Occurrence of cfr-Positive Linezolid-Susceptible Staphylococcus aureus and Non-aureus Staphylococcal Isolates from Pig Farms
Abstract
:1. Introduction
2. Results
2.1. Identification of cfr-Positive Linezolid-Susceptible Staphylococci in Pig Farms
2.2. Genetic Assessment of cfr ORF and Its Promoter in LZD-Susceptible Staphylococci
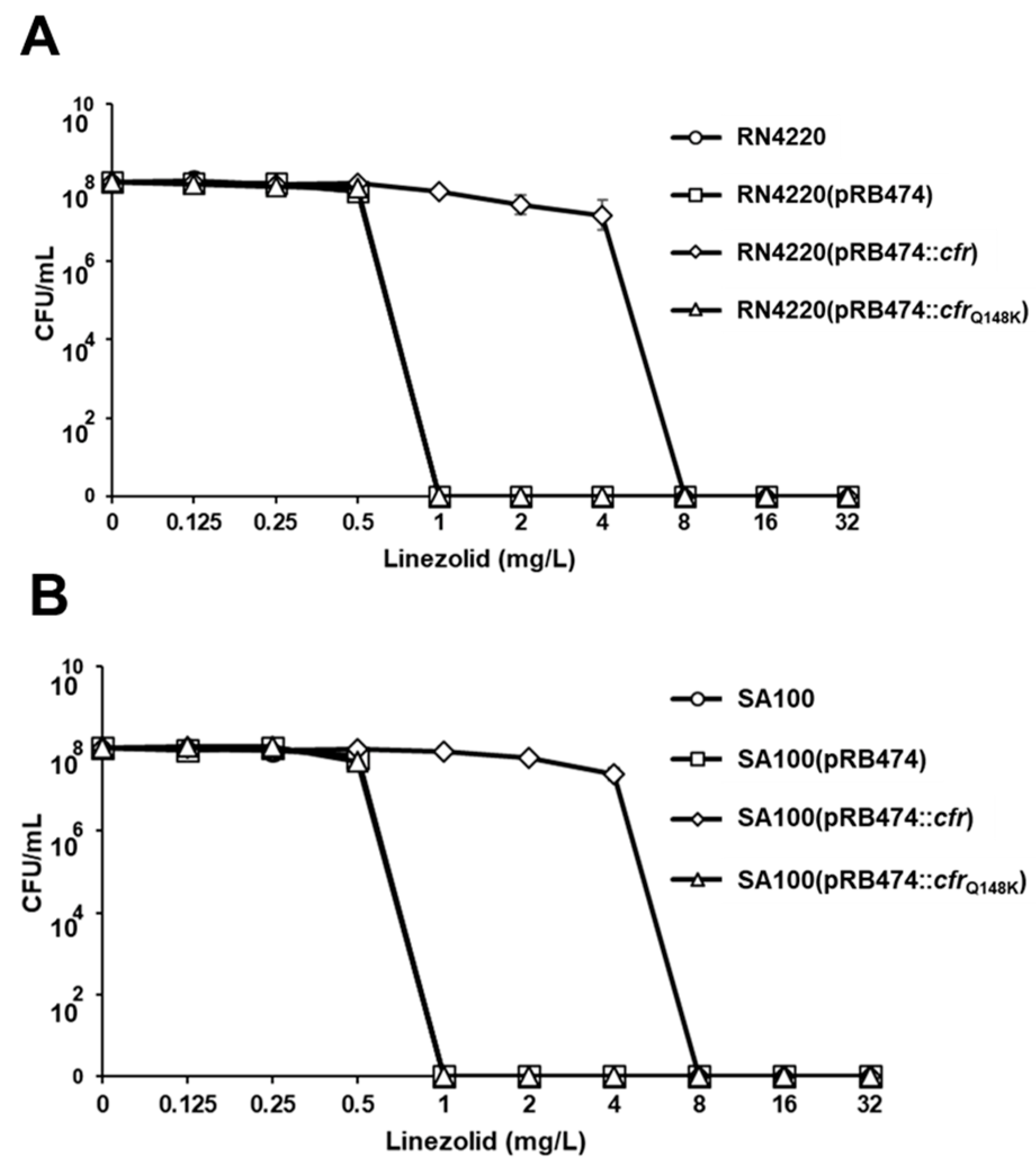
2.3. Impact of cfrQ148K on PhLOPSA Resistance Phenotype
2.4. Linezolid Population Analysis Profiles
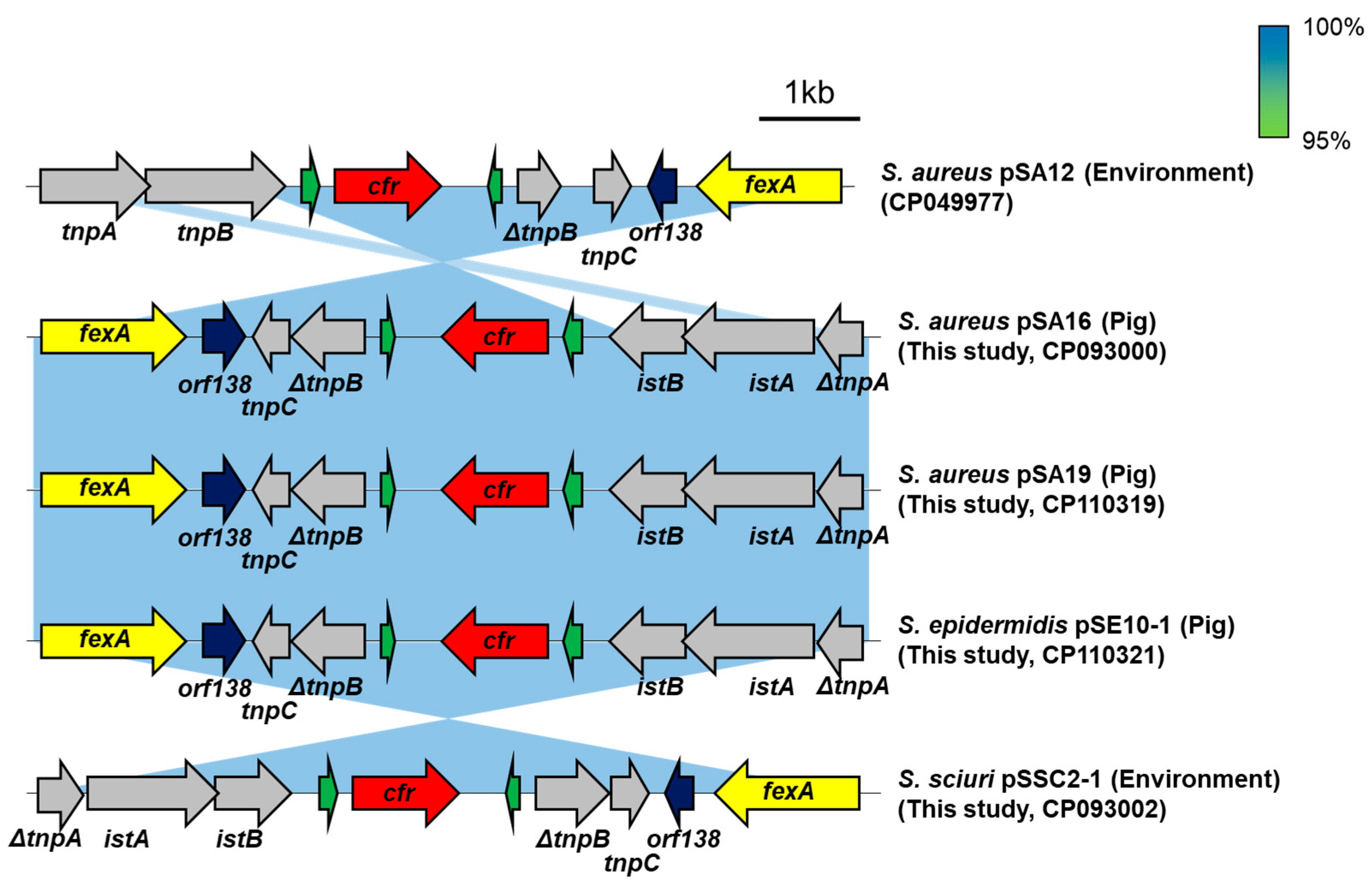
2.5. Genetic Environment of cfrQ148K
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. DNA Isolation and cfr Sequencing
4.3. Sequencing of 23S rRNA and Ribosomal Protein Genes
4.4. Genetic Manipulations and cfr Cloning
4.5. Antimicrobial Susceptibility Testing
4.6. Population Analysis
4.7. Whole Genome Sequencing Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tsiodras, S.; Gold, H.S.; Sakoulas, G.; Eliopoulos, G.M.; Wennersten, C.; Venkataraman, L.; Moellering, R.C.; Ferraro, M.J. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet 2001, 358, 207–208. [Google Scholar] [CrossRef]
- Schwarz, S.; Zhang, W.; Du, X.D.; Kruger, H.; Feßler, A.T.; Ma, S.; Zhu, Y.; Wu, C.; Shen, J.; Wang, Y. Mobile oxazolidinone resistance genes in Gram-positive and Gram-negative bacteria. Clin. Microbiol. Rev. 2021, 34, e0018820. [Google Scholar] [CrossRef] [PubMed]
- Bender, J.; Strommenger, B.; Steglich, M.; Zimmermann, O.; Fenner, I.; Lensing, C.; Dagwadordsch, U.; Kekule, A.S.; Werner, G.; Layer, F. Linezolid resistance in clinical isolates of Staphylococcus epidermidis from German hospitals and characterization of two cfr-carrying plasmids. J. Antimicrob. Chemother. 2015, 70, 1630–1638. [Google Scholar] [CrossRef]
- Lee, G.Y.; Kim, G.B.; Yang, S.J. Co-occurrence of cfr-mediated linezolid-resistance in ST398 LA-MRSA and non-aureus staphylococci isolated from a pig farm. Vet. Microbiol. 2022, 266, 109336. [Google Scholar] [CrossRef]
- Ruiz-Ripa, L.; Belles-Belles, A.; Fernandez-Fernandez, R.; Garcia, M.; Vilaro, A.; Zarazaga, M.; Torres, C. Linezolid-resistant MRSA-CC398 carrying the cfr gene, and MRSA-CC9 isolates from pigs with signs of infection in Spain. J. Appl. Microbiol. 2021, 131, 615–622. [Google Scholar] [CrossRef]
- Timmermans, M.; Bogaerts, B.; Vanneste, K.; De Keersmaecker, S.C.J.; Roosens, N.H.C.; Kowalewicz, C.; Simon, G.; Argudin, M.A.; Deplano, A.; Hallin, M.; et al. Large diversity of linezolid-resistant isolates discovered in food-producing animals through linezolid selective monitoring in Belgium in 2019. J. Antimicrob. Chemother. 2021, 77, 49–57. [Google Scholar] [CrossRef]
- Cuny, C.; Arnold, P.; Hermes, J.; Eckmanns, T.; Mehraj, J.; Schoenfelder, S.; Ziebuhr, W.; Zhao, Q.; Wang, Y.; Feßler, A.T.; et al. Occurrence of cfr-mediated multiresistance in staphylococci from veal calves and pigs, from humans at the corresponding farms, and from veterinarians and their family members. Vet. Microbiol. 2017, 200, 88–94. [Google Scholar] [CrossRef]
- Gu, B.; Kelesidis, T.; Tsiodras, S.; Hindler, J.; Humphries, R.M. The emerging problem of linezolid-resistant Staphylococcus. J. Antimicrob. Chemother. 2013, 68, 4–11. [Google Scholar] [CrossRef]
- Tewhey, R.; Gu, B.; Kelesidis, T.; Charlton, C.; Bobenchik, A.; Hindler, J.; Schork, N.J.; Humphries, R.M. Mechanisms of linezolid resistance among coagulase-negative staphylococci determined by whole-genome sequencing. Mbio 2014, 5, e00894-14. [Google Scholar] [CrossRef]
- Stefani, S.; Bongiorno, D.; Mongelli, G.; Campanile, F. Linezolid resistance in staphylococci. Pharmaceuticals 2010, 3, 1988–2006. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, S.; Werckenthin, C.; Kehrenberg, C. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemother. 2000, 44, 2530–2533. [Google Scholar] [CrossRef] [PubMed]
- Toh, S.M.; Xiong, L.; Arias, C.A.; Villegas, M.V.; Lolans, K.; Quinn, J.; Mankin, A.S. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 2007, 64, 1506–1514. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.; Stojkovic, V.; Noda-Garcia, L.; Young, I.D.; Myasnikov, A.G.; Kleinman, J.; Palla, A.; Floor, S.N.; Frost, A.; Fraser, J.S.; et al. Directed evolution of the rRNA methylating enzyme Cfr reveals molecular basis of antibiotic resistance. Elife 2022, 11, e70017. [Google Scholar] [CrossRef]
- Long, K.S.; Poehlsgaard, J.; Kehrenberg, C.; Schwarz, S.; Vester, B. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 2006, 50, 2500–2505. [Google Scholar] [CrossRef]
- Ruiz-Ripa, L.; Feßler, A.T.; Hanke, D.; Sanz, S.; Olarte, C.; Mama, O.M.; Eichhorn, I.; Schwarz, S.; Torres, C. Coagulase-negative staphylococci carrying cfr and PVL genes, and MRSA/MSSA-CC398 in the swine farm environment. Vet. Microbiol. 2020, 243, 108631. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, W.; Wang, J.; Wu, C.; Shen, Z.; Fu, X.; Yan, Y.; Zhang, Q.; Schwarz, S.; Shen, J. Distribution of the multidrug resistance gene cfr in Staphylococcus species isolates from swine farms in China. Antimicrob. Agents Chemother. 2012, 56, 1485–1490. [Google Scholar] [CrossRef]
- Lee, G.Y.; Seong, H.J.; Sul, W.J.; Yang, S.J. Genomic information on linezolid-resistant sequence-type 398 livestock-associated methicillin-resistant Staphylococcus aureus isolated from a pig. Foodborne Pathog. Dis. 2021, 18, 378–387. [Google Scholar] [CrossRef]
- Kang, H.Y.; Moon, D.C.; Mechesso, A.F.; Choi, J.H.; Kim, S.J.; Song, H.J.; Kim, M.H.; Yoon, S.S.; Lim, S.K. Emergence of cfr-mediated linezolid resistance in Staphylococcus aureus isolated from pig carcasses. Antibiotics 2020, 9, 769. [Google Scholar] [CrossRef] [PubMed]
- Gales, A.C.; Deshpande, L.M.; de Souza, A.G.; Pignatari, A.C.; Mendes, R.E. MSSA ST398/t034 carrying a plasmid-mediated Cfr and Erm (B) in Brazil. J. Antimicrob. Chemother. 2015, 70, 303–305. [Google Scholar] [CrossRef]
- Kehrenberg, C.; Cuny, C.; Strommenger, B.; Schwarz, S.; Witte, W. Methicillin-resistant and -susceptible Staphylococcus aureus strains of clonal lineages ST398 and ST9 from swine carry the multidrug resistance gene cfr. Antimicrob. Agents Chemother. 2009, 53, 779–781. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Wang, H.; Zhu, F.; Jiang, S.; Sun, L.; Zhao, F.; Yu, Y.; Chen, Y. Characterization of an ST5-SCCmec II-t311 methicillin-resistant Staphylococcus aureus strain with a widespread cfr-positive plasmid. J. Infect. Chemother. 2020, 26, 699–705. [Google Scholar] [PubMed]
- Li, S.; Zhao, L.; Zheng, B.; Shen, P.; Ji, J.; Lv, J.; Li, L.; Xiao, Y. Identification and characterization of cfr-positive Staphylococcus aureus isolates from community-onset infectious patients in a county hospital in China. J. Med. Microbiol. 2015, 64, 910–915. [Google Scholar] [PubMed]
- Sun, C.; Zhang, P.; Ji, X.; Fan, R.; Chen, B.; Wang, Y.; Schwarz, S.; Wu, C. Presence and molecular characteristics of oxazolidinone resistance in staphylococci from household animals in rural China. J. Antimicrob. Chemother. 2018, 73, 1194–1200. [Google Scholar] [PubMed]
- Bender, J.K.; Fleige, C.; Klare, I.; Fiedler, S.; Mischnik, A.; Mutters, N.T.; Dingle, K.E.; Werner, G. Detection of a cfr (B) variant in German Enterococcus faecium clinical isolates and the impact on linezolid resistance in Enterococcus spp. PLoS ONE 2016, 11, e0167042. [Google Scholar]
- Liu, Y.; Wang, Y.; Schwarz, S.; Wang, S.L.; Chen, L.R.; Wu, C.M.; Shen, J.Z. Investigation of a multiresistance gene cfr that fails to mediate resistance to phenicols and oxazolidinones in Enterococcus faecalis. J. Antimicrob. Chemother. 2014, 69, 892–898. [Google Scholar]
- Fang, L.X.; Duan, J.H.; Chen, M.Y.; Deng, H.; Liang, H.Q.; Xiong, Y.Q.; Sun, J.; Liu, Y.H.; Liao, X.P. Prevalence of cfr in Enterococcus faecalis strains isolated from swine farms in China: Predominated cfr-carrying pCPPF5-like plasmids conferring “non-linezolid resistance” phenotype. Infect. Genet. Evol. 2018, 62, 188–192. [Google Scholar] [PubMed]
- Yang, S.J.; Mishra, N.N.; Rubio, A.; Bayer, A.S. Causal role of single nucleotide polymorphisms within the mprF gene of Staphylococcus aureus in daptomycin resistance. Antimicrob. Agents Chemother. 2013, 57, 5658–5664. [Google Scholar] [CrossRef]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Kreiswirth, B.N.; Lofdahl, S.; Betley, M.J.; Oreilly, M.; Schlievert, P.M.; Bergdoll, M.S.; Novick, R.P. The toxic shock syndrome exotoxin ttructural gene is not detectably transmitted by a prophage. Nature 1983, 305, 709–712. [Google Scholar] [CrossRef]
- Bruckner, R. A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene 1992, 122, 187–192. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standard Institute (CLSI). VET01S Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 5th ed.; Clinical Laboratory Standard Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Clinical and Laboratory Standard Institute (CLSI). M100 Performance Standards for Antimicrobial Susceptibility Testing, 32nd ed.; Clinical Laboratory Standard Institute: Wayne, PA, USA, 2022. [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 11.0; European Committee on Antimicrobial Susceptibility Testing: Växjö, Sweden, 2022. [Google Scholar]
- Bakthavatchalam, Y.D.; Vasudevan, K.; Neeravi, A.; Perumal, R.; Veeraraghavan, B. First draft genome sequence of linezolid and rifampicin resistant Staphylococcus haemolyticus. Jpn. J. Infect. Dis. 2020, 73, 296–299. [Google Scholar] [CrossRef]
- Li, S.M.; Zhou, Y.F.; Li, L.; Fang, L.X.; Duan, J.H.; Liu, F.R.; Liang, H.Q.; Wu, Y.T.; Gu, W.Q.; Liao, X.P.; et al. Characterization of the multi-drug resistance gene cfr in methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from animals and humans in China. Front. Microbiol. 2018, 9, 2925. [Google Scholar] [CrossRef]
- Fioriti, S.; Coccitto, S.N.; Morroni, G.; Simoni, S.; Cinthi, M.; Magi, G.; Sante, L.D.; Alvarez-Suarez, J.M.; Mingoia, M.; Giovanetti, E.; et al. Detection of a chromosomal truncated cfr gene in a linezolid-susceptible LA-MRSA ST398 isolate of porcine origin, Italy. J. Glob. Antimicrob. Resist. 2021, 26, 199–201. [Google Scholar] [CrossRef]
- Monk, I.R.; Foster, T.J. Genetic manipulation of staphylococci-breaking through the barrier. Front. Cell. Infect. Microbiol. 2012, 2, 49. [Google Scholar] [CrossRef] [PubMed]
- LaMarre, J.M.; Locke, J.B.; Shaw, K.J.; Mankin, A.S. Low fitness cost of the multidrug resistance gene cfr. Antimicrob. Agents Chemother. 2011, 55, 3714–3719. [Google Scholar] [CrossRef]
- Dubois, D.; Leyssene, D.; Chacornac, J.P.; Kostrzewa, M.; Schmit, P.O.; Talon, R.; Bonnet, R.; Delmas, J. Identification of a variety of Staphylococcus species by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2010, 48, 941–945. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.M.; Kim, M.S.; Park, K.U.; Song, J.; Kim, E.C. tuf gene sequence analysis has greater discriminatory power than 16S rRNA sequence analysis in identification of clinical Isolates of coagulase-negative staphylococci. J. Clin. Microbiol. 2011, 49, 4142–4149. [Google Scholar] [CrossRef] [PubMed]
- Schauer, B.; Szostak, M.P.; Ehricht, R.; Monecke, S.; Fessler, A.T.; Schwarz, S.; Spergser, J.; Krametter-Frotscher, R.; Loncaric, I. Diversity of methicillin-resistant coagulase-negative Staphylococcus spp. and methicillin-resistant Mammaliicoccus spp. isolated from ruminants and New World camelids. Vet. Microbiol. 2021, 254, 109005. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.C.; Vargas, M.R.; Miragaia, M.; Peacock, S.J.; Archer, G.L.; Enright, M.C. Improved multilocus sequence typing scheme for Staphylococcus epidermidis. J. Clin. Microbiol. 2007, 45, 616–619. [Google Scholar] [CrossRef]
- Enright, M.C.; Day, N.P.; Davies, C.E.; Peacock, S.J.; Spratt, B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000, 38, 1008–1015. [Google Scholar] [CrossRef]
- Kondo, Y.; Ito, T.; Ma, X.X.; Watanabe, S.; Kreiswirth, B.N.; Etienne, J.; Hiramatsu, K. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: Rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 2007, 51, 264–274. [Google Scholar] [CrossRef]
- Naccache, S.N.; Callan, K.; Burnham, C.A.; Wallace, M.A.; Westblade, L.F.; Dien Bard, J.; Staphylococcus Ad Hoc Working Group of the CLSI Antimicrobial Susceptibility Testing Subcommittee. Evaluation of oxacillin and cefoxitin disk fiffusion and microbroth dilution methods for detecting mecA-mediated beta-lactam resistance in contemporary Staphylococcus epidermidis isolates. J. Clin. Microbiol. 2019, 57, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kehrenberg, C.; Schwarz, S. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 2006, 50, 1156–1163. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ripa, L.; Fessler, A.T.; Hanke, D.; Eichhorn, I.; Azcona-Gutierrez, J.M.; Alonso, C.A.; Perez-Moreno, M.O.; Aspiroz, C.; Belles, A.; Schwarz, S.; et al. Mechanisms of linezolid resistance among clinical Staphylococcus spp. in Spain: Spread of methicillin- and linezolid-resistant S. epidermidis ST2. Microb. Drug Resist. 2021, 27, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Dyer, D.W.; Iandolo, J.J. Rapid isolation of DNA from Staphylococcus aureus. Appl. Environ. Microbiol. 1983, 46, 283–285. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Nojima, H.; Okayama, H. High efficiency transformation of Escherichia coli with plasmids. Gene 1990, 96, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Schenk, S.; Laddaga, R.A. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 1992, 73, 133–138. [Google Scholar] [CrossRef]
- Schneewind, O.; Missiakas, D. Genetic manipulation of Staphylococcus aureus. Curr. Protoc. Microbiol. 2014, 32, 9C.3.1–9C.3.19. [Google Scholar] [CrossRef]
- Ikeda-Dantsuji, Y.; Hanaki, H.; Nakae, T.; Takesue, Y.; Tomono, K.; Honda, J.; Yanagihara, K.; Mikamo, H.; Fukuchi, K.; Kaku, M.; et al. Emergence of linezolid-resistant mutants in a susceptible-cell population of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2011, 55, 2466–2468. [Google Scholar] [CrossRef] [Green Version]
| Species | Strain 1 | Origin | MLST-SCCmec | Methicillin Resistance 2 | Antimicrobial Resistance 3 | Positivity | MICs (mg/L) 4 | Reference | |
|---|---|---|---|---|---|---|---|---|---|
| cfr | fexA | LZD | |||||||
| CoPS S. aureus | SA2 | Pig | ST398-V | MR | AMP-CEF-CHL-CIP-CLI-ERY-GEN-LZD-SYN-TET-TIA | + | + | 12 | [17] |
| SA3 | Pig | ST398-V | MR | AMP-CEF-CHL-CIP-CLI-ERY-GEN-LZD-SYN-TET-TIA | + | + | 12 | [4] | |
| SA12 | Environ. | ST398-V | MR | AMP-CEF-CHL-CIP-CLI-ERY-GEN-LZD-SYN-TET-TIA | + | + | 16 | [4] | |
| SA16 | Pig | ST398 | MS | AMP-CHL-CIP-CLI-ERY-GEN-TET-TIA | + | + | 2 | In this study | |
| SA17 | Pig | ST398 | MS | AMP-CHL-CIP-CLI-ERY-GEN-TET-TIA | + | + | 2 | In this study | |
| SA18 | Pig | ST398 | MS | AMP-CHL-CIP-CLI-ERY-GEN-TET-TIA | + | + | 2 | In this study | |
| SA19 | Pig | ST398 | MS | AMP-CHL-CIP-CLI-ERY-GEN-TET-TIA | + | + | 2 | In this study | |
| SA20 | Environ. | ST398 | MS | AMP-CHL-CIP-CLI-ERY-GEN-TET-TIA | + | + | 2 | In this study | |
| SA21 | Environ. | ST398 | MS | AMP-CHL-CIP-CLI-ERY-GEN-TET-TIA | + | + | 2 | In this study | |
| CoNS S. epidermidis | SE7 | Pig | ST570 | MS | AMP-CHL-CLI-ERY-GEN-LZD-SYN-TET-TIA | + | + | 48 | [4] |
| SE9 | Pig | ST570 | MS | AMP-CHL-CLI-ERY-GEN-SYN-TIA | + | + | 2 | In this study | |
| SE10 | Pig | ST570 | MS | AMP-CHL-CLI-TIA | + | + | 4 | In this study | |
| S. sciuri | SSC1 | Environ. | NT-NT | MR | AMP-CEF-CHL-CLI-SYN-TET-TIA | + | + | 2 | In this study |
| SSC2 | Environ. | ST85-NT | MR | AMP-CEF-CHL-CLI-TET-TIA | + | + | 4 | In this study | |
| S. simulans | SSM1 | Pig | - | MS | CHL-CLI-GEN-TIA | + | + | 0.75 | In this study |
| Strain or Plasmid | Genotypic and Phenotypic Characteristics | Reference |
|---|---|---|
| E. coli | ||
| DH5α | F-Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17(rk−, mk+) phoA supE44 thi-1 gyrA96 relA1 λ-; Host stain for transformation of plasmid constructs | [28] |
| S. aureus | ||
| RN4220 | 8325-4, laboratory strain; accepts for foreign DNA | [29] |
| SA12 | ST398-SCCmec V MRSA strain carrying cfr; LZDr | [4] |
| SA16 | ST398 MSSA strain carrying cfrQ148K; LZDs | In this study |
| SA100 | ST398-SCCmec V MRSA; cfr and fexA-negative, LZDs | In this study |
| Plasmid | ||
| pRB474 | E. coli-S. aureus shuttle vector; AMPr and CHLr | [30] |
| pRB474::cfr | The wild-type cfr from SA12 cloned into pRB474 | In this study |
| pRB474::cfrQ148K | Point-mutated cfr from SA16 cloned into pRB474 | In this study |
| Antimicrobial Agents (MIC) | MICs (mg/L) 1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SA12 2 | SA16 3 | RN4220 | RN4220 (pRB474) | RN4220 (pRB474::cfr) | RN4220 (pRB474::cfrQ148K) | SA100 | SA100 (pRB474) | SA100 (pRB474::cfr) | SA100 (pRB474::cfrQ148K) | |
| Florfenicol (>8) | 128 | 256 | 8 | 8 | 256 | 16 | 8 | 8 | 256 | 8 |
| Chloramphenicol (≥32) | 256 | 256 | 16 | 32 | 256 | 32 | 16 | 32 | 256 | 32 |
| Clindamycin (≥4) | 256 | 256 | 0.125 | 0.125 | 256 | 0.125 | 0.125 | 0.125 | 256 | 0.125 |
| Linezolid (≥8) | 16 | 2 | 2 | 2 | 8 | 2 | 2 | 2 | 8 | 2 |
| Tiamulin (>2) | 128 | 256 | 0.5 | 0.5 | 128 | 0.5 | 1 | 1 | 256 | 1 |
| Quinupristin/dalfopristin (>4) | >32 | 2 | 0.38 | 0.38 | 2 | 0.38 | 0.38 | 0.38 | 2 | 0.5 |
| Vancomycin (≥16) | 1 | 1.5 | 1 | 1 | 1 | 1 | 1.5 | 1.5 | 1.5 | 1.5 |
| Tetracycline (≥16) | >256 | >256 | 0.25 | 0.25 | 0.25 | 0.25 | >256 | >256 | >256 | >256 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, G.Y.; Yang, S.-J. Occurrence of cfr-Positive Linezolid-Susceptible Staphylococcus aureus and Non-aureus Staphylococcal Isolates from Pig Farms. Antibiotics 2023, 12, 359. https://doi.org/10.3390/antibiotics12020359
Lee GY, Yang S-J. Occurrence of cfr-Positive Linezolid-Susceptible Staphylococcus aureus and Non-aureus Staphylococcal Isolates from Pig Farms. Antibiotics. 2023; 12(2):359. https://doi.org/10.3390/antibiotics12020359
Chicago/Turabian StyleLee, Gi Yong, and Soo-Jin Yang. 2023. "Occurrence of cfr-Positive Linezolid-Susceptible Staphylococcus aureus and Non-aureus Staphylococcal Isolates from Pig Farms" Antibiotics 12, no. 2: 359. https://doi.org/10.3390/antibiotics12020359
APA StyleLee, G. Y., & Yang, S.-J. (2023). Occurrence of cfr-Positive Linezolid-Susceptible Staphylococcus aureus and Non-aureus Staphylococcal Isolates from Pig Farms. Antibiotics, 12(2), 359. https://doi.org/10.3390/antibiotics12020359






