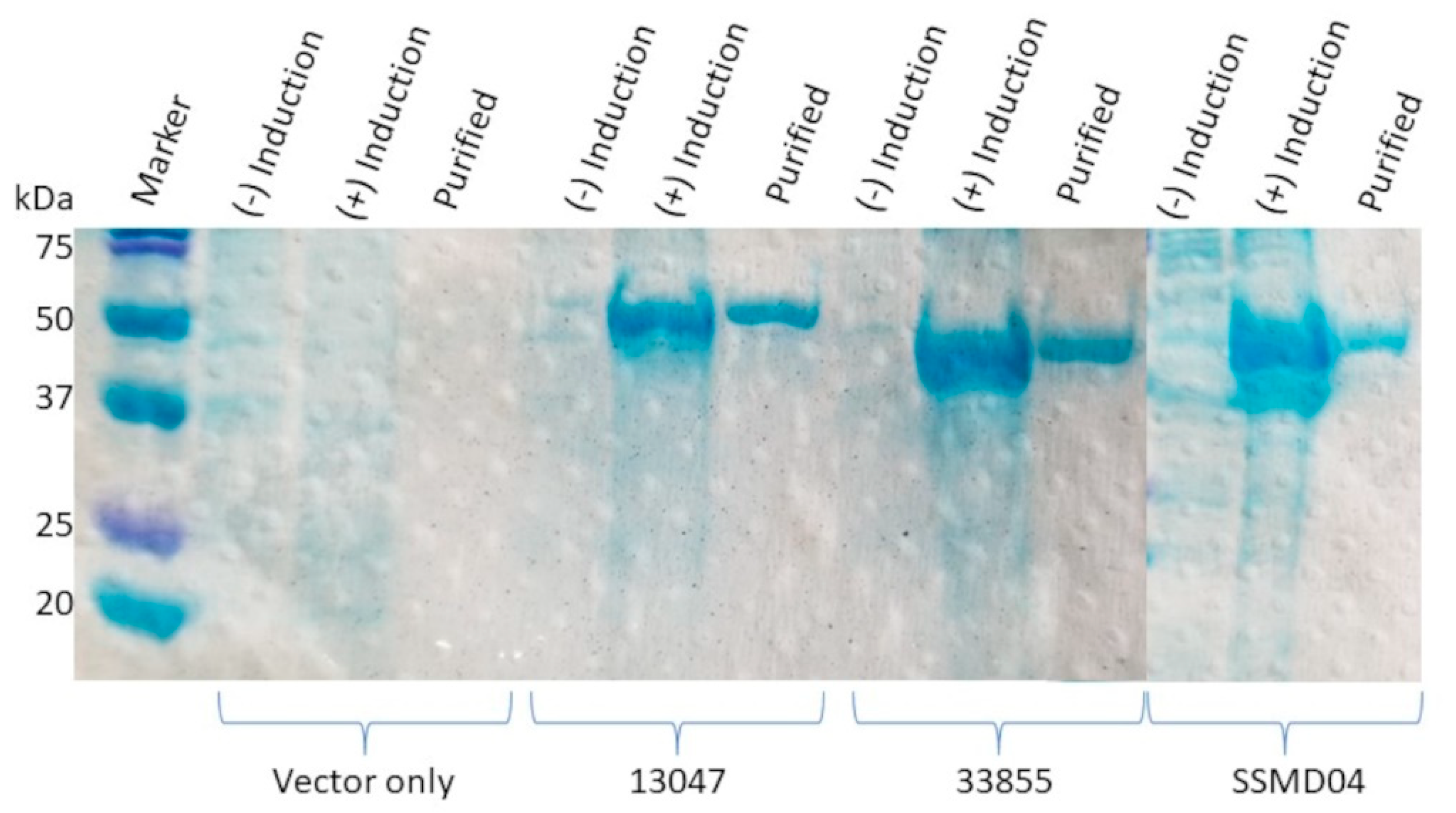
2.2. Overexpression and Purification of rAmpC Proteins
2.3. Determination of Extended-Spectrum AmpC β-Lactamase (ESAC) and Class B Metallo-β-Lactamase Activity
2.4. Enzymatic Activity and Kinetic Analysis of the rAmpC Proteins
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Chemicals, and Supplies
4.2. Cloning of the AmpC Genes
4.3. Expression and Purification of AmpC Proteins
4.4. Kinetic Analysis of the rAmpC Proteins
4.5. Assay for Extended-Spectrum AmpC β-Lactamase (ESAC) and Class B Metallo-β-Lactamase Activity
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grimont, P.; Grimont, F.; Farmer, J., III; Asbury, M. Cedecea davisae gen. nov., sp. nov. and Cedecea lapagei sp. nov., New Enterobacteriaceae from Clinical Specimens. Int. J. Syst. Bacteriol. 1981, 31, 317–326. [Google Scholar] [CrossRef] [Green Version]
- Brenner, D.J.; Krieg, N.R.; Staley, J.R. Bergey’s Manual of Systematic Bacteriology. In The proteobacteria: Part B: The Gammaproteobacteria, 2nd ed.; Garrity, G., Ed.; Springer: New York, NY, USA, 2005; Volume 2, p. 683. [Google Scholar]
- Thompson, D.K.; Sharkady, S.M. Expanding spectrum of opportunistic Cedecea infections: Current clinical status and multidrug resistance. Int. J. Infect. Dis. 2020, 100, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Ginn, P.S.; Tart, S.B.; Sharkady, S.M.; Thompson, D.K. Urinary Catheter Colonization by Multidrug-Resistant. Case Rep. Infect. Dis. 2018, 2018, 7520527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, H.; Masroor, T.; Parmar, S.A.; Panigrahi, D. Urinary tract infection by a rare pathogen Cedecea neteri in a pregnant female with Polyhydramnios: Rare case report from UAE. BMC Infect. Dis. 2021, 21, 637. [Google Scholar] [CrossRef]
- Aguilera, A.; Pascual, J.; Loza, E.; Lopez, J.; Garcia, G.; Liano, F.; Quereda, C.; Ortuno, J. Bacteraemia with Cedecea neteri in a patient with systemic lupus erythematosus. Postgrad. Med. J. 1995, 71, 179–180. [Google Scholar] [CrossRef] [Green Version]
- Chavez Herrera, V.R.; Rosas De Silva, M.F.; Orendain Alcaraz, H.; Ceja Espiritu, G.; Carrazco Peña, K.; Melnikov, V. Death related to Cedecea lapagei in a soft tissue bullae infection: A case report. J. Med. Case Rep. 2018, 12, 328. [Google Scholar] [CrossRef]
- Bush, K. Past and Present Perspectives on β-Lactamases. Antimicrob. Agents Chemother. 2018, 62, e01076-18. [Google Scholar] [CrossRef] [Green Version]
- Bush, K.; Jacoby, G.A.; Medeiros, A.A. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 1995, 39, 1211–1233. [Google Scholar] [CrossRef] [Green Version]
- Ambler, R.P. The structure of beta-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1980, 289, 321–331. [Google Scholar] [CrossRef]
- Tooke, C.L.; Hinchliffe, P.; Bragginton, E.C.; Colenso, C.K.; Hirvonen, V.H.A.; Takebayashi, Y.; Spencer, J. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 2019, 431, 3472–3500. [Google Scholar] [CrossRef]
- Jacoby, G.A. AmpC beta-lactamases. Clin. Microbiol. Rev. 2009, 22, 161–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ammenouche, N.; Dupont, H.; Mammeri, H. Characterization of a novel AmpC β-lactamase produced by a carbapenem-resistant Cedecea davisae clinical isolate. Antimicrob. Agents Chemother. 2014, 58, 6942–6945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Notter, J.; Seiffert, S.N.; Zimmermann-Kogadeeva, M.; Bösch, A.; Wenger, R.; Strahm, C.; Frischknecht, M.; Livermore, D.M.; Babouee Flury, B. AmpC hyperproduction in a Cedecea davisae implant-associated bone infection during treatment: A case report and therapeutic implications. BMC Infect. Dis. 2022, 22, 33. [Google Scholar] [CrossRef]
- Thompson, D.K.; Sharkady, S.M. Genomic Insights into Drug Resistance Determinants in. Microorganisms 2021, 9, 1741. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.G.; Tan, K.H.; Yin, W.F.; Tan, J.Y. Complete Genome Sequence of Cedecea neteri Strain SSMD04, a Bacterium Isolated from Pickled Mackerel Sashimi. Genome Announc. 2014, 2, e01339-14. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Joris, B.; Ghuysen, J.M.; Dive, G.; Renard, A.; Dideberg, O.; Charlier, P.; Frère, J.M.; Kelly, J.A.; Boyington, J.C.; Moews, P.C. The active-site-serine penicillin-recognizing enzymes as members of the Streptomyces R61 DD-peptidase family. Biochem. J. 1988, 250, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Matagne, A.; Lamotte-Brasseur, J.; Frère, J.M. Catalytic properties of class A beta-lactamases: Efficiency and diversity. Biochem. J. 1998, 330 Pt 2, 581–598. [Google Scholar] [CrossRef]
- Srivastava, A.; Singhal, N.; Goel, M.; Virdi, J.S.; Kumar, M. Identification of family specific fingerprints in β-lactamase families. Sci. World J. 2014, 2014, 980572. [Google Scholar] [CrossRef] [Green Version]
- Davin-Regli, A.; Lavigne, J.P.; Pagès, J.M. Enterobacter spp.: Update on Taxonomy, Clinical Aspects, and Emerging Antimicrobial Resistance. Clin. Microbiol. Rev. 2019, 32, e00002-19. [Google Scholar] [CrossRef]
- Simpson, R.J.; Peter, D.; Adams, P.D.; Golemis, E. (Eds.) Basic Methods in Protein Purification and Analysis: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2009. [Google Scholar]
- Gudeta, D.D.; Pollini, S.; Docquier, J.D.; Bortolaia, V.; Rossolini, G.M.; Guardabassi, L. Biochemical Characterization of CPS-1, a Subclass B3 Metallo-β-Lactamase from a Chryseobacterium piscium Soil Isolate. Antimicrob. Agents Chemother. 2015, 60, 1869–1873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mojica, M.F.; Bonomo, R.A.; Fast, W. B1-Metallo-β-Lactamases: Where Do We Stand? Curr. Drug Targets 2016, 17, 1029–1050. [Google Scholar] [CrossRef] [PubMed]
- Sawa, T.; Kooguchi, K.; Moriyama, K. Molecular diversity of extended-spectrum β-lactamases and carbapenemases, and antimicrobial resistance. J. Intensive Care 2020, 8, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Philippon, A.; Arlet, G.; Labia, R.; Iorga, B.I. Class C β-Lactamases: Molecular Characteristics. Clin. Microbiol. Rev. 2022, 35, e0015021. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Mammeri, H. Extended-spectrum cephalosporinases: Structure, detection and epidemiology. Future Microbiol. 2007, 2, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum beta-lactamases: A clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galleni, M.; Amicosante, G.; Frère, J.M. A survey of the kinetic parameters of class C beta-lactamases. Cephalosporins and other beta-lactam compounds. Biochem. J. 1988, 255, 123–129. [Google Scholar] [CrossRef]
- Hugonnet, J.E.; Blanchard, J.S. Irreversible inhibition of the Mycobacterium tuberculosis beta-lactamase by clavulanate. Biochemistry 2007, 46, 11998–12004. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Yang, G.-L.; Li, W.-P.; Li, M.-C.; Bao, X.-Y. Antimicrobial resistance and epidemiology of extended spectrum-β-lactamases (ESBL)-producing Escherichia coli and Enterobacter cloacae isolates from intensive care units at obstetrics & gynaecology departments: A retrospective analysis. CEOG 2021, 48, 820–827. [Google Scholar] [CrossRef]
- Gekenidis, M.T.; Kläui, A.; Smalla, K.; Drissner, D. Transferable Extended-Spectrum β-Lactamase (ESBL) Plasmids in. Microorganisms 2020, 8, 978. [Google Scholar] [CrossRef]
- Mohamudha Parveen, R.; Harish, B.N.; Parija, S.C. Ampc Beta lactamases among gram negative clinical isolates from a tertiary hospital, South India. Braz. J. Microbiol. 2010, 41, 596–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papp-Wallace, K.M.; Mallo, S.; Bethel, C.R.; Taracila, M.A.; Hujer, A.M.; Fernández, A.; Gatta, J.A.; Smith, K.M.; Xu, Y.; Page, M.G.; et al. A kinetic analysis of the inhibition of FOX-4 β-lactamase, a plasmid-mediated AmpC cephalosporinase, by monocyclic β-lactams and carbapenems. J. Antimicrob. Chemother. 2014, 69, 682–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacoby, G.A.; Medeiros, A.A. More extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 1991, 35, 1697–1704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotsakis, S.D.; Papagiannitsis, C.C.; Tzelepi, E.; Tzouvelekis, L.S.; Miriagou, V. Extended-spectrum properties of CMY-30, a Val211Gly mutant of CMY-2 cephalosporinase. Antimicrob. Agents Chemother. 2009, 53, 3520–3523. [Google Scholar] [CrossRef] [Green Version]
- Nukaga, M.; Kumar, S.; Nukaga, K.; Pratt, R.F.; Knox, J.R. Hydrolysis of third-generation cephalosporins by class C beta-lactamases. Structures of a transition state analog of cefotoxamine in wild-type and extended spectrum enzymes. J. Biol. Chem. 2004, 279, 9344–9352. [Google Scholar] [CrossRef] [Green Version]
- Walker, J.M. The bicinchoninic acid (BCA) assay for protein quantitation. Methods Mol. Biol. 1994, 32, 5–8. [Google Scholar] [CrossRef]
- Gallagher, S.; Sasse, J. Protein analysis by SDS-PAGE and detection by Coomassie blue or silver staining. Curr. Protoc. Pharmacol. 2001, 2, A.3B.1–A.3B.10. [Google Scholar] [CrossRef]
- Faheem, M.; Rehman, M.T.; Danishuddin, M.; Khan, A.U. Biochemical characterization of CTX-M-15 from Enterobacter cloacae and designing a novel non-β-lactam-β-lactamase inhibitor. PLoS ONE 2013, 8, e56926. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. β-Lactams and β-Lactamase Inhibitors: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a006262. [Google Scholar] [CrossRef]
- Lauretti, L.; Riccio, M.L.; Mazzariol, A.; Cornaglia, G.; Amicosante, G.; Fontana, R.; Rossolini, G.M. Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 1999, 43, 1584–1590. [Google Scholar] [CrossRef]

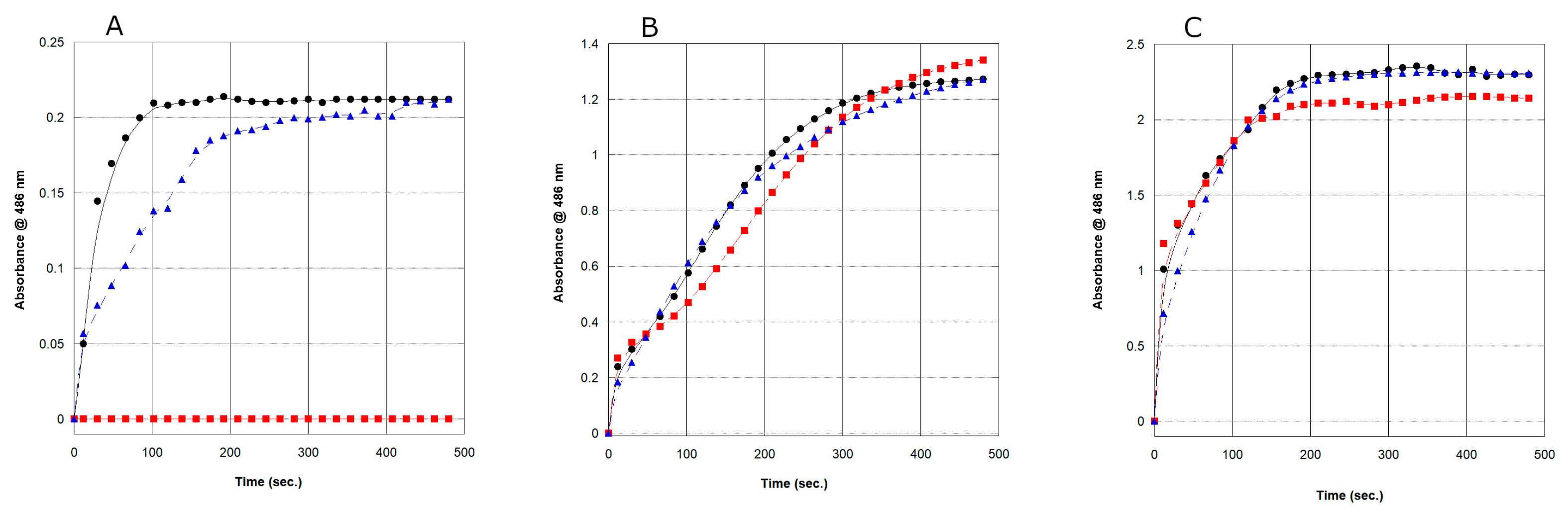
| Strain | Km (µM) | kcat (s−1) | kcat/Km (mM−1s−1) |
|---|---|---|---|
| E. cloacae (ATCC 13047) | 11 ± 2 | 0.83 ± 0.04 | 7.5 |
| C. neteri (SSMD04) | 46 ± 5 | 1.93 ± 0.16 | 41.9 |
| C. neteri (ATCC 33855) | 133 ± 11 | 3.13 ± 0.27 | 23.5 |
| Km,apparent (µM) 1 | |||
|---|---|---|---|
| ATCC 13047 | SSMD04 | ATCC 33855 | |
| Antibiotic | |||
| Ampicillin | 7 | 72 | 12 |
| Penicillin | 33 | 58 | 11 |
| Cefazolin | 0.43 | 67 | 100 |
| Cephalexin | 6.8 | 2 | 5 |
| Cefoxitin | 12 | 48 | 54 |
| Cefuroxime | 0.35 | 5 | 4 |
| Imipenem | 0.85 | 5 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharkady, S.M.; Bailey, B.; Thompson, D.K. Characterization of Two Novel AmpC Beta-Lactamases from the Emerging Opportunistic Pathogen, Cedecea neteri. Antibiotics 2023, 12, 219. https://doi.org/10.3390/antibiotics12020219
Sharkady SM, Bailey B, Thompson DK. Characterization of Two Novel AmpC Beta-Lactamases from the Emerging Opportunistic Pathogen, Cedecea neteri. Antibiotics. 2023; 12(2):219. https://doi.org/10.3390/antibiotics12020219
Chicago/Turabian StyleSharkady, Stephen M., Brandon Bailey, and Dorothea K. Thompson. 2023. "Characterization of Two Novel AmpC Beta-Lactamases from the Emerging Opportunistic Pathogen, Cedecea neteri" Antibiotics 12, no. 2: 219. https://doi.org/10.3390/antibiotics12020219
APA StyleSharkady, S. M., Bailey, B., & Thompson, D. K. (2023). Characterization of Two Novel AmpC Beta-Lactamases from the Emerging Opportunistic Pathogen, Cedecea neteri. Antibiotics, 12(2), 219. https://doi.org/10.3390/antibiotics12020219







