Cloning and Molecular Characterization of the phlD Gene Involved in the Biosynthesis of “Phloroglucinol”, a Compound with Antibiotic Properties from Plant Growth Promoting Bacteria Pseudomonas spp.
Abstract
:1. Introduction
2. Materials and Methods
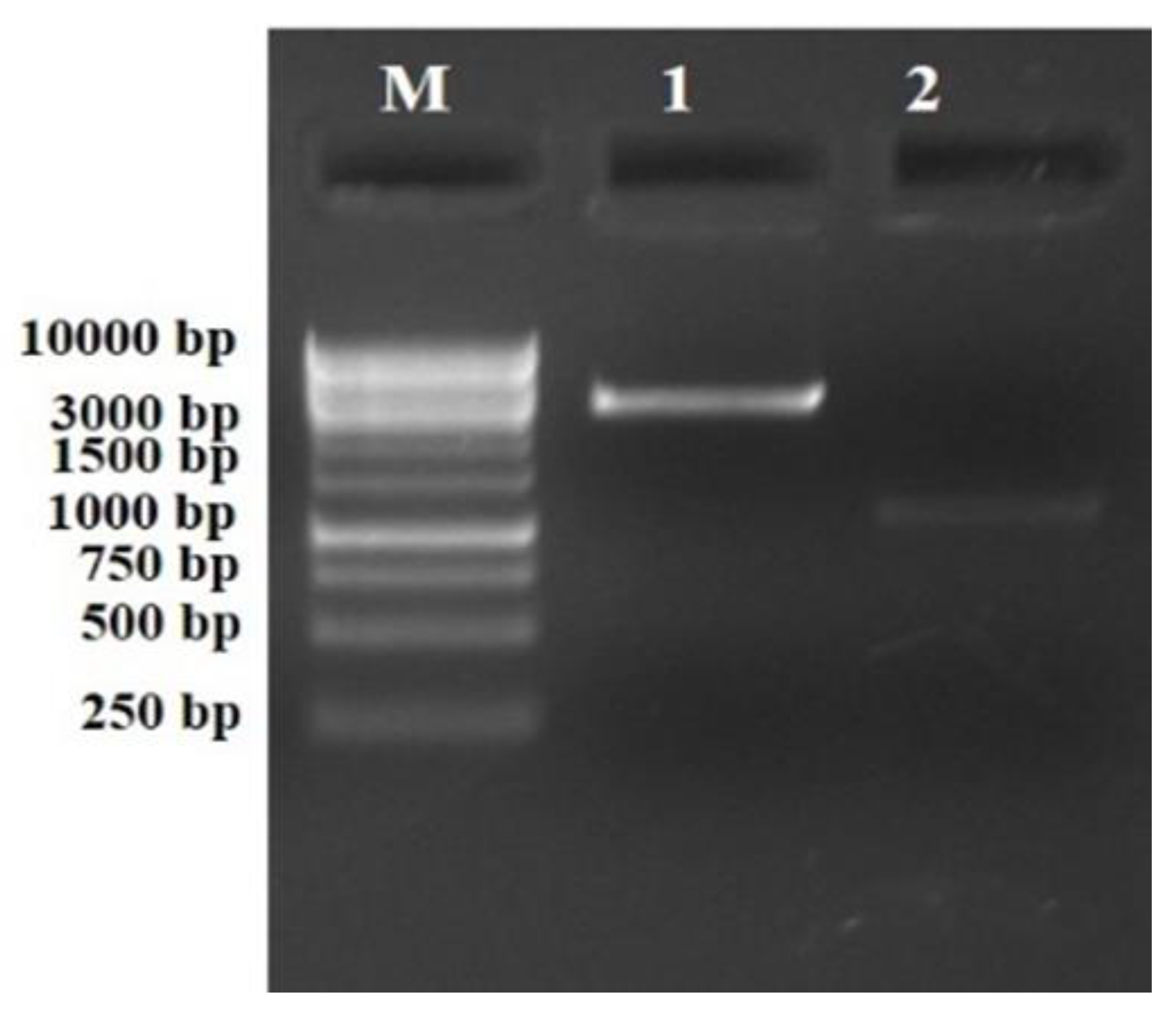
2.1. Genomic DNA Isolation and PCR Amplification of the phlD Gene
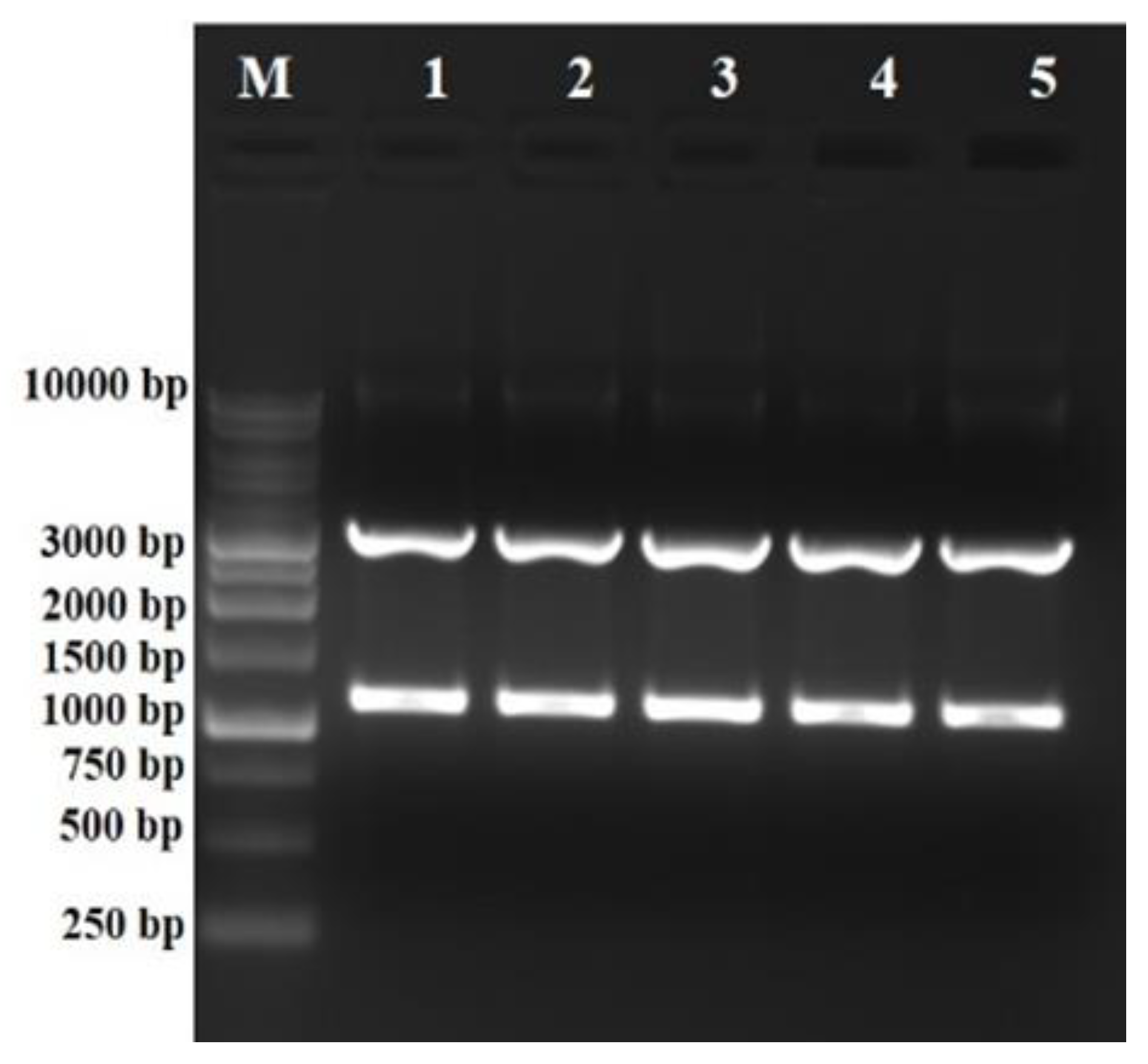
2.2. Cloning of the phlD Gene in pBluescript (SK+) Vector
2.3. Confirmation of Cloning and Sequencing
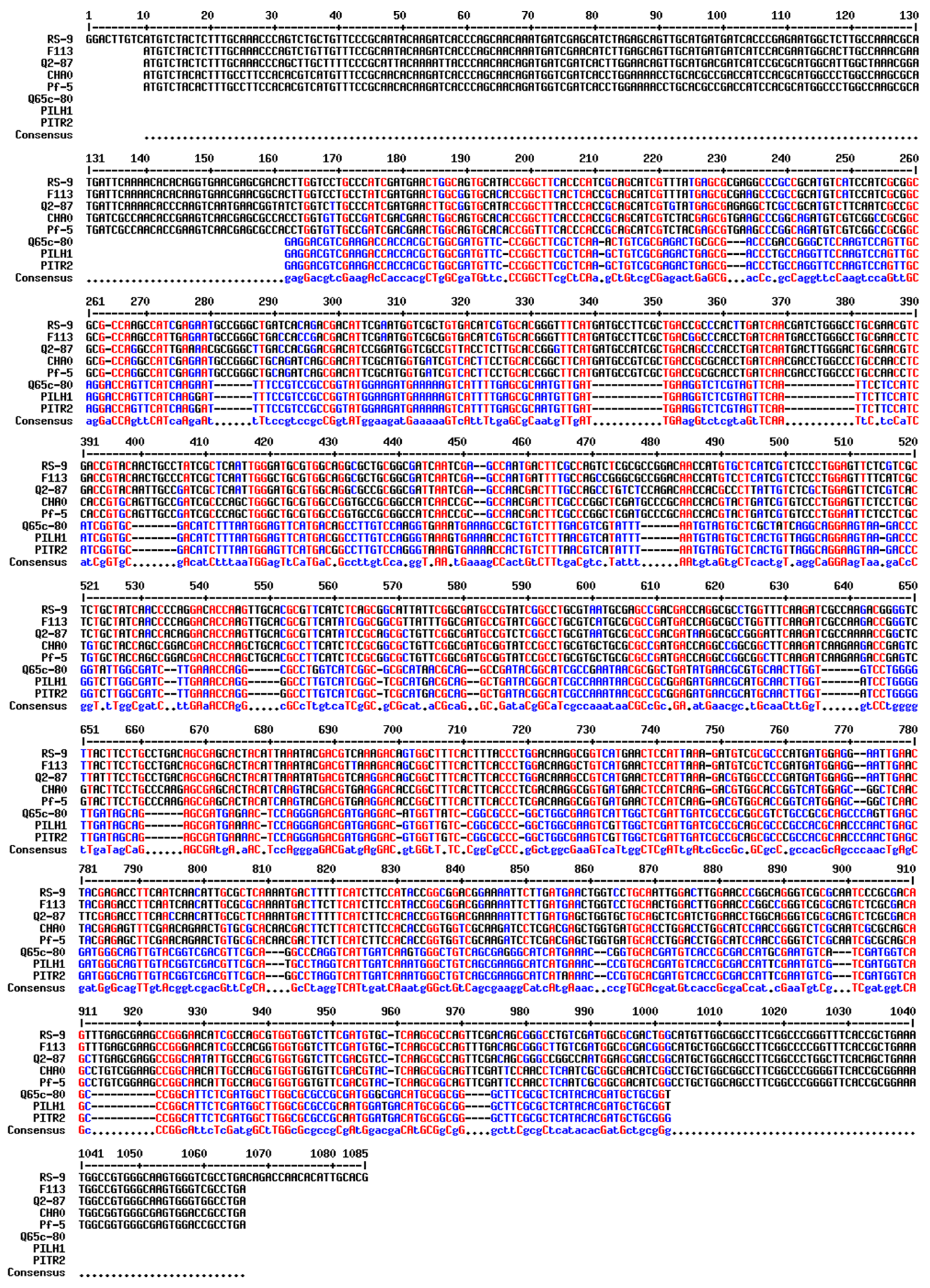
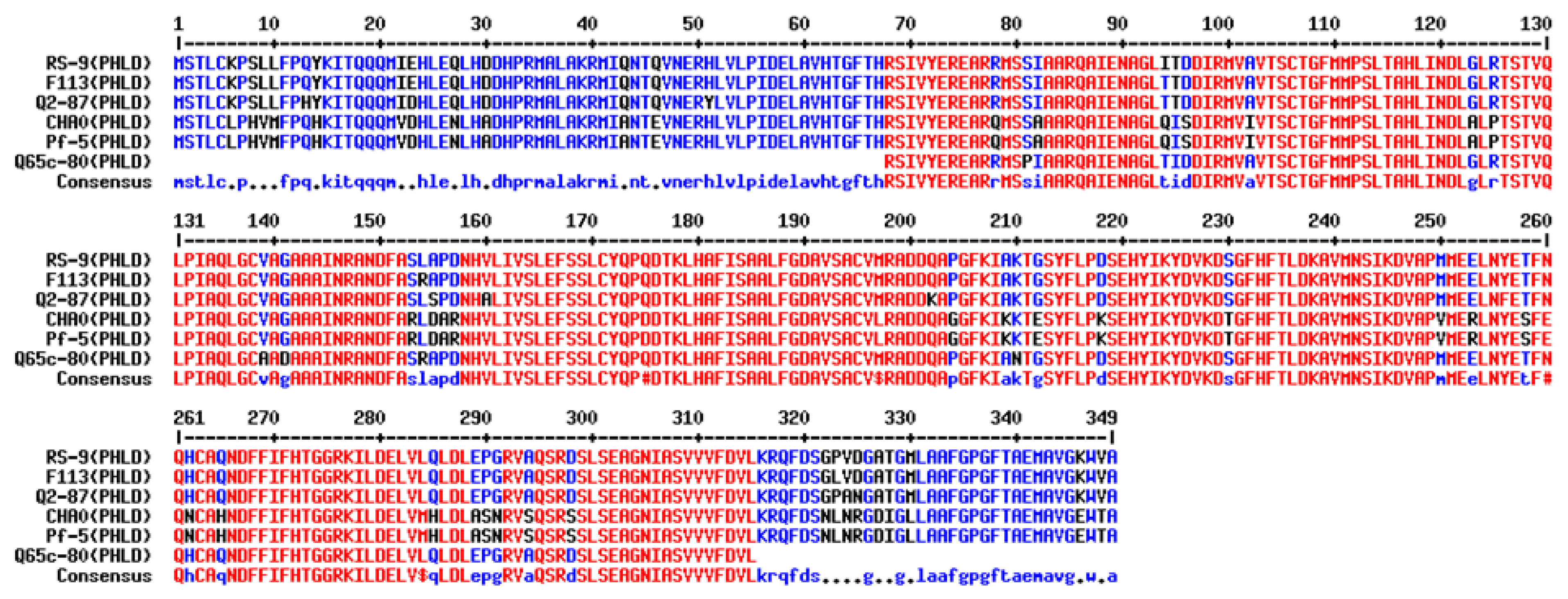
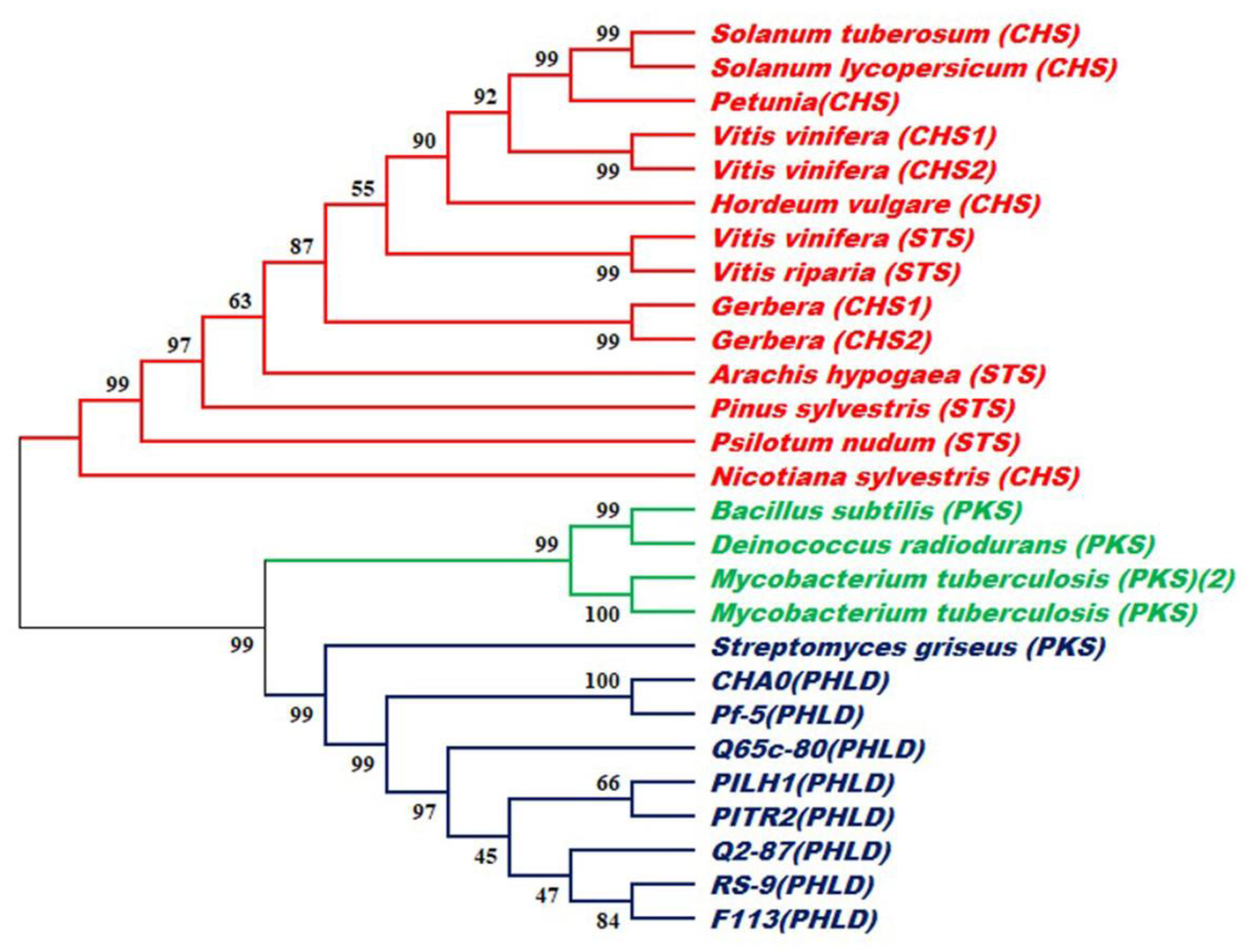
2.4. Phylogenetic Analysis
2.5. Structure Prediction
2.6. Ramachandran Plot Analysis
2.7. Validation and Visualization of Modeled Structure
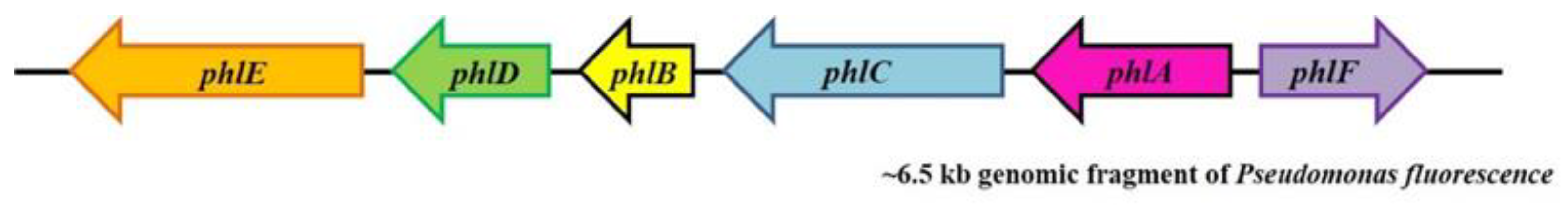
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kloepper, J.; Lifshitz, R.; Schroth, M. Pseudomonas Inoculants to Benefit Plant Protection; Institute for Scientific Information: Philadelphia, PA, USA, 1988. [Google Scholar]
- Troppens, D.; Moynihan, J.; Barret, M.; O’Gara, F.; Morrissey, J. Molecular Microbial Ecology of the Rhizosphere; John Wiley and Sons, Ltd.: Hoboken, NJ, USA, 2013. [Google Scholar]
- Mandryk-Litvinkovich, M.N.; Muratova, A.A.; Nosonova, T.L.; Evdokimova, O.V.; Valentovich, L.N.; Titok, M.A.; Kolomiets, E.I. Molecular Genetic Analysis of Determinants Defining Synthesis of 2,4-Diacetylphloroglucinol by Pseudomonas Brassicacearum BIM B-446 Bacteria. Appl. Biochem. Microbiol. 2017, 53, 31–39. [Google Scholar] [CrossRef]
- Thomashow, L.; Weller, D. Current Concepts in the Use of Introduced Bacteria for Biological Disease Control: Mechanisms and Antifungal Metabolites. In Plant-Microbe Interactions; Springer US: Boston, MA, USA, 1996; pp. 187–235. [Google Scholar]
- van Loon, L.C.; Bakker, P.A.H.M.; Pieterse, C.M.J. Systemic Resistance Induced By Rhizosphere Bacteria. Annu. Rev. Phytopathol. 1998, 36, 453–483. [Google Scholar] [CrossRef] [Green Version]
- Almario, J.; Moënne-Loccoz, Y.; Muller, D. Monitoring of the Relation between 2,4-Diacetylphloroglucinol-Producing Pseudomonas and Thielaviopsis Basicola Populations by Real-Time PCR in Tobacco Black Root-Rot Suppressive and Conducive Soils. Soil Biol. Biochem. 2013, 57, 144–155. [Google Scholar] [CrossRef]
- Haichar, F.; Fochesato, S.; Achouak, W. Host Plant Specific Control of 2,4-Diacetylphloroglucinol Production in the Rhizosphere. Agronomy 2013, 3, 621–631. [Google Scholar] [CrossRef]
- Zhou, T.-T.; Li, C.-Y.; Chen, D.; Wu, K.; Shen, Q.-R.; Shen, B. PhlF− Mutant of Pseudomonas Fluorescens J2 Improved 2,4-DAPG Biosynthesis and Biocontrol Efficacy against Tomato Bacterial Wilt. Biol. Control 2014, 78, 1–8. [Google Scholar] [CrossRef]
- Bonsall, R.F.; Weller, D.M.; Thomashow, L.S. Quantification of 2,4-Diacetylphloroglucinol Produced by Fluorescent Pseudomonas Spp. In Vitro and in the Rhizosphere of Wheat. Appl. Environ. Microbiol. 1997, 63, 951–955. [Google Scholar] [CrossRef] [Green Version]
- Duffy, B.K.; Défago, G. Zinc Improves Biocontrol of Fusarium Crown and Root Rot of Tomato by Pseudomonas fluorescens and Represses the Production of Pathogen Metabolites Inhibitory to Bacterial Antibiotic Biosynthesis. Phytopathology 1997, 87, 1250–1257. [Google Scholar] [CrossRef] [Green Version]
- Raaijmakers, J.M.; Weller, D.M. Natural Plant Protection by 2,4-Diacetylphloroglucinol-Producing Pseudomonas Spp. in Take-All Decline Soils. Mol. Plant-Microbe Interact. 1998, 11, 144–152. [Google Scholar] [CrossRef] [Green Version]
- Sulaiman, M.; Jannat, K.; Nissapatorn, V.; Rahmatullah, M.; Paul, A.K.; de Lourdes Pereira, M.; Rajagopal, M.; Suleiman, M.; Butler, M.S.; bin Break, M.K.; et al. Antibacterial and Antifungal Alkaloids from Asian Angiosperms: Distribution, Mechanisms of Action, Structure-Activity, and Clinical Potentials. Antibiotics 2022, 11, 1146. [Google Scholar] [CrossRef]
- Hansanant, N.; Smith, L. Occidiofungin: Actin Binding as a Novel Mechanism of Action in an Antifungal Agent. Antibiotics 2022, 11, 1143. [Google Scholar] [CrossRef]
- Delany, I.; Sheehan, M.M.; Fenton, A.; Bardin, S.; Aarons, S.; O’Gara, F. Regulation of Production of the Antifungal Metabolite 2,4-Diacetylphloroglucinol in Pseudomonas Fluorescens F113: Genetic Analysis of PhlF as a Transcriptional Repressor The GenBank Accession Number for the Sequence Reported in This Paper Is AF129856. Microbiology 2000, 146, 537–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keel, C. Suppression of Root Diseases by Pseudomonas fluorescens CHA0: Importance of the Bacterial Secondary Metabolite 2,4-Diacetylphloroglucinol. Mol. Plant-Microbe Interact. 1992, 5, 4. [Google Scholar] [CrossRef] [Green Version]
- Mavrodi, O.V.; McSpadden Gardener, B.B.; Mavrodi, D.V.; Bonsall, R.F.; Weller, D.M.; Thomashow, L.S. Genetic Diversity of PhlD from 2,4-Diacetylphloroglucinol-Producing Fluorescent Pseudomonas spp. Phytopathology 2001, 91, 35–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voisard, C.; Bull, C.; Keel, C.; Laville, J.; Maurhofer, M.; Schnider, U.; Dfago, G.; Haas, D. Biocontrol of Root Diseases ByPseudomonas Fluorescens CHA0: Current Concepts and Experimental Approaches. In Molecular Ecology of Rhizosphere Microorganisms; Wiley-VCH Verlag GmbH: Weinheim, Germany, 1994; pp. 67–89. [Google Scholar]
- Whistler, C.A.; Corbell, N.A.; Sarniguet, A.; Ream, W.; Loper, J.E. The Two-Component Regulators GacS and GacA Influence Accumulation of the Stationary-Phase Sigma Factor ς S and the Stress Response in Pseudomonas Fluorescens Pf-5. J. Bacteriol. 1998, 180, 6635–6641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Z.; Cang, S.; Matsufuji, M.; Nakata, K.; Nagamatsu, Y.; Yoshimoto, A. High Production of Pyoluteorin and 2,4-Diacetylphloroglucinol by Pseudomonas Fluorescens S272 Grown on Ethanol as a Sole Carbon Source. J. Ferment. Bioeng. 1998, 86, 559–563. [Google Scholar] [CrossRef]
- Duffy, B.K.; Défago, G. Environmental Factors Modulating Antibiotic and Siderophore Biosynthesis by Pseudomonas Fluorescens Biocontrol Strains. Appl. Environ. Microbiol. 1999, 65, 2429–2438. [Google Scholar] [CrossRef] [Green Version]
- Shanahan, P.; O’Sullivan, D.J.; Simpson, P.; Glennon, J.D.; O’Gara, F. Isolation of 2,4-Diacetylphloroglucinol from a Fluorescent Pseudomonad and Investigation of Physiological Parameters Influencing Its Production. Appl. Environ. Microbiol. 1992, 58, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Biessy, A.; Filion, M. Phloroglucinol Derivatives in Plant-Beneficial Pseudomonas spp.: Biosynthesis, Regulation, and Functions. Metabolites 2021, 11, 182. [Google Scholar] [CrossRef]
- Shimizu, Y.; Ogata, H.; Goto, S. Type III Polyketide Synthases: Functional Classification and Phylogenomics. ChemBioChem 2017, 18, 50–65. [Google Scholar] [CrossRef]
- Achkar, J.; Xian, M.; Zhao, H.; Frost, J.W. Biosynthesis of Phloroglucinol. J. Am. Chem. Soc. 2005, 127, 5332–5333. [Google Scholar] [CrossRef]
- Zha, W.; Rubin-Pitel, S.B.; Zhao, H. Characterization of the Substrate Specificity of PhlD, a Type III Polyketide Synthase from Pseudomonas Fluorescens. J. Biol. Chem. 2006, 281, 32036–32047. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Rai, R.; Dash, P.K. Isolation, Cloning and Characterization of PhlA Gene from an Indigenous Pseudomonas Strain from Indian Soil. Int. J. Trop. Agric. 2015, 33, 3195–3200. [Google Scholar]
- Dash, P.; Gupta, P.; Panwar, B.S.; Rai, R. Isolation, Cloning and Characterization of PhlB Gene from an Indian Strain of Gram Negative Soil Bacteria Pseudomonas Fluorescens. Indian J. Exp. Biol. 2020, 58, 412–419. [Google Scholar]
- Gupta, P.; Karthik, K.; Sreevathsa, R.; Rai, R.; Dash, P.K. Cloning and Characterization of Phloroglucinol Biosynthetic Gene PhlC from An Indian Strain of Pseudomonas Fluorescens. Indian J. Exp. Biol. 2022, 60, 607–614. [Google Scholar] [CrossRef]
- Rai, R.; Srinivasamurthy, R.; Dash, P.K.; Gupta, P. Isolation, Characterization and Evaluation of the Biocontrol Potential of Pseudomonas Protegens RS-9 against Ralstonia Solanacearum in Tomato. Indian J. Exp. Biol. 2017, 55, 595–603. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Robert, X.; Gouet, P. Deciphering Key Features in Protein Structures with the New ENDscript Server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [Green Version]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-Scale Protein Function Classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [Green Version]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y. I-TASSER: Fully Automated Protein Structure Prediction in CASP8. Proteins Struct. Funct. Bioinform. 2009, 77, 100–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lovell, S.C.; Davis, I.W.; Arendall, W.B.; de Bakker, P.I.W.; Word, J.M.; Prisant, M.G.; Richardson, J.S.; Richardson, D.C. Structure Validation by Cα Geometry: ϕ,ψ and Cβ Deviation. Proteins Struct. Funct. Bioinform. 2003, 50, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Fiser, A.; Sali, A. ModLoop: Automated Modeling of Loops in Protein Structures. Bioinformatics 2003, 19, 2500–2501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laskowski, R.A. PDBsum More: New Summaries and Analyses of the Known 3D Structures of Proteins and Nucleic Acids. Nucleic Acids Res. 2004, 33, D266–D268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Castrignano, T. The PMDB Protein Model Database. Nucleic Acids Res. 2006, 34, D306–D309. [Google Scholar] [CrossRef]
- Gokulan, K.; O’Leary, S.E.; Russell, W.K.; Russell, D.H.; Lalgondar, M.; Begley, T.P.; Ioerger, T.R.; Sacchettini, J.C. Crystal Structure of Mycobacterium Tuberculosis Polyketide Synthase 11 (PKS11) Reveals Intermediates in the Synthesis of Methyl-Branched Alkylpyrones. J. Biol. Chem. 2013, 288, 16484–16494. [Google Scholar] [CrossRef] [Green Version]
- Richardson, J.S. The Anatomy and Taxonomy of Protein Structure. Adv. Protein Chem. 1981, 34, 167–339. [Google Scholar]
- Sibanda, B.L.; Blundell, T.L.; Thornton, J.M. Conformation of β-Hairpins in Protein Structures. J. Mol. Biol. 1989, 206, 759–777. [Google Scholar] [CrossRef]
- Hutchinson, C.R.; Fujii, I. POLYKETIDE SYNTHASE GENE MANIPULATION: A Structure-Function Approach in Engineering Novel Antibiotics. Annu. Rev. Microbiol. 1995, 49, 201–238. [Google Scholar] [CrossRef] [PubMed]
- Cook, R.J.; Thomashow, L.S.; Weller, D.M.; Fujimoto, D.; Mazzola, M.; Bangera, G.; Kim, D.S. Molecular Mechanisms of Defense by Rhizobacteria against Root Disease. Proc. Natl. Acad. Sci. USA 1995, 92, 4197–4201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fenton, A.M.; Stephens, P.M.; Crowley, J.; O’Callaghan, M.; O’Gara, F. Exploitation of Gene(s) Involved in 2,4-Diacetylphloroglucinol Biosynthesis to Confer a New Biocontrol Capability to a Pseudomonas Strain. Appl. Environ. Microbiol. 1992, 58, 3873–3878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharifi-Tehrani, A.; Zala, M.; Natsch, A.; Moënne-Loccoz, Y.; Défago, G. Biocontrol of Soil-Borne Fungal Plant Diseases by 2,4- Diacetylphloroglucinol-Producing Fluorescent Pseudomonads with Different Restriction Profiles of Amplified 16S RDNA. Eur. J. Plant Pathol. 1998, 104, 631–643. [Google Scholar] [CrossRef]
- Stutz, E.W. Naturally Occurring Fluorescent Pseudomonads Involved in Suppression of Black Root Rot of Tobacco. Phytopathology 1986, 76, 181. [Google Scholar] [CrossRef]
- Tamietti, G.; Ferraris, L.; Matta, A.; Abbattista Gentile, I. Physiological Responses of Tomato Plants Grown in Fusarium Suppressive Soil. J. Phytopathol. 1993, 138, 66–76. [Google Scholar] [CrossRef]
- Suresh, P.; Rekha, M.; Gomathinayagam, S.; Ramamoorthy, V.; Sharma, M.P.; Sakthivel, P.; Sekar, K.; Valan Arasu, M.; Shanmugaiah, V. Characterization and Assessment of 2, 4-Diacetylphloroglucinol (DAPG)-Producing Pseudomonas Fluorescens VSMKU3054 for the Management of Tomato Bacterial Wilt Caused by Ralstonia Solanacearum. Microorganisms 2022, 10, 1508. [Google Scholar] [CrossRef]
- Dash, P.K.; Cao, Y.; Jailani, A.K.; Gupta, P.; Venglat, P.; Xiang, D.; Rai, R.; Sharma, R.; Thirunavukkarasu, N.; Abdin, M.Z.; et al. Genome-Wide Analysis of Drought Induced Gene Expression Changes in Flax (Linum usitatissimum). GM Crops Food 2014, 5, 106–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Hobson, N.; Galindo, L.; Zhu, S.; Shi, D.; McDill, J.; Yang, L.; Hawkins, S.; Neutelings, G.; Datla, R.; et al. The Genome of Flax ( Linum Usitatissimum ) Assembled de Novo from Short Shotgun Sequence Reads. Plant J. 2012, 72, 461–473. [Google Scholar] [CrossRef] [Green Version]
- Dash, P.K.; Rai, R.; Mahato, A.K.; Gaikwad, K.; Singh, N.K. Transcriptome Landscape at Different Developmental Stages of a Drought Tolerant Cultivar of Flax (Linum Usitatissimum). Front. Chem. 2017, 5, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dash, P.K.; Gupta, P.; Pradhan, S.K.; Shasany, A.K.; Rai, R. Analysis of Homologous Regions of Small RNAs MIR397 and MIR408 Reveals the Conservation of Microsynteny among Rice Crop-Wild Relatives. Cells 2022, 11, 3461. [Google Scholar] [CrossRef] [PubMed]
- Shivakumara, T.N.; Sreevathsa, R.; Dash, P.K.; Sheshshayee, M.S.; Papolu, P.K.; Rao, U.; Tuteja, N.; UdayaKumar, M. Overexpression of Pea DNA Helicase 45 (PDH45) Imparts Tolerance to Multiple Abiotic Stresses in Chili (Capsicum annuum L.). Sci. Rep. 2017, 7, 2760. [Google Scholar] [CrossRef] [PubMed]
- Kesiraju, K.; Tyagi, S.; Mukherjee, S.; Rai, R.; Singh, N.K.; Sreevathsa, R.; Dash, P.K. An Apical Meristem-Targeted in Planta Transformation Method for the Development of Transgenics in Flax (Linum Usitatissimum): Optimization and Validation. Front. Plant Sci. 2021, 11, 562056. [Google Scholar] [CrossRef]
- Gupta, P.; Dash, P.K. Precise Method of in Situ Drought Stress Induction in Flax (Linum usitatissimum) for RNA Isolation towards down-Stream Analysis. Ann. Agric. Res. 2015, 36, 10–17. [Google Scholar]
- Bastia, R.; Pandit, E.; Sanghamitra, P.; Barik, S.R.; Nayak, D.K.; Sahoo, A.; Moharana, A.; Meher, J.; Dash, P.K.; Raj, R.; et al. Association Mapping for Quantitative Trait Loci Controlling Superoxide Dismutase, Flavonoids, Anthocyanins, Carotenoids, γ-Oryzanol and Antioxidant Activity in Rice. Agronomy 2022, 12, 3036. [Google Scholar] [CrossRef]
- Pradhan, K.C.; Pandit, E.; Mohanty, S.P.; Moharana, A.; Sanghamitra, P.; Meher, J.; Jena, B.K.; Dash, P.K.; Behera, L.; Mohapatra, P.M.; et al. Development of Broad Spectrum and Durable Bacterial Blight Resistant Variety through Pyramiding of Four Resistance Genes in Rice. Agronomy 2022, 12, 1903. [Google Scholar] [CrossRef]
- Sedeek, K.E.M.; Mahas, A.; Mahfouz, M. Plant Genome Engineering for Targeted Improvement of Crop Traits. Front. Plant Sci. 2019, 10, 114. [Google Scholar] [CrossRef] [Green Version]
- Pradhan, S.K.; Pandit, E.; Pawar, S.; Bharati, B.; Chatopadhyay, K.; Singh, S.; Dash, P.; Reddy, J.N. Association Mapping Reveals Multiple QTLs for Grain Protein Content in Rice Useful for Biofortification. Mol. Genet. Genom. 2019, 294, 963–983. [Google Scholar] [CrossRef]
- Yang, F.; Cao, Y. Biosynthesis of Phloroglucinol Compounds in Microorganisms—Review. Appl. Microbiol. Biotechnol. 2012, 93, 487–495. [Google Scholar] [CrossRef]
- Rao, G.; Lee, J.-K.; Zhao, H. Directed Evolution of Phloroglucinol Synthase PhlD with Increased Stability for Phloroglucinol Production. Appl. Microbiol. Biotechnol. 2013, 97, 5861–5867. [Google Scholar] [CrossRef]
- Yu, S.; Guo, L.; Zhao, L.; Chen, Z.; Huo, Y. Metabolic Engineering of E. Coli for Producing Phloroglucinol from Acetate. Appl. Microbiol. Biotechnol. 2020, 104, 7787–7799. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; Weller, D.M.; Thomashow, L.S. Frequency of Antibiotic-Producing Pseudomonas Spp. in Natural Environments. Appl. Environ. Microbiol. 1997, 63, 881–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picard, C.; di Cello, F.; Ventura, M.; Fani, R.; Guckert, A. Frequency and Biodiversity of 2,4-Diacetylphloroglucinol-Producing Bacteria Isolated from the Maize Rhizosphere at Different Stages of Plant Growth. Appl. Environ. Microbiol. 2000, 66, 948–955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donadio, S.; Staver, M.; McAlpine, J.; Swanson, S.; Katz, L. Modular Organization of Genes Required for Complex Polyketide Biosynthesis. Science (1979) 1991, 252, 675–679. [Google Scholar] [CrossRef]
- Katz, L.; Donadio, S. POLYKETIDE SYNTHESIS: Prospects for Hybrid Antibiotics. Annu. Rev. Microbiol. 1993, 47, 875–912. [Google Scholar] [CrossRef] [PubMed]
- Nowak-Thompson, B.; Gould, S.J.; Loper, J.E. Identification and Sequence Analysis of the Genes Encoding a Polyketide Synthase Required for Pyoluteorin Biosynthesis in Pseudomonas Fluorescens Pf-5. Gene 1997, 204, 17–24. [Google Scholar] [CrossRef]
- McDaniel, R.; Ebert-Khosla, S.; Hopwood, D.A.; Khosla, C. Engineered Biosynthesis of Novel Polyketides. Science (1979) 1993, 262, 1546–1550. [Google Scholar] [CrossRef]
- Bangera, M.G.; Weller, D.M.; Thomshaw, L.S. Genetic Analysis of the 2,4-Diacetylphloroglucinol Biosynthetic Locus from Pseudomonas Fluorescens Q2-87. In Advances in Molecular Genetics of Plant-Microbe Interactions; Daniels, M., Downie, J., Osbourn, A., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1994. [Google Scholar]
- Bangera, M.G.; Thomashow, L.S. Identification and Characterization of a Gene Cluster for Synthesis of the Polyketide Antibiotic 2,4-Diacetylphloroglucinol from Pseudomonas fluorescens Q2-87. J. Bacteriol. 1999, 181, 3155–3163. [Google Scholar] [CrossRef] [Green Version]
- Ramette, A.; Moënne-Loccoz, Y.; Défago, G. Polymorphism of the Polyketide Synthase Gene PhlD in Biocontrol Fluorescent Pseudomonads Producing 2,4-Diacetylphloroglucinol and Comparison of PhlD with Plant Polyketide Synthases. Mol. Plant-Microbe Interact. 2001, 14, 639–652. [Google Scholar] [CrossRef] [Green Version]
- Tropf, S.; Lanz, T.; Rensing, S.A.; Schrder, J.; Schrder, G. Evidence That Stilbene Synthases Have Developed from Chalcone Synthases Several Times in the Course of Evolution. J. Mol. Evol. 1994, 38, 610–618. [Google Scholar] [CrossRef]
- de Luca, D.; Lauritano, C. In Silico Identification of Type III PKS Chalcone and Stilbene Synthase Homologs in Marine Photosynthetic Organisms. Biology 2020, 9, 110. [Google Scholar] [CrossRef] [PubMed]
- Höfte, M. The Use of Pseudomonas spp. as Bacterial Biocontrol Agents to Control Plant Disease; Burleigh Dodds: Cambridge, UK, 2021. [Google Scholar]
- Panpatte, D.G.; Jhala, Y.K.; Shelat, H.N.; Vyas, R.V. Pseudomonas Fluorescens: A Promising Biocontrol Agent and PGPR for Sustainable Agriculture. In Microbial Inoculants in Sustainable Agricultural Productivity; Springer India: New Delhi, India, 2016; pp. 257–270. [Google Scholar]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, P.; Dash, P.K.; Sanjay, T.D.; Pradhan, S.K.; Sreevathsa, R.; Rai, R. Cloning and Molecular Characterization of the phlD Gene Involved in the Biosynthesis of “Phloroglucinol”, a Compound with Antibiotic Properties from Plant Growth Promoting Bacteria Pseudomonas spp. Antibiotics 2023, 12, 260. https://doi.org/10.3390/antibiotics12020260
Gupta P, Dash PK, Sanjay TD, Pradhan SK, Sreevathsa R, Rai R. Cloning and Molecular Characterization of the phlD Gene Involved in the Biosynthesis of “Phloroglucinol”, a Compound with Antibiotic Properties from Plant Growth Promoting Bacteria Pseudomonas spp. Antibiotics. 2023; 12(2):260. https://doi.org/10.3390/antibiotics12020260
Chicago/Turabian StyleGupta, Payal, Prasanta K. Dash, Tenkabailu Dharmanna Sanjay, Sharat Kumar Pradhan, Rohini Sreevathsa, and Rhitu Rai. 2023. "Cloning and Molecular Characterization of the phlD Gene Involved in the Biosynthesis of “Phloroglucinol”, a Compound with Antibiotic Properties from Plant Growth Promoting Bacteria Pseudomonas spp." Antibiotics 12, no. 2: 260. https://doi.org/10.3390/antibiotics12020260
APA StyleGupta, P., Dash, P. K., Sanjay, T. D., Pradhan, S. K., Sreevathsa, R., & Rai, R. (2023). Cloning and Molecular Characterization of the phlD Gene Involved in the Biosynthesis of “Phloroglucinol”, a Compound with Antibiotic Properties from Plant Growth Promoting Bacteria Pseudomonas spp. Antibiotics, 12(2), 260. https://doi.org/10.3390/antibiotics12020260







