Horizontal Gene Transfer of Antibiotic Resistance Genes in Biofilms
Abstract
:1. Introduction
1.1. Methodology
1.2. Biofilm Development and Occurrence
1.3. Biofilms as Reservoir for Antibiotic Resistance Genes
1.4. Horizontal Gene Transfer in Biofilms
1.5. Environmental Factors Influencing Biofilm Formation
1.6. Abiotic and Biotic Factors Influence Horizontal Gene Transfer in Biofilms
1.7. State of the Art of Biofilm Studies
1.8. Culture-Independent Approaches
2. Transformation
2.1. Transformation of Gram-negative Bacteria in Biofilms
Methodology to Assess the Transformation Rate in Biofilms
2.2. Transformation of Gram-positive Bacteria in Biofilms
Higher Transformation Rates in Early Biofilms Than in Planktonic Cultures
3. Transduction
3.1. Generalized Transduction
3.2. Specialized Transduction
3.3. Lateral Transduction
3.4. Lack of Transduction Experiments in Biofilms
3.5. Potential of Phages in ARG Dissemination
3.5.1. Occurrence of Phages in Aquatic Environments
3.5.2. Phage Host Range and Its Contribution to the Spread of ARGs
3.6. Challenges of Studying Transduction in Biofilms
4. Conjugation
4.1. Conjugative Plasmids
Methodology to Assess Conjugative Plasmid Transfer Rates in situ
4.2. Integrative and Conjugative Elements (ICEs)
Methodology to Assess ICE Transfer Rates in Biofilms
5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rieu, A.; Aoudia, N.; Jego, G.; Chluba, J.; Yousfi, N.; Briandet, R.; Deschamps, J.; Gasquet, B.; Monedero, V.; Garrido, C.; et al. The biofilm mode of life boosts the anti-inflammatory properties of Lactobacillus. Cell Microbiol. 2014, 16, 1836–1853. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wuertz, S. Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 2019, 17, 247–260. [Google Scholar] [CrossRef]
- Borsodi, A.K.; Anda, D.; Makk, J.; Krett, G.; Dobosy, P.; Büki, G.; Erőss, A.; Mádl-Szőnyi, J. Biofilm forming bacteria and archaea in thermal karst springs of Gellért Hill discharge area (Hungary). J. Basic Microbiol. 2018, 58, 928–937. [Google Scholar] [CrossRef]
- Kochetkova, T.V.; Toshchakov, S.V.; Zayulina, K.S.; Elcheninov, A.G.; Zavarzina, D.G.; Lavrushin, V.Y.; Bonch-Osmolovskaya, E.A.; Kublanov, I.V. Hot in Cold: Microbial Life in the Hottest Springs in Permafrost. Microorganisms 2020, 8, 1308. [Google Scholar] [CrossRef]
- Bowen, W.H.; Burne, R.A.; Wu, H.; Koo, H. Oral Biofilms: Pathogens, Matrix, and Polymicrobial Interactions in Microenvironments. Trends Microbiol. 2018, 26, 229–242. [Google Scholar] [CrossRef]
- Savio, D.; Stadler, P.; Reischer, G.H.; Kirschner, A.K.T.; Demeter, K.; Linke, R.; Blaschke, A.P.; Sommer, R.; Szewzyk, U.; Wilhartitz, I.C.; et al. Opening the black box of spring water microbiology from alpine karst aquifers to support proactive drinking water resource management. WIREs Water 2018, 5, e1282. [Google Scholar] [CrossRef]
- Pandit, A.; Adholeya, A.; Cahill, D.; Brau, L.; Kochar, M. Microbial biofilms in nature: Unlocking their potential for agricultural applications. J. Appl. Microbiol. 2020, 129, 199–211. [Google Scholar] [CrossRef]
- Sjöberg, S.; Stairs, C.; Allard, B.; Hallberg, R.; Homa, F.; Martin, T.; Ettema, T.J.G.; Dupraz, C. Bubble biofilm: Bacterial colonization of air-air interface. Biofilm 2020, 2, 100030. [Google Scholar] [CrossRef]
- Paytubi, S.; Cansado, C.; Madrid, C.; Balsalobre, C. Nutrient Composition Promotes Switching between Pellicle and Bottom Biofilm in Salmonella. Front. Microbiol. 2017, 8, 2160. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Baveye, P.; Neu, T.R.; Stoodley, P.; Szewzyk, U.; Wingender, J.; Wuertz, S. Who put the film in biofilm? The migration of a term from wastewater engineering to medicine and beyond. NPJ Biofilms Microbiomes 2021, 7, 10. [Google Scholar] [CrossRef]
- Sauer, K.; Stoodley, P.; Goeres, D.M.; Hall-Stoodley, L.; Burmølle, M.; Stewart, P.S.; Bjarnsholt, T. The biofilm life cycle: Expanding the conceptual model of biofilm formation. Nat. Rev. Microbiol. 2022, 20, 608–620. [Google Scholar] [CrossRef]
- Motta, J.-P.; Wallace, J.L.; Buret, A.G.; Deraison, C.; Vergnolle, N. Gastrointestinal biofilms in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 314–334. [Google Scholar] [CrossRef]
- Mishra, S.; Huang, Y.; Li, J.; Wu, X.; Zhou, Z.; Lei, Q.; Bhatt, P.; Chen, S. Biofilm-mediated bioremediation is a powerful tool for the removal of environmental pollutants. Chemosphere 2022, 294, 133609. [Google Scholar] [CrossRef]
- Hayta, E.N.; Ertelt, M.J.; Kretschmer, M.; Lieleg, O. Bacterial Materials: Applications of Natural and Modified Biofilms. Adv. Mater. Interfaces 2021, 8, 2101024. [Google Scholar] [CrossRef]
- Little, B.J.; Blackwood, D.J.; Hinks, J.; Lauro, F.M.; Marsili, E.; Okamoto, A.; Rice, S.A.; Wade, S.A.; Flemming, H.-C. Microbially influenced corrosion—Any progress? Corros. Sci. 2020, 170, 108641. [Google Scholar] [CrossRef]
- Curtin, A.M.; Buckley, H.L. Biofouling detection methods that are widely applicable and useful across disciplines: A mini-review. Biofouling 2021, 37, 494–505. [Google Scholar] [CrossRef]
- Cámara, M.; Green, W.; MacPhee, C.E.; Rakowska, P.D.; Raval, R.; Richardson, M.C.; Slater-Jefferies, J.; Steventon, K.; Webb, J.S. Economic significance of biofilms: A multidisciplinary and cross-sectoral challenge. NPJ Biofilms Microbiomes 2022, 8, 42. [Google Scholar] [CrossRef]
- Bahamondez-Canas, T.F.; Heersema, L.A.; Smyth, H.D.C. Current Status of In Vitro Models and Assays for Susceptibility Testing for Wound Biofilm Infections. Biomedicines 2019, 7, 34. [Google Scholar] [CrossRef]
- Vyas, H.K.N.; Xia, B.; Mai-Prochnow, A. Clinically relevant in vitro biofilm models: A need to mimic and recapitulate the host environment. Biofilm 2022, 4, 100069. [Google Scholar] [CrossRef]
- Dunny, G.M.; Hancock, L.E.; Shankar, N. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection: Enterococcal Biofilm Structure and Role in Colonization and Disease; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Schilcher, K.; Horswill, A.R. Staphylococcal Biofilm Development: Structure, Regulation, and Treatment Strategies. Microbiol. Mol. Biol. Rev. 2020, 84, e00026-19. [Google Scholar] [CrossRef]
- Sauer, K.; Camper, A.K.; Ehrlich, G.D.; Costerton, J.W.; Davies, D.G. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 2002, 184, 1140–1154. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.W.K.; Periasamy, S.; Mukherjee, M.; Xie, C.; Kjelleberg, S.; Rice, S.A. Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. ISME J. 2014, 8, 894–907. [Google Scholar] [CrossRef] [PubMed]
- Kviatkovski, I.; Mamane, H.; Lakretz, A.; Sherman, I.; Beno-Moualem, D.; Minz, D. Resistance of a multiple-isolate marine culture to ultraviolet C irradiation: Inactivation vs biofilm formation. Lett. Appl. Microbiol. 2018, 67, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Costa-Orlandi, C.B.; Bila, N.M.; Vaso, C.O.; da Silva Pires, A.C.M.; de Matos Silva, S.; Medina Alarcón, K.P.; Marcos, C.M.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J.S. Polymicrobial biofilms: Impact on fungal pathogenesis. In Understanding Microbial Biofilms; Elsevier: Amsterdam, The Netherlands, 2023; pp. 521–567. ISBN 9780323999779. [Google Scholar]
- Hall, C.W.; Mah, T.-F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Kaplan, J.B.; Mlynek, K.D.; Hettiarachchi, H.; Alamneh, Y.A.; Biggemann, L.; Zurawski, D.V.; Black, C.C.; Bane, C.E.; Kim, R.K.; Granick, M.S. Extracellular polymeric substance (EPS)-degrading enzymes reduce staphylococcal surface attachment and biocide resistance on pig skin in vivo. PLoS ONE 2018, 13, e0205526. [Google Scholar] [CrossRef]
- Hernández-Jiménez, E.; Del Campo, R.; Toledano, V.; Vallejo-Cremades, M.T.; Muñoz, A.; Largo, C.; Arnalich, F.; García-Rio, F.; Cubillos-Zapata, C.; López-Collazo, E. Biofilm vs. planktonic bacterial mode of growth: Which do human macrophages prefer? Biochem. Biophys. Res. Commun. 2013, 441, 947–952. [Google Scholar] [CrossRef]
- Uruén, C.; Chopo-Escuin, G.; Tommassen, J.; Mainar-Jaime, R.C.; Arenas, J. Biofilms as Promoters of Bacterial Antibiotic Resistance and Tolerance. Antibiotics 2020, 10, 3. [Google Scholar] [CrossRef]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef]
- Davidson, D.J.; Spratt, D.; Liddle, A.D. Implant materials and prosthetic joint infection: The battle with the biofilm. EFORT Open Rev. 2019, 4, 633–639. [Google Scholar] [CrossRef]
- Khoddami, S.; Chew, B.H.; Lange, D. Problems and solutions of stent biofilm and encrustations: A review of literature. Turk. J. Urol. 2020, 46, S11–S18. [Google Scholar] [CrossRef]
- Köves, B.; Magyar, A.; Tenke, P. Spectrum and antibiotic resistance of catheter-associated urinary tract infections. GMS Infect. Dis. 2017, 5, Doc06. [Google Scholar] [CrossRef]
- Qiang, L.; Cheng, J.; Mirzoyan, S.; Kerkhof, L.J.; Häggblom, M.M. Characterization of Microplastic-Associated Biofilm Development along a Freshwater-Estuarine Gradient. Environ. Sci. Technol. 2021, 55, 16402–16412. [Google Scholar] [CrossRef] [PubMed]
- Vishwakarma, V. Impact of environmental biofilms: Industrial components and its remediation. J. Basic Microbiol. 2020, 60, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Pompilio, A.; Scribano, D.; Sarshar, M.; Di Bonaventura, G.; Palamara, A.T.; Ambrosi, C. Gram-Negative Bacteria Holding Together in a Biofilm: The Acinetobacter baumannii Way. Microorganisms 2021, 9, 1353. [Google Scholar] [CrossRef]
- Cendra, M.D.M.; Torrents, E. Pseudomonas aeruginosa biofilms and their partners in crime. Biotechnol. Adv. 2021, 49, 107734. [Google Scholar] [CrossRef] [PubMed]
- Guerra, M.E.S.; Destro, G.; Vieira, B.; Lima, A.S.; Ferraz, L.F.C.; Hakansson, A.P.; Darrieux, M.; Converso, T.R. Klebsiella pneumoniae Biofilms and Their Role in Disease Pathogenesis. Front. Cell. Infect. Microbiol. 2022, 12, 877995. [Google Scholar] [CrossRef] [PubMed]
- Folliero, V.; Franci, G.; Dell’Annunziata, F.; Giugliano, R.; Foglia, F.; Sperlongano, R.; de Filippis, A.; Finamore, E.; Galdiero, M. Evaluation of Antibiotic Resistance and Biofilm Production among Clinical Strain Isolated from Medical Devices. Int. J. Microbiol. 2021, 2021, 9033278. [Google Scholar] [CrossRef]
- Yaita, K.; Gotoh, K.; Nakano, R.; Iwahashi, J.; Sakai, Y.; Horita, R.; Yano, H.; Watanabe, H. Biofilm-Forming by Carbapenem Resistant Enterobacteriaceae May Contribute to the Blood Stream Infection. Int. J. Mol. Sci. 2019, 20, 5954. [Google Scholar] [CrossRef]
- Gemba, M.; Rosiak, E.; Nowak-Życzyńska, Z.; Kałęcka, P.; Łodykowska, E.; Kołożyn-Krajewska, D. Factors Influencing Biofilm Formation by Salmonella enterica sv. Typhimurium, E. cloacae, E. hormaechei, Pantoea spp., and Bacillus spp. Isolated from Human Milk Determined by PCA Analysis. Foods 2022, 11, 3862. [Google Scholar] [CrossRef]
- Ramić, D.; Ogrizek, J.; Bucar, F.; Jeršek, B.; Jeršek, M.; Možina, S.S. Campylobacter jejuni Biofilm Control with Lavandin Essential Oils and By-Products. Antibiotics 2022, 11, 854. [Google Scholar] [CrossRef]
- Chen, P.; Wang, J.J.; Hong, B.; Tan, L.; Yan, J.; Zhang, Z.; Liu, H.; Pan, Y.; Zhao, Y. Characterization of Mixed-Species Biofilm Formed by Vibrio parahaemolyticus and Listeria monocytogenes. Front. Microbiol. 2019, 10, 2543. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, L. Antibiofilm effect and mechanism of protocatechuic aldehyde against Vibrio parahaemolyticus. Front. Microbiol. 2022, 13, 1060506. [Google Scholar] [CrossRef]
- De Brito, F.A.E.; de Freitas, A.P.P.; Nascimento, M.S. Multidrug-Resistant Biofilms (MDR): Main Mechanisms of Tolerance and Resistance in the Food Supply Chain. Pathogens 2022, 11, 1416. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal Biofilms. Microbiol. Spectr. 2018, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Idrees, M.; Sawant, S.; Karodia, N.; Rahman, A. Staphylococcus aureus Biofilm: Morphology, Genetics, Pathogenesis and Treatment Strategies. Int. J. Environ. Res. Public Health 2021, 18, 7602. [Google Scholar] [CrossRef] [PubMed]
- Barnes, A.M.T.; Frank, K.L.; Dale, J.L.; Manias, D.A.; Powers, J.L.; Dunny, G.M. Enterococcus faecalis colonizes and forms persistent biofilm microcolonies on undamaged endothelial surfaces in a rabbit endovascular infection model. FEMS Microbes 2021, 2, xtab014. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.T.; Andam, C.P. Extensive Horizontal Gene Transfer within and between Species of Coagulase-Negative Staphylococcus. Genome Biol. Evol. 2021, 13, evab206. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Ren, B.; Zhou, X.; Zhang, L.; Xu, X. Cross-kingdom interaction between Candida albicans and oral bacteria. Front. Microbiol. 2022, 13, 911623. [Google Scholar] [CrossRef]
- Van Dyck, K.; Viela, F.; Mathelié-Guinlet, M.; Demuyser, L.; Hauben, E.; Jabra-Rizk, M.A.; Velde, G.V.; Dufrêne, Y.F.; Krom, B.P.; van Dijck, P. Adhesion of Staphylococcus aureus to Candida albicans During Co-Infection Promotes Bacterial Dissemination Through the Host Immune Response. Front. Cell. Infect. Microbiol. 2020, 10, 624839. [Google Scholar] [CrossRef] [PubMed]
- Palencia, S.L.; García, A.; Palencia, M. Multiple surface interaction mechanisms direct the anchoring, co-aggregation and formation of dual-species biofilm between Candida albicans and Helicobacter pylori. J. Adv. Res. 2022, 35, 169–185. [Google Scholar] [CrossRef] [PubMed]
- Tsui, C.; Kong, E.F.; Jabra-Rizk, M.A. Pathogenesis of Candida albicans biofilm. Pathog. Dis. 2016, 74, ftw018. [Google Scholar] [CrossRef]
- Cangui-Panchi, S.P.; Ñacato-Toapanta, A.L.; Enríquez-Martínez, L.J.; Reyes, J.; Garzon-Chavez, D.; Machado, A. Biofilm-forming microorganisms causing hospital-acquired infections from intravenous catheter: A systematic review. Curr. Res. Microb. Sci. 2022, 3, 100175. [Google Scholar] [CrossRef]
- Rumbaugh, K.P.; Sauer, K. Biofilm dispersion. Nat. Rev. Microbiol. 2020, 18, 571–586. [Google Scholar] [CrossRef]
- Caldara, M.; Belgiovine, C.; Secchi, E.; Rusconi, R. Environmental, Microbiological, and Immunological Features of Bacterial Biofilms Associated with Implanted Medical Devices. Clin. Microbiol. Rev. 2022, 35, e0022120. [Google Scholar] [CrossRef]
- Stewart, P.S.; Bjarnsholt, T. Risk factors for chronic biofilm-related infection associated with implanted medical devices. Clin. Microbiol. Infect. 2020, 26, 1034–1038. [Google Scholar] [CrossRef]
- Pietrocola, G.; Campoccia, D.; Motta, C.; Montanaro, L.; Arciola, C.R.; Speziale, P. Colonization and Infection of Indwelling Medical Devices by Staphylococcus aureus with an Emphasis on Orthopedic Implants. Int. J. Mol. Sci. 2022, 23, 5958. [Google Scholar] [CrossRef] [PubMed]
- Krukiewicz, K.; Kazek-Kęsik, A.; Brzychczy-Włoch, M.; Łos, M.J.; Ateba, C.N.; Mehrbod, P.; Ghavami, S.; Shyntum, D.Y. Recent Advances in the Control of Clinically Important Biofilms. Int. J. Mol. Sci. 2022, 23, 9526. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, R.; Yousefimashouf, R.; Arabestani, M.R.; Sedighi, I.; Alikhani, M.Y. The issue beyond resistance: Methicillin-resistant Staphylococcus epidermidis biofilm formation is induced by subinhibitory concentrations of cloxacillin, cefazolin, and clindamycin. PLoS ONE 2022, 17, e0277287. [Google Scholar] [CrossRef]
- Josephs-Spaulding, J.; Singh, O.V. Medical Device Sterilization and Reprocessing in the Era of Multidrug-Resistant (MDR) Bacteria: Issues and Regulatory Concepts. Front. Med. Technol. 2020, 2, 587352. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Alam, M.-U.; Luies, S.K.; Kamal, A.; Ferdous, S.; Lin, A.; Sharior, F.; Khan, R.; Rahman, Z.; Parvez, S.M.; et al. Contamination of Fresh Produce with Antibiotic-Resistant Bacteria and Associated Risks to Human Health: A Scoping Review. Int. J. Environ. Res. Public Health 2021, 19, 360. [Google Scholar] [CrossRef] [PubMed]
- Blau, K.; Jacquiod, S.; Sørensen, S.J.; Su, J.-Q.; Zhu, Y.-G.; Smalla, K.; Jechalke, S. Manure and Doxycycline Affect the Bacterial Community and Its Resistome in Lettuce Rhizosphere and Bulk Soil. Front. Microbiol. 2019, 10, 725. [Google Scholar] [CrossRef]
- Kaviani Rad, A.; Astaykina, A.; Streletskii, R.; Afsharyzad, Y.; Etesami, H.; Zarei, M.; Balasundram, S.K. An Overview of Antibiotic Resistance and Abiotic Stresses Affecting Antimicrobial Resistance in Agricultural Soils. Int. J. Environ. Res. Public Health 2022, 19, 4666. [Google Scholar] [CrossRef]
- Lima, T.; Domingues, S.; Da Silva, G.J. Manure as a Potential Hotspot for Antibiotic Resistance Dissemination by Horizontal Gene Transfer Events. Vet. Sci. 2020, 7, 110. [Google Scholar] [CrossRef]
- Li, H.; Zheng, X.; Tan, L.; Shao, Z.; Cao, H.; Xu, Y. The vertical migration of antibiotic-resistant genes and pathogens in soil and vegetables after the application of different fertilizers. Environ. Res. 2022, 203, 111884. [Google Scholar] [CrossRef]
- Sharma, R.; Bisaria, V.S.; Sharma, S. Rhizosphere: A Home for Human Pathogens. In Plant Biotic Interactions; Varma, A., Tripathi, S., Prasad, R., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 113–127. ISBN 978-3-030-26656-1. [Google Scholar]
- Schlech, W.F. Epidemiology and Clinical Manifestations of Listeria monocytogenes Infection. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- He, Y.; Yuan, Q.; Mathieu, J.; Stadler, L.; Senehi, N.; Sun, R.; Alvarez, P.J.J. Antibiotic resistance genes from livestock waste: Occurrence, dissemination, and treatment. Npj Clean Water 2020, 3, 4. [Google Scholar] [CrossRef]
- Mestrovic, T.; Robles Aguilar, G.; Swetschinski, L.R.; Ikuta, K.S.; Gray, A.P.; Davis Weaver, N.; Han, C.; Wool, E.E.; Gershberg Hayoon, A.; Hay, S.I.; et al. The burden of bacterial antimicrobial resistance in the WHO European region in 2019: A cross-country systematic analysis. Lancet Public Health 2022, 7, e897–e913. [Google Scholar] [CrossRef]
- Rahmoun, L.A.; Azrad, M.; Peretz, A. Antibiotic Resistance and Biofilm Production Capacity in Clostridioides difficile. Front. Cell. Infect. Microbiol. 2021, 11, 683464. [Google Scholar] [CrossRef] [PubMed]
- Lozano, C.; López, M.; Rojo-Bezares, B.; Sáenz, Y. Antimicrobial Susceptibility Testing in Pseudomonas aeruginosa Biofilms: One Step Closer to a Standardized Method. Antibiotics 2020, 9, 880. [Google Scholar] [CrossRef]
- Wu, H.; Moser, C.; Wang, H.-Z.; Høiby, N.; Song, Z.-J. Strategies for combating bacterial biofilm infections. Int. J. Oral Sci. 2015, 7, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Choudhary, M.I.; Römling, U.; Nadeem, F.; Bilal, H.M.; Zafar, M.; Jahan, H.; Ur-Rahman, A. Innovative Strategies to Overcome Antimicrobial Resistance and Tolerance. Microorganisms 2022, 11, 16. [Google Scholar] [CrossRef] [PubMed]
- Sotto, A.; Laurent, F.; Schuldiner, S.; Vouillarmet, J.; Corvec, S.; Bemer, P.; Boutoille, D.; Dunyach-Rémy, C.; Lavigne, J.-P. Evaluation of the Use of Antibiofilmogram Technology in the Clinical Evolution of Foot Ulcers Infected by Staphylococcus aureus in Persons Living with Diabetes: A Pilot Study. J. Clin. Med. 2021, 10, 5928. [Google Scholar] [CrossRef] [PubMed]
- Tasse, J.; Croisier, D.; Badel-Berchoux, S.; Chavanet, P.; Bernardi, T.; Provot, C.; Laurent, F. Preliminary results of a new antibiotic susceptibility test against biofilm installation in device-associated infections: The Antibiofilmogram®. Pathog. Dis. 2016, 74, ftw057. [Google Scholar] [CrossRef]
- Udaondo, Z.; Matilla, M.A. Mining for novel antibiotics in the age of antimicrobial resistance. Microb. Biotechnol. 2020, 13, 1702–1704. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.-P.; Yang, Y.; Lu, D.-P.; Niu, Z.-S.; Feng, J.-N.; Chen, Y.-R.; Tou, F.-Y.; Garner, E.; Xu, J.; Liu, M.; et al. Biofilms as a sink for antibiotic resistance genes (ARGs) in the Yangtze Estuary. Water Res. 2018, 129, 277–286. [Google Scholar] [CrossRef]
- Jian, Z.; Zeng, L.; Xu, T.; Sun, S.; Yan, S.; Yang, L.; Huang, Y.; Jia, J.; Dou, T. Antibiotic resistance genes in bacteria: Occurrence, spread, and control. J. Basic Microbiol. 2021, 61, 1049–1070. [Google Scholar] [CrossRef]
- Stanley, D.; Batacan, R.; Bajagai, Y.S. Rapid growth of antimicrobial resistance: The role of agriculture in the problem and the solutions. Appl. Microbiol. Biotechnol. 2022, 106, 6953–6962. [Google Scholar] [CrossRef]
- Hiller, C.X.; Hübner, U.; Fajnorova, S.; Schwartz, T.; Drewes, J.E. Antibiotic microbial resistance (AMR) removal efficiencies by conventional and advanced wastewater treatment processes: A review. Sci. Total Environ. 2019, 685, 596–608. [Google Scholar] [CrossRef]
- Abe, K.; Nomura, N.; Suzuki, S. Biofilms: Hot spots of horizontal gene transfer (HGT) in aquatic environments, with a focus on a new HGT mechanism. FEMS Microbiol. Ecol. 2020, 96, fiaa031. [Google Scholar] [CrossRef] [PubMed]
- Marti, E.; Variatza, E.; Balcazar, J.L. The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol. 2014, 22, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Domingues, S.; Nielsen, K.M. Membrane vesicles and horizontal gene transfer in prokaryotes. Curr. Opin. Microbiol. 2017, 38, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.M.; Grossman, A.D. Integrative and Conjugative Elements (ICEs): What They Do and How They Work. Annu. Rev. Genet. 2015, 49, 577–601. [Google Scholar] [CrossRef] [PubMed]
- Dell’Annunziata, F.; Folliero, V.; Giugliano, R.; de Filippis, A.; Santarcangelo, C.; Izzo, V.; Daglia, M.; Galdiero, M.; Arciola, C.R.; Franci, G. Gene Transfer Potential of Outer Membrane Vesicles of Gram-Negative Bacteria. Int. J. Mol. Sci. 2021, 22, 5985. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xiao, J.; Wang, S.; Zhou, J.; Qin, J.; Jia, Z.; Wang, Y.; Wang, Z.; Zhang, Y.; Hao, H. Research Progress on Bacterial Membrane Vesicles and Antibiotic Resistance. Int. J. Mol. Sci. 2022, 23, 11553. [Google Scholar] [CrossRef]
- Dell’Annunziata, F.; Dell’Aversana, C.; Doti, N.; Donadio, G.; Dal Piaz, F.; Izzo, V.; de Filippis, A.; Galdiero, M.; Altucci, L.; Boccia, G.; et al. Outer Membrane Vesicles Derived from Klebsiella pneumoniae Are a Driving Force for Horizontal Gene Transfer. Int. J. Mol. Sci. 2021, 22, 8732. [Google Scholar] [CrossRef]
- Rumbo, C.; Fernández-Moreira, E.; Merino, M.; Poza, M.; Mendez, J.A.; Soares, N.C.; Mosquera, A.; Chaves, F.; Bou, G. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: A new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011, 55, 3084–3090. [Google Scholar] [CrossRef]
- Li, C.; Wen, R.; Mu, R.; Chen, X.; Ma, P.; Gu, K.; Huang, Z.; Ju, Z.; Lei, C.; Tang, Y.; et al. Outer Membrane Vesicles of Avian Pathogenic Escherichia coli Mediate the Horizontal Transmission of blaCTX-M-55. Pathogens 2022, 11, 481. [Google Scholar] [CrossRef]
- Cao, Y.; Lin, H. Characterization and function of membrane vesicles in Gram-positive bacteria. Appl. Microbiol. Biotechnol. 2021, 105, 1795–1801. [Google Scholar] [CrossRef]
- Seike, S.; Kobayashi, H.; Ueda, M.; Takahashi, E.; Okamoto, K.; Yamanaka, H. Outer Membrane Vesicles Released From Aeromonas Strains Are Involved in the Biofilm Formation. Front. Microbiol. 2020, 11, 613650. [Google Scholar] [CrossRef]
- Zhao, Z.; Wang, L.; Miao, J.; Zhang, Z.; Ruan, J.; Xu, L.; Guo, H.; Zhang, M.; Qiao, W. Regulation of the formation and structure of biofilms by quorum sensing signal molecules packaged in outer membrane vesicles. Sci. Total Environ. 2022, 806, 151403. [Google Scholar] [CrossRef] [PubMed]
- Cooke, A.C.; Florez, C.; Dunshee, E.B.; Lieber, A.D.; Terry, M.L.; Light, C.J.; Schertzer, J.W. Pseudomonas Quinolone Signal-Induced Outer Membrane Vesicles Enhance Biofilm Dispersion in Pseudomonas aeruginosa. mSphere 2020, 5, e01109-20. [Google Scholar] [CrossRef]
- Baumgarten, T.; Sperling, S.; Seifert, J.; von Bergen, M.; Steiniger, F.; Wick, L.Y.; Heipieper, H.J. Membrane vesicle formation as a multiple-stress response mechanism enhances Pseudomonas putida DOT-T1E cell surface hydrophobicity and biofilm formation. Appl. Environ. Microbiol. 2012, 78, 6217–6224. [Google Scholar] [CrossRef]
- Yonezawa, H.; Osaki, T.; Kurata, S.; Fukuda, M.; Kawakami, H.; Ochiai, K.; Hanawa, T.; Kamiya, S. Outer membrane vesicles of Helicobacter pylori TK1402 are involved in biofilm formation. BMC Microbiol. 2009, 9, 197. [Google Scholar] [CrossRef]
- Altindis, E.; Fu, Y.; Mekalanos, J.J. Proteomic analysis of Vibrio cholerae outer membrane vesicles. Proc. Natl. Acad. Sci. USA 2014, 111, E1548–E1556. [Google Scholar] [CrossRef]
- Wang, Y.; Hoffmann, J.P.; Baker, S.M.; zu Bentrup, K.H.; Wimley, W.C.; Fuselier, J.A.; Bitoun, J.P.; Morici, L.A. Inhibition of Streptococcus mutans biofilms with bacterial-derived outer membrane vesicles. BMC Microbiol. 2021, 21, 234. [Google Scholar] [CrossRef] [PubMed]
- Alves, N.J.; Turner, K.B.; Medintz, I.L.; Walper, S.A. Protecting enzymatic function through directed packaging into bacterial outer membrane vesicles. Sci. Rep. 2016, 6, 24866. [Google Scholar] [CrossRef] [PubMed]
- Dubey, G.P.; Ben-Yehuda, S. Intercellular nanotubes mediate bacterial communication. Cell 2011, 144, 590–600. [Google Scholar] [CrossRef]
- Dubey, G.P.; Malli Mohan, G.B.; Dubrovsky, A.; Amen, T.; Tsipshtein, S.; Rouvinski, A.; Rosenberg, A.; Kaganovich, D.; Sherman, E.; Medalia, O.; et al. Architecture and Characteristics of Bacterial Nanotubes. Dev. Cell 2016, 36, 453–461. [Google Scholar] [CrossRef]
- Von Wintersdorff, C.J.H.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef] [Green Version]
- Glen, K.A.; Lamont, I.L. β-lactam Resistance in Pseudomonas aeruginosa: Current Status, Future Prospects. Pathogens 2021, 10, 1638. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Dorosky, R.J.; Han, C.S.; Lo, C.-C.; Dichosa, A.E.K.; Chain, P.S.; Yu, J.M.; Pierson, L.S.; Pierson, E.A. Adaptation genomics of a small-colony variant in a Pseudomonas chlororaphis 30-84 biofilm. Appl. Environ. Microbiol. 2015, 81, 890–899. [Google Scholar] [CrossRef]
- Chen, Y.; Li, P.; Huang, Y.; Yu, K.; Chen, H.; Cui, K.; Huang, Q.; Zhang, J.; Gin, K.Y.H.; He, Y. Environmental media exert a bottleneck in driving the dynamics of antibiotic resistance genes in modern aquatic environment. Water Res. 2019, 162, 127–138. [Google Scholar] [CrossRef]
- Haudiquet, M.; de Sousa, J.M.; Touchon, M.; Rocha, E.P.C. Selfish, promiscuous and sometimes useful: How mobile genetic elements drive horizontal gene transfer in microbial populations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2022, 377, 20210234. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Konkel, M.E.; Lu, X. Antimicrobial Resistance Gene Transfer from Campylobacter jejuni in Mono- and Dual-Species Biofilms. Appl. Environ. Microbiol. 2021, 87, e0065921. [Google Scholar] [CrossRef] [PubMed]
- Gu, T.; Meesrisom, A.; Luo, Y.; Dinh, Q.N.; Lin, S.; Yang, M.; Sharma, A.; Tang, R.; Zhang, J.; Jia, Z.; et al. Listeria monocytogenes biofilm formation as affected by stainless steel surface topography and coating composition. Food Control. 2021, 130, 108275. [Google Scholar] [CrossRef]
- Xu, L.-C.; Siedlecki, C.A. Submicron topography design for controlling staphylococcal bacterial adhesion and biofilm formation. J. Biomed. Mater. Res. A 2022, 110, 1238–1250. [Google Scholar] [CrossRef]
- Krsmanovic, M.; Biswas, D.; Ali, H.; Kumar, A.; Ghosh, R.; Dickerson, A.K. Hydrodynamics and surface properties influence biofilm proliferation. Adv. Colloid Interface Sci. 2021, 288, 102336. [Google Scholar] [CrossRef]
- Zheng, S.; Bawazir, M.; Dhall, A.; Kim, H.-E.; He, L.; Heo, J.; Hwang, G. Implication of Surface Properties, Bacterial Motility, and Hydrodynamic Conditions on Bacterial Surface Sensing and Their Initial Adhesion. Front. Bioeng. Biotechnol. 2021, 9, 643722. [Google Scholar] [CrossRef]
- Lee, S.W.; Phillips, K.S.; Gu, H.; Kazemzadeh-Narbat, M.; Ren, D. How microbes read the map: Effects of implant topography on bacterial adhesion and biofilm formation. Biomaterials 2021, 268, 120595. [Google Scholar] [CrossRef]
- Ishihama, H.; Ishii, K.; Nagai, S.; Kakinuma, H.; Sasaki, A.; Yoshioka, K.; Kuramoto, T.; Shiono, Y.; Funao, H.; Isogai, N.; et al. An antibacterial coated polymer prevents biofilm formation and implant-associated infection. Sci. Rep. 2021, 11, 3602. [Google Scholar] [CrossRef]
- Kloss, M.; Moerke, C.; Woitschach, F.; Wulf, K.; Illner, S.; Schulz, S.; Pauker, V.I.; Riedel, K.; Grabow, N.; Ince, H.; et al. Novel dalbavancin-PLLA implant coating prevents hematogenous Staphylococcus aureus infection in a minimally invasive mouse tail vein model. Front. Bioeng. Biotechnol. 2022, 10, 1021827. [Google Scholar] [CrossRef] [PubMed]
- Silva, V.; Pereira, J.E.; Maltez, L.; Poeta, P.; Igrejas, G. Influence of Environmental Factors on Biofilm Formation of Staphylococci Isolated from Wastewater and Surface Water. Pathogens 2022, 11, 1069. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, J.; Ji, Y. Environmental factors modulate biofilm formation by Staphylococcus aureus. Sci. Prog. 2020, 103, 36850419898659. [Google Scholar] [CrossRef] [PubMed]
- Behbahani, S.B.; Kiridena, S.D.; Wijayaratna, U.N.; Taylor, C.; Anker, J.N.; Tzeng, T.-R.J. pH variation in medical implant biofilms: Causes, measurements, and its implications for antibiotic resistance. Front. Microbiol. 2022, 13, 1028560. [Google Scholar] [CrossRef] [PubMed]
- Blanco, Y.; Rivas, L.A.; González-Toril, E.; Ruiz-Bermejo, M.; Moreno-Paz, M.; Parro, V.; Palacín, A.; Aguilera, Á.; Puente-Sánchez, F. Environmental parameters, and not phylogeny, determine the composition of extracellular polymeric substances in microbial mats from extreme environments. Sci. Total Environ. 2019, 650, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Sherman, E.; Bayles, K.; Moormeier, D.; Endres, J.; Wei, T. Observations of Shear Stress Effects on Staphylococcus aureus Biofilm Formation. mSphere 2019, 4, e00372-19. [Google Scholar] [CrossRef] [PubMed]
- Tsagkari, E.; Connelly, S.; Liu, Z.; McBride, A.; Sloan, W.T. The role of shear dynamics in biofilm formation. NPJ Biofilms Microbiomes 2022, 8, 33. [Google Scholar] [CrossRef]
- Bernardi, S.; Anderson, A.; Macchiarelli, G.; Hellwig, E.; Cieplik, F.; Vach, K.; Al-Ahmad, A. Subinhibitory Antibiotic Concentrations Enhance Biofilm Formation of Clinical Enterococcus faecalis Isolates. Antibiotics 2021, 10, 874. [Google Scholar] [CrossRef]
- Pinilla-Redondo, R.; Riber, L.; Sørensen, S.J. Fluorescence Recovery Allows the Implementation of a Fluorescence Reporter Gene Platform Applicable for the Detection and Quantification of Horizontal Gene Transfer in Anoxic Environments. Appl. Environ. Microbiol. 2018, 84, e02507-17. [Google Scholar] [CrossRef] [Green Version]
- Skolimowski, M.; Nielsen, M.W.; Emnéus, J.; Molin, S.; Taboryski, R.; Sternberg, C.; Dufva, M.; Geschke, O. Microfluidic dissolved oxygen gradient generator biochip as a useful tool in bacterial biofilm studies. Lab Chip 2010, 10, 2162–2169. [Google Scholar] [CrossRef] [PubMed]
- Nagasawa, R.; Sato, T.; Nomura, N.; Nakamura, T.; Senpuku, H. Potential Risk of Spreading Resistance Genes within Extracellular-DNA-Dependent Biofilms of Streptococcus mutans in Response to Cell Envelope Stress Induced by Sub-MICs of Bacitracin. Appl. Environ. Microbiol. 2020, 86, e00770-20. [Google Scholar] [CrossRef] [PubMed]
- Chow, L.K.M.; Ghaly, T.M.; Gillings, M.R. A survey of sub-inhibitory concentrations of antibiotics in the environment. J. Environ. Sci. 2021, 99, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Christensen, B.B.; Sternberg, C.; Andersen, J.B.; Eberl, L.; Moller, S.; Givskov, M.; Molin, S. Establishment of new genetic traits in a microbial biofilm community. Appl. Environ. Microbiol. 1998, 64, 2247–2255. [Google Scholar] [CrossRef]
- Król, J.E.; Nguyen, H.D.; Rogers, L.M.; Beyenal, H.; Krone, S.M.; Top, E.M. Increased transfer of a multidrug resistance plasmid in Escherichia coli biofilms at the air-liquid interface. Appl. Environ. Microbiol. 2011, 77, 5079–5088. [Google Scholar] [CrossRef] [PubMed]
- Benz, F.; Huisman, J.S.; Bakkeren, E.; Herter, J.A.; Stadler, T.; Ackermann, M.; Diard, M.; Egli, A.; Hall, A.R.; Hardt, W.-D.; et al. Plasmid- and strain-specific factors drive variation in ESBL-plasmid spread in vitro and in vivo. ISME J. 2021, 15, 862–878. [Google Scholar] [CrossRef]
- Haudiquet, M.; Buffet, A.; Rendueles, O.; Rocha, E.P.C. Interplay between the cell envelope and mobile genetic elements shapes gene flow in populations of the nosocomial pathogen Klebsiella pneumoniae. PLoS Biol. 2021, 19, e3001276. [Google Scholar] [CrossRef]
- Cook, L.; Chatterjee, A.; Barnes, A.; Yarwood, J.; Hu, W.-S.; Dunny, G. Biofilm growth alters regulation of conjugation by a bacterial pheromone. Mol. Microbiol. 2011, 81, 1499–1510. [Google Scholar] [CrossRef]
- Cook, L.C.; Dunny, G.M. Effects of biofilm growth on plasmid copy number and expression of antibiotic resistance genes in Enterococcus faecalis. Antimicrob. Agents Chemother. 2013, 57, 1850–1856. [Google Scholar] [CrossRef]
- Alderliesten, J.B.; Duxbury, S.J.N.; Zwart, M.P.; de Visser, J.A.G.M.; Stegeman, A.; Fischer, E.A.J. Effect of donor-recipient relatedness on the plasmid conjugation frequency: A meta-analysis. BMC Microbiol. 2020, 20, 135. [Google Scholar] [CrossRef]
- Stalder, T.; Top, E. Plasmid transfer in biofilms: A perspective on limitations and opportunities. NPJ Biofilms Microbiomes 2016, 2, 16022. [Google Scholar] [CrossRef] [PubMed]
- Røder, H.L.; Trivedi, U.; Russel, J.; Kragh, K.N.; Herschend, J.; Thalsø-Madsen, I.; Tolker-Nielsen, T.; Bjarnsholt, T.; Burmølle, M.; Madsen, J.S. Biofilms can act as plasmid reserves in the absence of plasmid specific selection. NPJ Biofilms Microbiomes 2021, 7, 78. [Google Scholar] [CrossRef] [PubMed]
- Metzger, G.A.; Ridenhour, B.J.; France, M.; Gliniewicz, K.; Millstein, J.; Settles, M.L.; Forney, L.J.; Stalder, T.; Top, E.M. Biofilms preserve the transmissibility of a multi-drug resistance plasmid. NPJ Biofilms Microbiomes 2022, 8, 95. [Google Scholar] [CrossRef]
- Beaudoin, D.L.; Bryers, J.D.; Cunningham, A.B.; Peretti, S.W. Mobilization of broad host range plasmid from Pseudomonas putida to established biofilm of Bacillus azotoformans. II. Modeling. Biotechnol. Bioeng. 1998, 57, 280–286. [Google Scholar] [CrossRef]
- Angles, M.L.; Marshall, K.C.; Goodman, A.E. Plasmid Transfer between Marine Bacteria in the Aqueous Phase and Biofilms in Reactor Microcosms. Appl. Environ. Microbiol. 1993, 59, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Savage, V.J.; Chopra, I.; O’Neill, A.J. Staphylococcus aureus biofilms promote horizontal transfer of antibiotic resistance. Antimicrob. Agents Chemother. 2013, 57, 1968–1970. [Google Scholar] [CrossRef]
- Tanner, W.D.; Atkinson, R.M.; Goel, R.K.; Toleman, M.A.; Benson, L.S.; Porucznik, C.A.; VanDerslice, J.A. Horizontal transfer of the blaNDM-1 gene to Pseudomonas aeruginosa and Acinetobacter baumannii in biofilms. FEMS Microbiol. Lett. 2017, 364, fnx048. [Google Scholar] [CrossRef]
- Arias-Andres, M.; Klümper, U.; Rojas-Jimenez, K.; Grossart, H.-P. Microplastic pollution increases gene exchange in aquatic ecosystems. Environ. Pollut. 2018, 237, 253–261. [Google Scholar] [CrossRef]
- Li, B.; Qiu, Y.; Zhang, J.; Huang, X.; Shi, H.; Yin, H. Real-Time Study of Rapid Spread of Antibiotic Resistance Plasmid in Biofilm Using Microfluidics. Environ. Sci. Technol. 2018, 52, 11132–11141. [Google Scholar] [CrossRef]
- Huisman, J.S.; Benz, F.; Duxbury, S.J.N.; de Visser, J.A.G.M.; Hall, A.R.; Fischer, E.A.J.; Bonhoeffer, S. Estimating plasmid conjugation rates: A new computational tool and a critical comparison of methods. Plasmid 2022, 121, 102627. [Google Scholar] [CrossRef]
- Simonsen, L. Dynamics of plasmid transfer on surfaces. J. Gen. Microbiol. 1990, 136, 1001–1007. [Google Scholar] [CrossRef]
- Guzmán-Soto, I.; McTiernan, C.; Gonzalez-Gomez, M.; Ross, A.; Gupta, K.; Suuronen, E.J.; Mah, T.-F.; Griffith, M.; Alarcon, E.I. Mimicking biofilm formation and development: Recent progress in in vitro and in vivo biofilm models. iScience 2021, 24, 102443. [Google Scholar] [CrossRef] [PubMed]
- Gaston, J.R.; Andersen, M.J.; Johnson, A.O.; Bair, K.L.; Sullivan, C.M.; Guterman, L.B.; White, A.N.; Brauer, A.L.; Learman, B.S.; Flores-Mireles, A.L.; et al. Enterococcus faecalis Polymicrobial Interactions Facilitate Biofilm Formation, Antibiotic Recalcitrance, and Persistent Colonization of the Catheterized Urinary Tract. Pathogens 2020, 9, 835. [Google Scholar] [CrossRef]
- Allkja, J.; Goeres, D.M.; Azevedo, A.S.; Azevedo, N.F. Interactions of microorganisms within a urinary catheter polymicrobial biofilm model. Biotechnol. Bioeng. 2023, 120, 239–249. [Google Scholar] [CrossRef]
- Azeredo, J.; Azevedo, N.F.; Briandet, R.; Cerca, N.; Coenye, T.; Costa, A.R.; Desvaux, M.; Di Bonaventura, G.; Hébraud, M.; Jaglic, Z.; et al. Critical review on biofilm methods. Crit. Rev. Microbiol. 2017, 43, 313–351. [Google Scholar] [CrossRef]
- Lebeaux, D.; Chauhan, A.; Rendueles, O.; Beloin, C. From in vitro to in vivo Models of Bacterial Biofilm-Related Infections. Pathogens 2013, 2, 288–356. [Google Scholar] [CrossRef] [PubMed]
- Coenye, T.; Goeres, D.; van Bambeke, F.; Bjarnsholt, T. Should standardized susceptibility testing for microbial biofilms be introduced in clinical practice? Clin. Microbiol. Infect. 2018, 24, 570–572. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, H.; Sibley, C.D.; Surette, M.G.; Lewenza, S. Drosophila melanogaster as an animal model for the study of Pseudomonas aeruginosa biofilm infections in vivo. PLoS Pathog. 2011, 7, e1002299. [Google Scholar] [CrossRef] [PubMed]
- Kamareddine, L.; Wong, A.C.N.; Vanhove, A.S.; Hang, S.; Purdy, A.E.; Kierek-Pearson, K.; Asara, J.M.; Ali, A.; Morris, J.G.; Watnick, P.I. Activation of Vibrio cholerae quorum sensing promotes survival of an arthropod host. Nat. Microbiol. 2018, 3, 243–252. [Google Scholar] [CrossRef]
- Desai, S.K.; Padmanabhan, A.; Harshe, S.; Zaidel-Bar, R.; Kenney, L.J. Salmonella biofilms program innate immunity for persistence in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2019, 116, 12462–12467. [Google Scholar] [CrossRef] [Green Version]
- Van Meervenne, E.; de Weirdt, R.; van Coillie, E.; Devlieghere, F.; Herman, L.; Boon, N. Biofilm models for the food industry: Hot spots for plasmid transfer? Pathog. Dis. 2014, 70, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Bryers, J.D. Non-invasive determination of conjugative transfer of plasmids bearing antibiotic-resistance genes in biofilm-bound bacteria: Effects of substrate loading and antibiotic selection. Appl. Microbiol. Biotechnol. 2013, 97, 317–328. [Google Scholar] [CrossRef]
- Yaffe, E.; Relman, D.A. Tracking microbial evolution in the human gut using Hi-C reveals extensive horizontal gene transfer, persistence and adaptation. Nat. Microbiol. 2020, 5, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Kent, A.G.; Vill, A.C.; Shi, Q.; Satlin, M.J.; Brito, I.L. Widespread transfer of mobile antibiotic resistance genes within individual gut microbiomes revealed through bacterial Hi-C. Nat. Commun. 2020, 11, 4379. [Google Scholar] [CrossRef]
- Munck, C.; Sheth, R.U.; Freedberg, D.E.; Wang, H.H. Recording mobile DNA in the gut microbiota using an Escherichia coli CRISPR-Cas spacer acquisition platform. Nat. Commun. 2020, 11, 95. [Google Scholar] [CrossRef]
- McGinn, J.; Marraffini, L.A. Molecular mechanisms of CRISPR-Cas spacer acquisition. Nat. Rev. Microbiol. 2019, 17, 7–12. [Google Scholar] [CrossRef]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Stalder, T.; Press, M.O.; Sullivan, S.; Liachko, I.; Top, E.M. Linking the resistome and plasmidome to the microbiome. ISME J. 2019, 13, 2437–2446. [Google Scholar] [CrossRef]
- Baudry, L.; Foutel-Rodier, T.; Thierry, A.; Koszul, R.; Marbouty, M. MetaTOR: A Computational Pipeline to Recover High-Quality Metagenomic Bins From Mammalian Gut Proximity-Ligation (meta3C) Libraries. Front. Genet. 2019, 10, 753. [Google Scholar] [CrossRef]
- Chen, I.; Christie, P.J.; Dubnau, D. The ins and outs of DNA transfer in bacteria. Science 2005, 310, 1456–1460. [Google Scholar] [CrossRef] [Green Version]
- Griffith, F. The Significance of Pneumococcal Types. Epidemiol. Infect. 1928, 27, 113–159. [Google Scholar] [CrossRef] [PubMed]
- Blokesch, M. Natural competence for transformation. Curr. Biol. 2016, 26, R1126–R1130. [Google Scholar] [CrossRef] [PubMed]
- Johnston, C.; Martin, B.; Fichant, G.; Polard, P.; Claverys, J.-P. Bacterial transformation: Distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 2014, 12, 181–196. [Google Scholar] [CrossRef] [PubMed]
- Losick, R.M. Bacillus subtilis: A bacterium for all seasons. Curr. Biol. 2020, 30, R1146–R1150. [Google Scholar] [CrossRef] [PubMed]
- Dubnau, D.; Blokesch, M. Mechanisms of DNA Uptake by Naturally Competent Bacteria. Annu. Rev. Genet. 2019, 53, 217–237. [Google Scholar] [CrossRef]
- Attaiech, L.; Charpentier, X. Silently transformable: The many ways bacteria conceal their built-in capacity of genetic exchange. Curr. Genet. 2017, 63, 451–455. [Google Scholar] [CrossRef] [PubMed]
- García-Curiel, L.; López-Cuellar, M.D.R.; Rodríguez-Hernández, A.I.; Chavarría-Hernández, N. Toward understanding the signals of bacteriocin production by Streptococcus spp. and their importance in current applications. World J. Microbiol. Biotechnol. 2021, 37, 15. [Google Scholar] [CrossRef]
- Maree, M.; Nguyen, T.L.T.; Ohniwa, R.L.; Higashide, M.; Msadek, T.; Morikawa, K. Natural transformation allows transfer of SCCmec-mediated methicillin resistance in Staphylococcus aureus biofilms. Nat. Commun. 2022, 13, 2477. [Google Scholar] [CrossRef]
- Marks, L.R.; Reddinger, R.M.; Hakansson, A.P. High levels of genetic recombination during nasopharyngeal carriage and biofilm formation in Streptococcus pneumoniae. mBio 2012, 3, e00200-12. [Google Scholar] [CrossRef]
- Trappetti, C.; Gualdi, L.; Di Meola, L.; Jain, P.; Korir, C.C.; Edmonds, P.; Iannelli, F.; Ricci, S.; Pozzi, G.; Oggioni, M.R. The impact of the competence quorum sensing system on Streptococcus pneumoniae biofilms varies depending on the experimental model. BMC Microbiol. 2011, 11, 75. [Google Scholar] [CrossRef] [Green Version]
- Averhoff, B.; Kirchner, L.; Pfefferle, K.; Yaman, D. Natural transformation in Gram-negative bacteria thriving in extreme environments: From genes and genomes to proteins, structures and regulation. Extremophiles 2021, 25, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Lerminiaux, N.A.; Cameron, A.D.S. Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 2019, 65, 34–44. [Google Scholar] [CrossRef]
- Mashburn-Warren, L.; Goodman, S.D.; Federle, M.J.; Prehna, G. The conserved mosaic prophage protein paratox inhibits the natural competence regulator ComR in Streptococcus. Sci. Rep. 2018, 8, 16535. [Google Scholar] [CrossRef]
- Buchrieser, C. Biodiversity of the species Listeria monocytogenes and the genus Listeria. Microbes Infect. 2007, 9, 1147–1155. [Google Scholar] [CrossRef] [PubMed]
- Borezee, E.; Msadek, T.; Durant, L.; Berche, P. Identification in Listeria monocytogenes of MecA, a homologue of the Bacillus subtilis competence regulatory protein. J. Bacteriol. 2000, 182, 5931–5934. [Google Scholar] [CrossRef] [PubMed]
- Meibom, K.L.; Blokesch, M.; Dolganov, N.A.; Wu, C.-Y.; Schoolnik, G.K. Chitin induces natural competence in Vibrio cholerae. Science 2005, 310, 1824–1827. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Bernardy, E.E.; Hammer, B.K.; Miyashiro, T. Competence and natural transformation in vibrios. Mol. Microbiol. 2013, 89, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Blokesch, M.; Schoolnik, G.K. Serogroup conversion of Vibrio cholerae in aquatic reservoirs. PLoS Pathog. 2007, 3, e81. [Google Scholar] [CrossRef]
- Kouzel, N.; Oldewurtel, E.R.; Maier, B. Gene Transfer Efficiency in Gonococcal Biofilms: Role of Biofilm Age, Architecture, and Pilin Antigenic Variation. J. Bacteriol. 2015, 197, 2422–2431. [Google Scholar] [CrossRef] [Green Version]
- Bae, J.; Oh, E.; Jeon, B. Enhanced transmission of antibiotic resistance in Campylobacter jejuni biofilms by natural transformation. Antimicrob. Agents Chemother. 2014, 58, 7573–7575. [Google Scholar] [CrossRef] [PubMed]
- Nolan, L.M.; Turnbull, L.; Katrib, M.; Osvath, S.R.; Losa, D.; Lazenby, J.J.; Whitchurch, C.B. Pseudomonas aeruginosa is capable of natural transformation in biofilms. Microbiology 2020, 166, 995–1003. [Google Scholar] [CrossRef]
- Hendrickx, L.; Hausner, M.; Wuertz, S. Natural genetic transformation in monoculture Acinetobacter sp. strain BD413 biofilms. Appl. Environ. Microbiol. 2003, 69, 1721–1727. [Google Scholar] [CrossRef]
- Perumbakkam, S.; Hess, T.F.; Crawford, R.L. A bioremediation approach using natural transformation in pure-culture and mixed-population biofilms. Biodegradation 2006, 17, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Kovach, M.E.; Elzer, P.H.; Hill, D.S.; Robertson, G.T.; Farris, M.A.; Roop, R.; Peterson, K.M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995, 166, 175–176. [Google Scholar] [CrossRef] [PubMed]
- Merod, R.T.; Wuertz, S. Extracellular polymeric substance architecture influences natural genetic transformation of Acinetobacter baylyi in biofilms. Appl. Environ. Microbiol. 2014, 80, 7752–7757. [Google Scholar] [CrossRef]
- Santala, V.; Karp, M.; Santala, S. Bioluminescence-based system for rapid detection of natural transformation. FEMS Microbiol. Lett. 2016, 363, fnw125. [Google Scholar] [CrossRef]
- Baur, B.; Hanselmann, K.; Schlimme, W.; Jenni, B. Genetic transformation in freshwater: Escherichia coli is able to develop natural competence. Appl. Environ. Microbiol. 1996, 62, 3673–3678. [Google Scholar] [CrossRef]
- Etchuuya, R.; Ito, M.; Kitano, S.; Shigi, F.; Sobue, R.; Maeda, S. Cell-to-cell transformation in Escherichia coli: A novel type of natural transformation involving cell-derived DNA and a putative promoting pheromone. PLoS ONE 2011, 6, e16355. [Google Scholar] [CrossRef]
- Maeda, S.; Ito, M.; Ando, T.; Ishimoto, Y.; Fujisawa, Y.; Takahashi, H.; Matsuda, A.; Sawamura, A.; Kato, S. Horizontal transfer of nonconjugative plasmids in a colony biofilm of Escherichia coli. FEMS Microbiol. Lett. 2006, 255, 115–120. [Google Scholar] [CrossRef] [Green Version]
- Maeda, S.; Sawamura, A.; Matsuda, A. Transformation of colonial Escherichia coli on solid media. FEMS Microbiol. Lett. 2004, 236, 61–64. [Google Scholar] [CrossRef] [PubMed]
- Ando, T.; Itakura, S.; Uchii, K.; Sobue, R.; Maeda, S. Horizontal transfer of non-conjugative plasmid in colony biofilm of Escherichia coli on food-based media. World J. Microbiol. Biotechnol. 2009, 25, 1865–1869. [Google Scholar] [CrossRef]
- Hashimoto, M.; Hasegawa, H.; Maeda, S. High temperatures promote cell-to-cell plasmid transformation in Escherichia coli. Biochem. Biophys. Res. Commun. 2019, 515, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Errington, J.; van der Aart, L.T. Microbe Profile: Bacillus subtilis: Model organism for cellular development, and industrial workhorse. Microbiology 2020, 166, 425–427. [Google Scholar] [CrossRef] [PubMed]
- She, Q.; Hunter, E.; Qin, Y.; Nicolau, S.; Zalis, E.A.; Wang, H.; Chen, Y.; Chai, Y. Negative Interplay between Biofilm Formation and Competence in the Environmental Strains of Bacillus subtilis. mSystems 2020, 5, e00539-20. [Google Scholar] [CrossRef]
- Larsen, T.; Fiehn, N.-E. Dental biofilm infections-an update. APMIS 2017, 125, 376–384. [Google Scholar] [CrossRef]
- Quintero, B.; Araque, M.; van der Jongh, C.G.; Escalona, F.; Correa, M.; Morillo-Puente, S.; Vielma, S.; Hermans, P.W.M. Epidemiology of Streptococcus pneumoniae and Staphylococcus aureus colonization in healthy Venezuelan children. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 7–19. [Google Scholar] [CrossRef]
- De Lastours, V.; Malosh, R.; Ramadugu, K.; Srinivasan, U.; Dawid, S.; Ohmit, S.; Foxman, B. Co-colonization by Streptococcus pneumoniae and Staphylococcus aureus in the throat during acute respiratory illnesses. Epidemiol. Infect. 2016, 144, 3507–3519. [Google Scholar] [CrossRef]
- Sempere, J.; Llamosí, M.; Román, F.; Lago, D.; González-Camacho, F.; Pérez-García, C.; Yuste, J.; Domenech, M. Clearance of mixed biofilms of Streptococcus pneumoniae and methicillin-susceptible/resistant Staphylococcus aureus by antioxidants N-acetyl-L-cysteine and cysteamine. Sci. Rep. 2022, 12, 6668. [Google Scholar] [CrossRef]
- Wang, B.Y.; Chi, B.; Kuramitsu, H.K. Genetic exchange between Treponema denticola and Streptococcus gordonii in biofilms. Oral Microbiol. Immunol. 2002, 17, 108–112. [Google Scholar] [CrossRef]
- Lattar, S.M.; Wu, X.; Brophy, J.; Sakai, F.; Klugman, K.P.; Vidal, J.E. A Mechanism of Unidirectional Transformation, Leading to Antibiotic Resistance, Occurs within Nasopharyngeal Pneumococcal Biofilm Consortia. mBio 2018, 9, e00561-18. [Google Scholar] [CrossRef] [PubMed]
- Perry, D.; Kuramitsu, H.K. Genetic transformation of Streptococcus mutans. Infect. Immun. 1981, 32, 1295–1297. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Lau, P.C.; Lee, J.H.; Ellen, R.P.; Cvitkovitch, D.G. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 2001, 183, 897–908. [Google Scholar] [CrossRef] [PubMed]
- Marks, L.R.; Mashburn-Warren, L.; Federle, M.J.; Hakansson, A.P. Streptococcus pyogenes biofilm growth in vitro and in vivo and its role in colonization, virulence, and genetic exchange. J. Infect. Dis. 2014, 210, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, K.; Takemura, A.J.; Inose, Y.; Tsai, M.; Le Nguyen, T.; Ohta, T.; Msadek, T. Expression of a cryptic secondary sigma factor gene unveils natural competence for DNA transformation in Staphylococcus aureus. PLoS Pathog. 2012, 8, e1003003. [Google Scholar] [CrossRef]
- Harrison, A.; Hardison, R.L.; Wallace, R.M.; Fitch, J.; Heimlich, D.R.; Bryan, M.O.; Dubois, L.; John-Williams, L.S.; Sebra, R.P.; White, P.; et al. Reprioritization of biofilm metabolism is associated with nutrient adaptation and long-term survival of Haemophilus influenzae. NPJ Biofilms Microbiomes 2019, 5, 33. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Håvarstein, L.S. Fratricide is essential for efficient gene transfer between pneumococci in biofilms. Appl. Environ. Microbiol. 2012, 78, 5897–5905. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, Y.; Li, Y.; Lu, Y.; Xiong, K.; Zhong, Q.; Wang, J. Bacteriophage-Resistant Mutant of Enterococcus faecalis Is Impaired in Biofilm Formation. Front. Microbiol. 2022, 13, 913023. [Google Scholar] [CrossRef]
- Calero-Cáceres, W.; Muniesa, M. Persistence of naturally occurring antibiotic resistance genes in the bacteria and bacteriophage fractions of wastewater. Water Res. 2016, 95, 11–18. [Google Scholar] [CrossRef]
- Hata, A.; Kitajima, M.; Katayama, H. Occurrence and reduction of human viruses, F-specific RNA coliphage genogroups and microbial indicators at a full-scale wastewater treatment plant in Japan. J. Appl. Microbiol. 2013, 114, 545–554. [Google Scholar] [CrossRef]
- Dennehy, J.J.; Abedon, S.T. Bacteriophage Ecology. In Bacteriophages; Harper, D.R., Abedon, S.T., Burrowes, B.H., McConville, M.L., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 253–294. ISBN 978-3-319-41985-5. [Google Scholar]
- Du, F.; Lv, X.; Duan, D.; Wang, L.; Huang, J. Characterization of a Linezolid- and Vancomycin-Resistant Streptococcus suis Isolate That Harbors optrA and vanG Operons. Front. Microbiol. 2019, 10, 2026. [Google Scholar] [CrossRef] [PubMed]
- Moon, K.; Jeon, J.H.; Kang, I.; Park, K.S.; Lee, K.; Cha, C.-J.; Lee, S.H.; Cho, J.-C. Freshwater viral metagenome reveals novel and functional phage-borne antibiotic resistance genes. Microbiome 2020, 8, 75. [Google Scholar] [CrossRef] [PubMed]
- Lekunberri, I.; Villagrasa, M.; Balcázar, J.L.; Borrego, C.M. Contribution of bacteriophage and plasmid DNA to the mobilization of antibiotic resistance genes in a river receiving treated wastewater discharges. Sci. Total Environ. 2017, 601–602, 206–209. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.N.; Penadés, J.R.; Chen, J. Genetic transduction by phages and chromosomal islands: The new and noncanonical. PLoS Pathog. 2019, 15, e1007878. [Google Scholar] [CrossRef] [PubMed]
- Penadés, J.R.; Chen, J.; Quiles-Puchalt, N.; Carpena, N.; Novick, R.P. Bacteriophage-mediated spread of bacterial virulence genes. Curr. Opin. Microbiol. 2015, 23, 171–178. [Google Scholar] [CrossRef]
- Fišarová, L.; Botka, T.; Du, X.; Mašlaňová, I.; Bárdy, P.; Pantůček, R.; Benešík, M.; Roudnický, P.; Winstel, V.; Larsen, J.; et al. Staphylococcus epidermidis Phages Transduce Antimicrobial Resistance Plasmids and Mobilize Chromosomal Islands. mSphere 2021, 6, e00223-21. [Google Scholar] [CrossRef]
- Uchiyama, J.; Takemura-Uchiyama, I.; Sakaguchi, Y.; Gamoh, K.; Kato, S.; Daibata, M.; Ujihara, T.; Misawa, N.; Matsuzaki, S. Intragenus generalized transduction in Staphylococcus spp. by a novel giant phage. ISME J. 2014, 8, 1949–1952. [Google Scholar] [CrossRef]
- Leclerc, Q.J.; Wildfire, J.; Gupta, A.; Lindsay, J.A.; Knight, G.M. Growth-Dependent Predation and Generalized Transduction of Antimicrobial Resistance by Bacteriophage. mSystems 2022, 7, e0013522. [Google Scholar] [CrossRef]
- Zhang, J.; He, X.; Shen, S.; Shi, M.; Zhou, Q.; Liu, J.; Wang, M.; Sun, Y. Effects of the Newly Isolated T4-like Phage on Transmission of Plasmid-Borne Antibiotic Resistance Genes via Generalized Transduction. Viruses 2021, 13, 2070. [Google Scholar] [CrossRef]
- Borodovich, T.; Shkoporov, A.N.; Ross, R.P.; Hill, C. Phage-mediated horizontal gene transfer and its implications for the human gut microbiome. Gastroenterol. Rep. 2022, 10, goac012. [Google Scholar] [CrossRef]
- Rodríguez-Rubio, L.; Blanco-Picazo, P.; Muniesa, M. Are Phages Parasites or Symbionts of Bacteria? In Biocommunication of Phages; Witzany, G., Ed.; Springer International Publishing: Cham, Switzerland, 2020; pp. 143–162. ISBN 978-3-030-45884-3. [Google Scholar]
- Chen, J.; Quiles-Puchalt, N.; Chiang, Y.N.; Bacigalupe, R.; Fillol-Salom, A.; Chee, M.S.J.; Fitzgerald, J.R.; Penadés, J.R. Genome hypermobility by lateral transduction. Science 2018, 362, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Bowring, J.Z.; Su, Y.; Alsaadi, A.; Svenningsen, S.L.; Parkhill, J.; Ingmer, H. Screening for Highly Transduced Genes in Staphylococcus aureus Revealed Both Lateral and Specialized Transduction. Microbiol. Spectr. 2022, 10, e0242321. [Google Scholar] [CrossRef] [PubMed]
- Pourcel, C.; Midoux, C.; Vergnaud, G.; Latino, L. The Basis for Natural Multiresistance to Phage in Pseudomonas aeruginosa. Antibiotics 2020, 9, 339. [Google Scholar] [CrossRef] [PubMed]
- Fillol-Salom, A.; Bacigalupe, R.; Humphrey, S.; Chiang, Y.N.; Chen, J.; Penadés, J.R. Lateral transduction is inherent to the life cycle of the archetypical Salmonella phage P22. Nat. Commun. 2021, 12, 6510. [Google Scholar] [CrossRef] [PubMed]
- Bearson, B.L.; Brunelle, B.W. Fluoroquinolone induction of phage-mediated gene transfer in multidrug-resistant Salmonella. Int. J. Antimicrob. Agents 2015, 46, 201–204. [Google Scholar] [CrossRef]
- Gunathilaka, G.U.; Tahlan, V.; Mafiz, A.I.; Polur, M.; Zhang, Y. Phages in urban wastewater have the potential to disseminate antibiotic resistance. Int. J. Antimicrob. Agents 2017, 50, 678–683. [Google Scholar] [CrossRef]
- Haaber, J.; Leisner, J.J.; Cohn, M.T.; Catalan-Moreno, A.; Nielsen, J.B.; Westh, H.; Penadés, J.R.; Ingmer, H. Bacterial viruses enable their host to acquire antibiotic resistance genes from neighbouring cells. Nat. Commun. 2016, 7, 13333. [Google Scholar] [CrossRef]
- Habets, A.; Antoine, C.; Wagemans, J.; Vermeersch, M.; Laforêt, F.; Diderich, J.; Lavigne, R.; Mainil, J.; Thiry, D. Impact of Shiga-toxin encoding gene transduction from O80:H2 Shiga toxigenic Escherichia coli (STEC) on non-STEC strains. Sci. Rep. 2022, 12, 21587. [Google Scholar] [CrossRef]
- Solheim, H.T.; Sekse, C.; Urdahl, A.M.; Wasteson, Y.; Nesse, L.L. Biofilm as an environment for dissemination of stx genes by transduction. Appl. Environ. Microbiol. 2013, 79, 896–900. [Google Scholar] [CrossRef]
- González, S.; Fernández, L.; Gutiérrez, D.; Campelo, A.B.; Rodríguez, A.; García, P. Analysis of Different Parameters Affecting Diffusion, Propagation and Survival of Staphylophages in Bacterial Biofilms. Front. Microbiol. 2018, 9, 2348. [Google Scholar] [CrossRef]
- Li, X.; Gonzalez, F.; Esteves, N.; Scharf, B.E.; Chen, J. Formation of phage lysis patterns and implications on co-propagation of phages and motile host bacteria. PLoS Comput. Biol. 2020, 16, e1007236. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, R.W.; Smith, M.C.; Burns, R.N.; Ford, M.E.; Hatfull, G.F. Evolutionary relationships among diverse bacteriophages and prophages: All the world’s a phage. Proc. Natl. Acad. Sci. USA 1999, 96, 2192–2197. [Google Scholar] [CrossRef] [PubMed]
- Mushegian, A.R. Are There 1031 Virus Particles on Earth, or More, or Fewer? J. Bacteriol. 2020, 202, e00052-20. [Google Scholar] [CrossRef] [PubMed]
- Wommack, K.E.; Colwell, R.R. Virioplankton: Viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 2000, 64, 69–114. [Google Scholar] [CrossRef]
- Yu, Z.; Schwarz, C.; Zhu, L.; Chen, L.; Shen, Y.; Yu, P. Hitchhiking Behavior in Bacteriophages Facilitates Phage Infection and Enhances Carrier Bacteria Colonization. Environ. Sci. Technol. 2021, 55, 2462–2472. [Google Scholar] [CrossRef]
- Morrison, W.D.; Miller, R.V.; Sayler, G.S. Frequency of F116-mediated transduction of Pseudomonas aeruginosa in a freshwater environment. Appl. Environ. Microbiol. 1978, 36, 724–730. [Google Scholar] [CrossRef]
- Ripp, S.; Miller, R.V. Effects of Suspended Particulates on the Frequency of Transduction among Pseudomonas aeruginosa in a Freshwater Environment. Appl. Environ. Microbiol. 1995, 61, 1214–1219. [Google Scholar] [CrossRef]
- Saye, D.J.; Ogunseitan, O.; Sayler, G.S.; Miller, R.V. Potential for transduction of plasmids in a natural freshwater environment: Effect of plasmid donor concentration and a natural microbial community on transduction in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 1987, 53, 987–995. [Google Scholar] [CrossRef]
- Saye, D.J.; Ogunseitan, O.A.; Sayler, G.S.; Miller, R.V. Transduction of linked chromosomal genes between Pseudomonas aeruginosa strains during incubation in situ in a freshwater habitat. Appl. Environ. Microbiol. 1990, 56, 140–145. [Google Scholar] [CrossRef]
- Koskella, B.; Meaden, S. Understanding bacteriophage specificity in natural microbial communities. Viruses 2013, 5, 806–823. [Google Scholar] [CrossRef] [Green Version]
- Raj, J.R.M.; Karunasagar, I. Phages amid antimicrobial resistance. Crit. Rev. Microbiol. 2019, 45, 701–711. [Google Scholar] [CrossRef]
- De Jonge, P.A.; Nobrega, F.L.; Brouns, S.J.J.; Dutilh, B.E. Molecular and Evolutionary Determinants of Bacteriophage Host Range. Trends Microbiol. 2019, 27, 51–63. [Google Scholar] [CrossRef]
- Cazares, D.; Cazares, A.; Figueroa, W.; Guarneros, G.; Edwards, R.A.; Vinuesa, P. A Novel Group of Promiscuous Podophages Infecting Diverse Gammaproteobacteria from River Communities Exhibits Dynamic Intergenus Host Adaptation. mSystems 2021, 6, e00773-20. [Google Scholar] [CrossRef]
- Enault, F.; Briet, A.; Bouteille, L.; Roux, S.; Sullivan, M.B.; Petit, M.-A. Phages rarely encode antibiotic resistance genes: A cautionary tale for virome analyses. ISME J. 2017, 11, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Calero-Cáceres, W.; Ye, M.; Balcázar, J.L. Bacteriophages as Environmental Reservoirs of Antibiotic Resistance. Trends Microbiol. 2019, 27, 570–577. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, E.; Bonnin, A.R.; Rocha, E.P. Phage-plasmids spread antibiotic resistance genes through infection and lysogenic conversion. mBio 2022, 13, e0185122. [Google Scholar] [CrossRef] [PubMed]
- Colavecchio, A.; Cadieux, B.; Lo, A.; Goodridge, L.D. Bacteriophages Contribute to the Spread of Antibiotic Resistance Genes among Foodborne Pathogens of the Enterobacteriaceae Family-A Review. Front. Microbiol. 2017, 8, 1108. [Google Scholar] [CrossRef]
- Brown-Jaque, M.; Rodriguez Oyarzun, L.; Cornejo-Sánchez, T.; Martín-Gómez, M.T.; Gartner, S.; de Gracia, J.; Rovira, S.; Alvarez, A.; Jofre, J.; González-López, J.J.; et al. Detection of Bacteriophage Particles Containing Antibiotic Resistance Genes in the Sputum of Cystic Fibrosis Patients. Front. Microbiol. 2018, 9, 856. [Google Scholar] [CrossRef]
- Kidambi, S.P.; Ripp, S.; Miller, R.V. Evidence for phage-mediated gene transfer among Pseudomonas aeruginosa strains on the phylloplane. Appl. Environ. Microbiol. 1994, 60, 496–500. [Google Scholar] [CrossRef]
- Goh, S.; Hussain, H.; Chang, B.J.; Emmett, W.; Riley, T.V.; Mullany, P. Phage ϕC2 mediates transduction of Tn6215, encoding erythromycin resistance, between Clostridium difficile strains. mBio 2013, 4, e00840-13. [Google Scholar] [CrossRef] [Green Version]
- Moon, B.Y.; Park, J.Y.; Hwang, S.Y.; Robinson, D.A.; Thomas, J.C.; Fitzgerald, J.R.; Park, Y.H.; Seo, K.S. Phage-mediated horizontal transfer of a Staphylococcus aureus virulence-associated genomic island. Sci. Rep. 2015, 5, 9784. [Google Scholar] [CrossRef] [PubMed]
- Xia, G.; Wolz, C. Phages of Staphylococcus aureus and their impact on host evolution. Infect. Genet. Evol. 2014, 21, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.N.; Porse, A.; Sommer, M.O.A.; Høiby, N.; Ciofu, O. Evolution of Antibiotic Resistance in Biofilm and Planktonic Pseudomonas aeruginosa Populations Exposed to Subinhibitory Levels of Ciprofloxacin. Antimicrob. Agents Chemother. 2018, 62, e00320-18. [Google Scholar] [CrossRef]
- Stanczak-Mrozek, K.I.; Laing, K.G.; Lindsay, J.A. Resistance gene transfer: Induction of transducing phage by sub-inhibitory concentrations of antimicrobials is not correlated to induction of lytic phage. J. Antimicrob. Chemother. 2017, 72, 1624–1631. [Google Scholar] [CrossRef] [PubMed]
- Wachino, J.-I.; Jin, W.; Kimura, K.; Arakawa, Y. Intercellular Transfer of Chromosomal Antimicrobial Resistance Genes between Acinetobacter baumannii Strains Mediated by Prophages. Antimicrob. Agents Chemother. 2019, 63, e00334-19. [Google Scholar] [CrossRef]
- Visnapuu, A.; van der Gucht, M.; Wagemans, J.; Lavigne, R. Deconstructing the Phage-Bacterial Biofilm Interaction as a Basis to Establish New Antibiofilm Strategies. Viruses 2022, 14, 1057. [Google Scholar] [CrossRef]
- Güemes, A.G.C.; Youle, M.; Cantú, V.A.; Felts, B.; Nulton, J.; Rohwer, F. Viruses as Winners in the Game of Life. Annu. Rev. Virol. 2016, 3, 197–214. [Google Scholar] [CrossRef]
- Martinez-Hernandez, F.; Fornas, O.; Gomez, M.L.; Bolduc, B.; de La Peña, M.J.C.; Martínez, J.M.; Anton, J.; Gasol, J.M.; Rosselli, R.; Rodriguez-Valera, F.; et al. Single-virus genomics reveals hidden cosmopolitan and abundant viruses. Nat. Commun. 2017, 8, 15892. [Google Scholar] [CrossRef]
- Turzynski, V.; Monsees, I.; Moraru, C.; Probst, A.J. Imaging Techniques for Detecting Prokaryotic Viruses in Environmental Samples. Viruses 2021, 13, 2126. [Google Scholar] [CrossRef]
- Kenzaka, T.; Tani, K.; Sakotani, A.; Yamaguchi, N.; Nasu, M. High-frequency phage-mediated gene transfer among Escherichia coli cells, determined at the single-cell level. Appl. Environ. Microbiol. 2007, 73, 3291–3299. [Google Scholar] [CrossRef] [Green Version]
- Kenzaka, T.; Tani, K.; Nasu, M. High-frequency phage-mediated gene transfer in freshwater environments determined at single-cell level. ISME J. 2010, 4, 648–659. [Google Scholar] [CrossRef]
- Papaianni, M.; Ricciardelli, A.; Casillo, A.; Corsaro, M.M.; Borbone, F.; Della Ventura, B.; Velotta, R.; Fulgione, A.; Woo, S.L.; Tutino, M.L.; et al. The Union Is Strength: The Synergic Action of Long Fatty Acids and a Bacteriophage against Xanthomonas campestris Biofilm. Microorganisms 2020, 9, 60. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, N.A.; Barnard, A.M.; Slater, H.; Simpson, N.J.; Salmond, G.P. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 2001, 25, 365–404. [Google Scholar] [CrossRef]
- Sørensen, S.J.; Bailey, M.; Hansen, L.H.; Kroer, N.; Wuertz, S. Studying plasmid horizontal transfer in situ: A critical review. Nat. Rev. Microbiol. 2005, 3, 700–710. [Google Scholar] [CrossRef]
- Christie, P.J. The Mosaic Type IV Secretion Systems. EcoSal Plus 2016, 7, 10. [Google Scholar] [CrossRef]
- Grohmann, E.; Christie, P.J.; Waksman, G.; Backert, S. Type IV secretion in Gram-negative and Gram-positive bacteria. Mol. Microbiol. 2018, 107, 455–471. [Google Scholar] [CrossRef] [PubMed]
- Kohler, V.; Vaishampayan, A.; Grohmann, E. Broad-host-range Inc18 plasmids: Occurrence, spread and transfer mechanisms. Plasmid 2018, 99, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Fischer, W.; Tegtmeyer, N.; Stingl, K.; Backert, S. Four Chromosomal Type IV Secretion Systems in Helicobacter pylori: Composition, Structure and Function. Front. Microbiol. 2020, 11, 1592. [Google Scholar] [CrossRef]
- Shen, Z.; Tang, C.M.; Liu, G.-Y. Towards a better understanding of antimicrobial resistance dissemination: What can be learnt from studying model conjugative plasmids? Mil. Med. Res. 2022, 9, 3. [Google Scholar] [CrossRef]
- Hinnekens, P.; Koné, K.M.; Fayad, N.; Leprince, A.; Mahillon, J. pXO16, the large conjugative plasmid from Bacillus thuringiensis serovar israelensis displays an extended host spectrum. Plasmid 2019, 102, 46–50. [Google Scholar] [CrossRef]
- Fercher, C.; Probst, I.; Kohler, V.; Goessweiner-Mohr, N.; Arends, K.; Grohmann, E.; Zangger, K.; Meyer, N.H.; Keller, W. VirB8-like protein TraH is crucial for DNA transfer in Enterococcus faecalis. Sci. Rep. 2016, 6, 24643. [Google Scholar] [CrossRef] [PubMed]
- Weiss, E.; Spicher, C.; Haas, R.; Fischer, W. Excision and transfer of an integrating and conjugative element in a bacterial species with high recombination efficiency. Sci. Rep. 2019, 9, 8915. [Google Scholar] [CrossRef] [PubMed]
- Kiss, J.; Szabó, M.; Hegyi, A.; Douard, G.; Praud, K.; Nagy, I.; Olasz, F.; Cloeckaert, A.; Doublet, B. Identification and Characterization of oriT and Two Mobilization Genes Required for Conjugative Transfer of Salmonella Genomic Island 1. Front. Microbiol. 2019, 10, 457. [Google Scholar] [CrossRef] [PubMed]
- Pinilla-Redondo, R.; Olesen, A.K.; Russel, J.; de Vries, L.E.; Christensen, L.D.; Musovic, S.; Nesme, J.; Sørensen, S.J. Broad Dissemination of Plasmids across Groundwater-Fed Rapid Sand Filter Microbiomes. mBio 2021, 12, e0306821. [Google Scholar] [CrossRef] [PubMed]
- Ehlers, L.J.; Bouwer, E.J. RP4 plasmid transfer among species of pseudomonas in a biofilm reactor. Water Sci. Technol. 1999, 39, 163–171. [Google Scholar] [CrossRef]
- Lampkowska, J.; Feld, L.; Monaghan, A.; Toomey, N.; Schjørring, S.; Jacobsen, B.; van der Voet, H.; Andersen, S.R.; Bolton, D.; Aarts, H.; et al. A standardized conjugation protocol to asses antibiotic resistance transfer between lactococcal species. Int. J. Food Microbiol. 2008, 127, 172–175. [Google Scholar] [CrossRef]
- Neil, K.; Allard, N.; Grenier, F.; Burrus, V.; Rodrigue, S. Highly efficient gene transfer in the mouse gut microbiota is enabled by the Incl2 conjugative plasmid TP114. Commun. Biol. 2020, 3, 523. [Google Scholar] [CrossRef]
- Bradley, D.E.; Taylor, D.E.; Cohen, D.R. Specification of surface mating systems among conjugative drug resistance plasmids in Escherichia coli K-12. J. Bacteriol. 1980, 143, 1466–1470. [Google Scholar] [CrossRef]
- Sabbagh, P.; Rajabnia, M.; Maali, A.; Ferdosi-Shahandashti, E. Integron and its role in antimicrobial resistance: A literature review on some bacterial pathogens. Iran. J. Basic Med. Sci. 2021, 24, 136–142. [Google Scholar] [CrossRef]
- Strugeon, E.; Tilloy, V.; Ploy, M.-C.; Da Re, S. The Stringent Response Promotes Antibiotic Resistance Dissemination by Regulating Integron Integrase Expression in Biofilms. mBio 2016, 7, e00868-16. [Google Scholar] [CrossRef] [Green Version]
- Pongchaikul, P.; Santanirand, P.; Antonyuk, S.; Winstanley, C.; Darby, A.C. AcGI1, a novel genomic island carrying antibiotic resistance integron In687 in multidrug resistant Achromobacter xylosoxidans in a teaching hospital in Thailand. FEMS Microbiol. Lett. 2020, 367, fnaa109. [Google Scholar] [CrossRef] [PubMed]
- Ghaly, T.M.; Gillings, M.R.; Penesyan, A.; Qi, Q.; Rajabal, V.; Tetu, S.G. The Natural History of Integrons. Microorganisms 2021, 9, 2212. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, J.P.; Kwong, S.M.; Murphy, R.J.T.; Yui Eto, K.; Price, K.J.; Nguyen, Q.T.; O’Brien, F.G.; Grubb, W.B.; Coombs, G.W.; Firth, N. An updated view of plasmid conjugation and mobilization in Staphylococcus. Mob. Genet. Elements 2016, 6, e1208317. [Google Scholar] [CrossRef] [PubMed]
- Smillie, C.; Garcillán-Barcia, M.P.; Francia, M.V.; Rocha, E.P.C.; de La Cruz, F. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 2010, 74, 434–452. [Google Scholar] [CrossRef]
- Binsker, U.; Käsbohrer, A.; Hammerl, J.A. Global colistin use: A review of the emergence of resistant Enterobacterales and the impact on their genetic basis. FEMS Microbiol. Rev. 2022, 46, fuab049. [Google Scholar] [CrossRef]
- Mukherjee, S.; Mitra, S.; Dutta, S.; Basu, S. Neonatal Sepsis: The Impact of Carbapenem-Resistant and Hypervirulent Klebsiella pneumoniae. Front. Med. 2021, 8, 634349. [Google Scholar] [CrossRef]
- Fang, L.-X.; Chen, C.; Cui, C.-Y.; Li, X.-P.; Zhang, Y.; Liao, X.-P.; Sun, J.; Liu, Y.-H. Emerging High-Level Tigecycline Resistance: Novel Tetracycline Destructases Spread via the Mobile Tet(X). Bioessays 2020, 42, e2000014. [Google Scholar] [CrossRef]
- Che, Y.; Yang, Y.; Xu, X.; Břinda, K.; Polz, M.F.; Hanage, W.P.; Zhang, T. Conjugative plasmids interact with insertion sequences to shape the horizontal transfer of antimicrobial resistance genes. Proc. Natl. Acad. Sci. USA 2021, 118, e2008731118. [Google Scholar] [CrossRef]
- Li, B.; Qiu, Y.; Zhang, J.; Liang, P.; Huang, X. Conjugative potential of antibiotic resistance plasmids to activated sludge bacteria from wastewater treatment plants. Int. Biodeterior. Biodegrad. 2019, 138, 33–40. [Google Scholar] [CrossRef]
- Li, L.; Dechesne, A.; He, Z.; Madsen, J.S.; Nesme, J.; Sørensen, S.J.; Smets, B.F. Estimating the Transfer Range of Plasmids Encoding Antimicrobial Resistance in a Wastewater Treatment Plant Microbial Community. Environ. Sci. Technol. Lett. 2018, 5, 260–265. [Google Scholar] [CrossRef] [Green Version]
- Valle, A.A.-D.; León-Sampedro, R.; Rodríguez-Beltrán, J.; DelaFuente, J.; Hernández-García, M.; Ruiz-Garbajosa, P.; Cantón, R.; Peña-Miller, R.; San Millán, A. Variability of plasmid fitness effects contributes to plasmid persistence in bacterial communities. Nat. Commun. 2021, 12, 2653. [Google Scholar] [CrossRef] [PubMed]
- Zaman, T.U.; Alrodayyan, M.; Albladi, M.; Aldrees, M.; Siddique, M.I.; Aljohani, S.; Balkhy, H.H. Clonal diversity and genetic profiling of antibiotic resistance among multidrug/carbapenem-resistant Klebsiella pneumoniae isolates from a tertiary care hospital in Saudi Arabia. BMC Infect. Dis. 2018, 18, 205. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yu, J.; Chen, F.; Yu, J.; Simner, P.; Tamma, P.; Liu, Y.; Shen, L. Emergence and establishment of KPC-2-producing ST11 Klebsiella pneumoniae in a general hospital in Shanghai, China. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.M.; Grinberg, I.; Eldar, A.; Grossman, A.D. A mobile genetic element increases bacterial host fitness by manipulating development. Elife 2021, 10, e65924. [Google Scholar] [CrossRef]
- Król, J.E.; Wojtowicz, A.J.; Rogers, L.M.; Heuer, H.; Smalla, K.; Krone, S.M.; Top, E.M. Invasion of E. coli biofilms by antibiotic resistance plasmids. Plasmid 2013, 70, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Maheshwari, M.; Ahmad, I.; Althubiani, A.S. Multidrug resistance and transferability of blaCTX-M among extended-spectrum β-lactamase-producing enteric bacteria in biofilm. J. Glob. Antimicrob. Resist. 2016, 6, 142–149. [Google Scholar] [CrossRef]
- Mo, S.S.; Sunde, M.; Ilag, H.K.; Langsrud, S.; Heir, E. Transfer Potential of Plasmids Conferring Extended-Spectrum-Cephalosporin Resistance in Escherichia coli from Poultry. Appl. Environ. Microbiol. 2017, 83, e00654-17. [Google Scholar] [CrossRef]
- Hausner, M.; Wuertz, S. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl. Environ. Microbiol. 1999, 65, 3710–3713. [Google Scholar] [CrossRef]
- Nancharaiah, Y.V.; Wattiau, P.; Wuertz, S.; Bathe, S.; Mohan, S.V.; Wilderer, P.A.; Hausner, M. Dual labeling of Pseudomonas putida with fluorescent proteins for in situ monitoring of conjugal transfer of the TOL plasmid. Appl. Environ. Microbiol. 2003, 69, 4846–4852. [Google Scholar] [CrossRef]
- Aspray, T.J.; Hansen, S.K.; Burns, R.G. A soil-based microbial biofilm exposed to 2,4-D: Bacterial community development and establishment of conjugative plasmid pJP4. FEMS Microbiol. Ecol. 2005, 54, 317–327. [Google Scholar] [CrossRef]
- Christensen, B.B.; Sternberg, C.; Molin, S. Bacterial plasmid conjugation on semi-solid surfaces monitored with the green fluorescent protein (GFP) from Aequorea victoria as a marker. Gene 1996, 173, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Dahlberg, C.; Bergström, M.; Hermansson, M. In Situ Detection of High Levels of Horizontal Plasmid Transfer in Marine Bacterial Communities. Appl. Environ. Microbiol. 1998, 64, 2670–2675. [Google Scholar] [CrossRef] [PubMed]
- Klümper, U.; Riber, L.; Dechesne, A.; Sannazzarro, A.; Hansen, L.H.; Sørensen, S.J.; Smets, B.F. Broad host range plasmids can invade an unexpectedly diverse fraction of a soil bacterial community. ISME J. 2015, 9, 934–945. [Google Scholar] [CrossRef]
- Ma, H.; Bryers, J.D. Non-invasive method to quantify local bacterial concentrations in a mixed culture biofilm. J. Ind. Microbiol. Biotechnol. 2010, 37, 1081–1089. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.; Franklin, M.J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 2008, 6, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.C.; Palmer, R.J.; Udsen, C.; White, D.C.; Molin, S. Assessment of GFP fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology 2001, 147, 1383–1391. [Google Scholar] [CrossRef] [PubMed]
- Chia, H.E.; Marsh, E.N.G.; Biteen, J.S. Extending fluorescence microscopy into anaerobic environments. Curr. Opin. Chem. Biol. 2019, 51, 98–104. [Google Scholar] [CrossRef]
- Li, B.; Qiu, Y.; Song, Y.; Lin, H.; Yin, H. Dissecting horizontal and vertical gene transfer of antibiotic resistance plasmid in bacterial community using microfluidics. Environ. Int. 2019, 131, 105007. [Google Scholar] [CrossRef]
- Qiu, Y.; Zhang, J.; Li, B.; Wen, X.; Liang, P.; Huang, X. A novel microfluidic system enables visualization and analysis of antibiotic resistance gene transfer to activated sludge bacteria in biofilm. Sci. Total Environ. 2018, 642, 582–590. [Google Scholar] [CrossRef]
- Wuertz, S.; Hendrickx, L.; Kuehn, M.; Rodenacker, K.; Hausner, M. In situ quantification of gene transfer in biofilms. Methods Enzymol. 2001, 336, 129–143. [Google Scholar] [CrossRef]
- Bellanger, X.; Payot, S.; Leblond-Bourget, N.; Guédon, G. Conjugative and mobilizable genomic islands in bacteria: Evolution and diversity. FEMS Microbiol. Rev. 2014, 38, 720–760. [Google Scholar] [CrossRef] [PubMed]
- Libante, V.; Nombre, Y.; Coluzzi, C.; Staub, J.; Guédon, G.; Gottschalk, M.; Teatero, S.; Fittipaldi, N.; Leblond-Bourget, N.; Payot, S. Chromosomal Conjugative and Mobilizable Elements in Streptococcus suis: Major Actors in the Spreading of Antimicrobial Resistance and Bacteriocin Synthesis Genes. Pathogens 2019, 9, 22. [Google Scholar] [CrossRef]
- Guédon, G.; Libante, V.; Coluzzi, C.; Payot, S.; Leblond-Bourget, N. The Obscure World of Integrative and Mobilizable Elements, Highly Widespread Elements that Pirate Bacterial Conjugative Systems. Genes 2017, 8, 337. [Google Scholar] [CrossRef] [PubMed]
- Lao, J.; Lacroix, T.; Guédon, G.; Coluzzi, C.; Payot, S.; Leblond-Bourget, N.; Chiapello, H. ICEscreen: A tool to detect Firmicute ICEs and IMEs, isolated or enclosed in composite structures. NAR Genom. Bioinform. 2022, 4, lqac079. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, O.S.; de Assis, J.C.S.; Santana, M.F. Breaking the ICE: An easy workflow for identifying and analyzing integrative and conjugative elements in bacterial genomes. Funct. Integr. Genom. 2022, 22, 1139–1145. [Google Scholar] [CrossRef] [PubMed]
- Guglielmini, J.; Quintais, L.; Garcillán-Barcia, M.P.; de La Cruz, F.; Rocha, E.P.C. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 2011, 7, e1002222. [Google Scholar] [CrossRef]
- Grindley, N.D.F.; Whiteson, K.L.; Rice, P.A. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006, 75, 567–605. [Google Scholar] [CrossRef] [PubMed]
- Delavat, F.; Miyazaki, R.; Carraro, N.; Pradervand, N.; van der Meer, J.R. The hidden life of integrative and conjugative elements. FEMS Microbiol. Rev. 2017, 41, 512–537. [Google Scholar] [CrossRef]
- Botelho, J.; Schulenburg, H. The Role of Integrative and Conjugative Elements in Antibiotic Resistance Evolution. Trends Microbiol. 2021, 29, 8–18. [Google Scholar] [CrossRef]
- Roberts, A.P.; Mullany, P. Tn916-like genetic elements: A diverse group of modular mobile elements conferring antibiotic resistance. FEMS Microbiol. Rev. 2011, 35, 856–871. [Google Scholar] [CrossRef] [Green Version]
- Botelho, J.; Grosso, F.; Peixe, L. ICEs Are the Main Reservoirs of the Ciprofloxacin-Modifying crpP Gene in Pseudomonas aeruginosa. Genes 2020, 11, 889. [Google Scholar] [CrossRef] [PubMed]
- Johannessen, H.; Anthonisen, I.L.; Zecic, N.; Hegstad, K.; Ranheim, T.E.; Skaare, D. Characterization and Fitness Cost of Tn7100, a Novel Integrative and Conjugative Element Conferring Multidrug Resistance in Haemophilus influenzae. Front. Microbiol. 2022, 13, 945411. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Kuang, X.; Han, R.-J.; Zhai, Y.-J.; He, D.-D.; Zhao, J.-F.; Liu, J.-H.; Hu, G.-Z. Characterization of a Novel Linezolid Resistance Gene optrA and Bacitracin Resistance Locus-Carrying Multiple Antibiotic Resistant Integrative and Conjugative Element ICESsu1112S in Streptococccus Suis. Microbiol. Spectr. 2022, 10, e0196321. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.P.; Davis, I.J.; Seville, L.; Villedieu, A.; Mullany, P. Characterization of the ends and target site of a novel tetracycline resistance-encoding conjugative transposon from Enterococcus faecium 664.1H1. J. Bacteriol. 2006, 188, 4356–4361. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Song, Y.; Zhang, Q.; Zhang, A.; Jin, M. Characterisation of a novel integrative and conjugative element ICESsD9 carrying erm(B) and tet(O) resistance determinants in Streptococcus suis, and the distribution of ICESsD9-like elements in clinical isolates. J. Glob. Antimicrob. Resist. 2016, 7, 13–18. [Google Scholar] [CrossRef]
- Auchtung, J.M.; Lee, C.A.; Monson, R.E.; Lehman, A.P.; Grossman, A.D. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc. Natl. Acad. Sci. USA 2005, 102, 12554–12559. [Google Scholar] [CrossRef]
- Waldor, M.K.; Tschäpe, H.; Mekalanos, J.J. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J. Bacteriol. 1996, 178, 4157–4165. [Google Scholar] [CrossRef]
- Franke, A.E.; Clewell, D.B. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of "conjugal" transfer in the absence of a conjugative plasmid. J. Bacteriol. 1981, 145, 494–502. [Google Scholar] [CrossRef]
- Shoemaker, N.B.; Barber, R.D.; Salyers, A.A. Cloning and characterization of a Bacteroides conjugal tetracycline-erythromycin resistance element by using a shuttle cosmid vector. J. Bacteriol. 1989, 171, 1294–1302. [Google Scholar] [CrossRef]
- Burrus, V.; Roussel, Y.; Decaris, B.; Guédon, G. Characterization of a novel integrative element, ICESt1, in the lactic acid bacterium Streptococcus thermophilus. Appl. Environ. Microbiol. 2000, 66, 1749–1753. [Google Scholar] [CrossRef] [Green Version]
- Pavlovic, G.; Burrus, V.; Gintz, B.; Decaris, B.; Guédon, G. Evolution of genomic islands by deletion and tandem accretion by site-specific recombination: ICESt1-related elements from Streptococcus thermophilus. Microbiology 2004, 150, 759–774. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, J.P.; Sullivan, J.T.; Stuart, G.S.; Lamont, I.L.; Ronson, C.W. Excision and transfer of the Mesorhizobium loti R7A symbiosis island requires an integrase IntS, a novel recombination directionality factor RdfS, and a putative relaxase RlxS. Mol. Microbiol. 2006, 62, 723–734. [Google Scholar] [CrossRef] [PubMed]
- Bellanger, X.; Roberts, A.P.; Morel, C.; Choulet, F.; Pavlovic, G.; Mullany, P.; Decaris, B.; Guédon, G. Conjugative transfer of the integrative conjugative elements ICESt1 and ICESt3 from Streptococcus thermophilus. J. Bacteriol. 2009, 191, 2764–2775. [Google Scholar] [CrossRef]
- Lécuyer, F.; Bourassa, J.-S.; Gélinas, M.; Charron-Lamoureux, V.; Burrus, V.; Beauregard, P.B. Biofilm Formation Drives Transfer of the Conjugative Element ICEBs1 in Bacillus subtilis. mSphere 2018, 3, e00473-18. [Google Scholar] [CrossRef] [PubMed]
- Bourassa, J.-S.; Jeannotte, G.; Lebel-Beaucage, S.; Beauregard, P.B. Second-Generation Transfer Mediates Efficient Propagation of ICEBs1 in Biofilms. J. Bacteriol. 2022, 204, e0018122. [Google Scholar] [CrossRef]
- Babic, A.; Berkmen, M.B.; Lee, C.A.; Grossman, A.D. Efficient gene transfer in bacterial cell chains. mBio 2011, 2, e00027-11. [Google Scholar] [CrossRef] [Green Version]
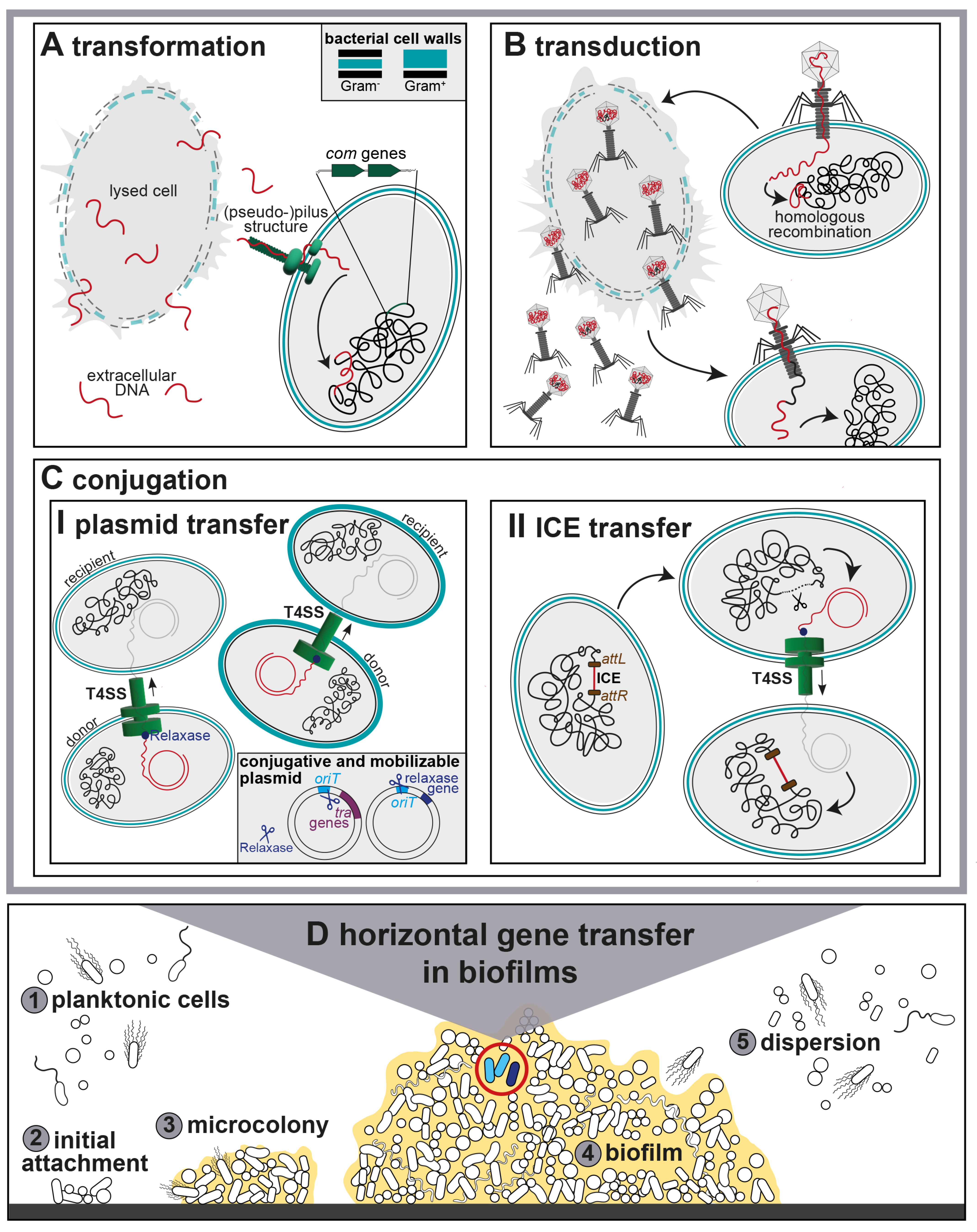
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michaelis, C.; Grohmann, E. Horizontal Gene Transfer of Antibiotic Resistance Genes in Biofilms. Antibiotics 2023, 12, 328. https://doi.org/10.3390/antibiotics12020328
Michaelis C, Grohmann E. Horizontal Gene Transfer of Antibiotic Resistance Genes in Biofilms. Antibiotics. 2023; 12(2):328. https://doi.org/10.3390/antibiotics12020328
Chicago/Turabian StyleMichaelis, Claudia, and Elisabeth Grohmann. 2023. "Horizontal Gene Transfer of Antibiotic Resistance Genes in Biofilms" Antibiotics 12, no. 2: 328. https://doi.org/10.3390/antibiotics12020328
APA StyleMichaelis, C., & Grohmann, E. (2023). Horizontal Gene Transfer of Antibiotic Resistance Genes in Biofilms. Antibiotics, 12(2), 328. https://doi.org/10.3390/antibiotics12020328





