Prevalence of Antimicrobial Resistance and Clonal Relationship in ESBL/AmpC-Producing Proteus mirabilis Isolated from Meat Products and Community-Acquired Urinary Tract Infection (UTI-CA) in Southern Brazil
Abstract
1. Introduction
2. Results
2.1. Phenotypic Resistance to Antimicrobials
2.2. Detection of Antimicrobial Resistance Genes
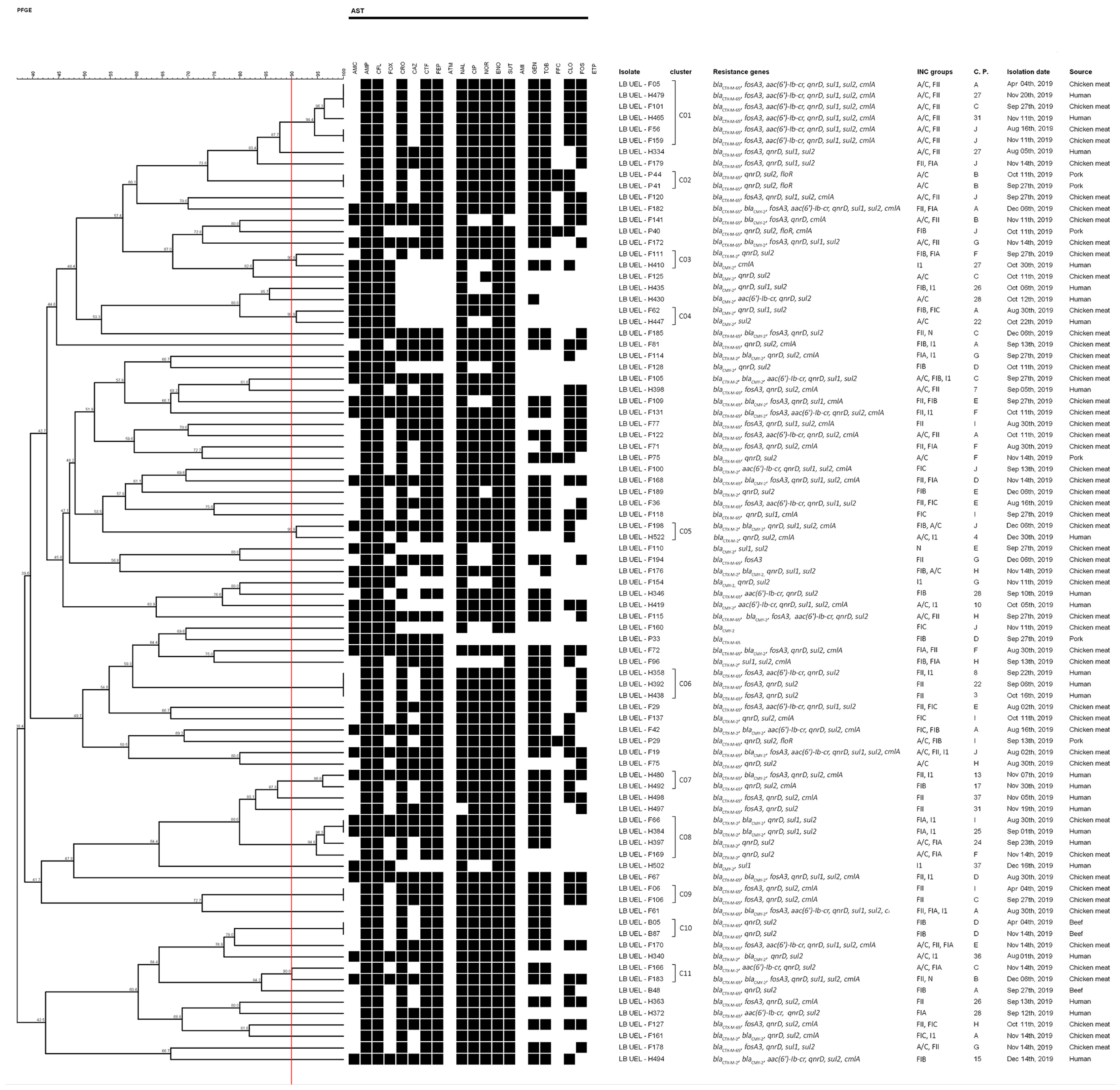
2.3. Molecular Typing of Isolates
3. Discussion
4. Materials and Methods
4.1. Bacterial Isolates
4.2. Antimicrobial Susceptibility Test
4.3. Detection of Resistance Genes
4.4. Pulsed-Field Gel Electrophoresis (PFGE)
4.5. Sequencing of ESBL and blaCMY
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Drzewiecka, D. Significance and roles of Proteus spp. bacteria in natural environments. Microb. Ecol. 2016, 72, 741–758. [Google Scholar] [CrossRef]
- Schaffer, J.N.; Pearson, M.M. Urinary tract infections: Molecular pathogenesis and clinical management. In Urinary Tract Infections, 2nd ed.; Mulvey, M.A., Klumpp, D.J., Stapleton, A.E., Eds.; ASM Press: Washington, DC, USA, 2017; pp. 383–434. [Google Scholar]
- Armbruster, C.E.; Mobley, H.L.T.; Pearson, M.M. Pathogenesis of Proteus mirabilis infection. EcoSal. Plus 2018, 8, 01–73. [Google Scholar] [CrossRef]
- Gajdács, M.; Urbán, E. Comparative epidemiology and resistance trends of Proteae in urinary tract infections of inpatients and outpatients: A 10-year retrospective study. Antibiotics 2019, 8, 91. [Google Scholar] [CrossRef] [PubMed]
- Reu, C.E.; Volanski, W.; Prediger, K.C.; Picheth, G.; Fadel-Picheth, C.M.T. Epidemiology of pathogens causing urinary tract infections in an urban community in southern Brazil. Braz. J. Infect. Dis. 2018, 22, 505–507. [Google Scholar] [CrossRef]
- Yu, Z.; Joossens, M.; Van den Abeele, A.-M.; Kerkhof, P.-J.; Houf, K. Isolation, characterization and antibiotic resistance of Proteus mirabilis from belgian broiler carcasses at retail and human stool. Food. Microbiol. 2021, 96, 103724. [Google Scholar] [CrossRef]
- Tesson, V.; Federighi, M.; Cummins, E.; de Oliveira Mota, J.; Guillou, S.; Boué, G. A systematic review of beef meat quantitative microbial risk assessment models. Int. J. Environ. Res. Public Health 2020, 17, 688. [Google Scholar] [CrossRef]
- Baéza, E.; Guillier, L.; Petracci, M. Review: Production factors affecting poultry carcass and meat quality attributes. Animal 2022, 16, 100331. [Google Scholar] [CrossRef]
- Guo, S.; Aung, K.T.; Tay, M.Y.F.; Seow, K.L.G.; Ng, L.C.; Schlundt, J. Extended-spectrum β-lactamase-producing Proteus mirabilis with multidrug resistance isolated from raw chicken in Singapore: Genotypic and phenotypic analysis. J. Glob. Antimicrob. Resist. 2019, 19, 252–254. [Google Scholar] [CrossRef] [PubMed]
- Sanches, M.S.; Baptista, A.A.S.; de Souza, M.; Menck-Costa, M.F.; Koga, V.L.; Kobayashi, R.K.T.; Rocha, S.P.D. Genotypic and phenotypic profiles of virulence factors and antimicrobial resistance of Proteus mirabilis isolated from chicken carcasses: Potential zoonotic risk. Braz. J. Microbiol. 2019, 50, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Sanches, M.S.; Rodrigues da Silva, C.; Silva, L.C.; Montini, V.H.; Lopes Barboza, M.G.; Guidone, G.H.M.; Dias de Oliva, B.H.; Nishio, E.K.; Faccin Galhardi, L.C.; Vespero, E.C.; et al. Proteus mirabilis from community-acquired urinary tract infections (UTI-CA) shares genetic similarity and virulence factors with isolates from chicken, beef and pork meat. Microb. Pathog. 2021, 158, 105098. [Google Scholar] [CrossRef] [PubMed]
- Antimicrobial Resistance: Global Report on Surveillance. Available online: https://apps.who.int/iris/bitstream/handle/10665/112642/9789241564748_eng.pdf;jsessionid=9BD9CFC92083023155E1028C22D10F1F?sequence=1 (accessed on 12 November 2021).
- Di, K.N.; Pham, D.T.; Tee, T.S.; Binh, Q.A.; Nguyen, T.C. Antibiotic usage and resistance in animal production in Vietnam: A review of existing literature. Trop. Anim. Health Prod. 2021, 53, 340. [Google Scholar] [CrossRef] [PubMed]
- Kasimanickam, V.; Kasimanickam, M.; Kasimanickam, R. Antibiotics use in food animal production: Escalation of antimicrobial resistance: Where are we now in combating AMR? Med. Sci. 2021, 9, 14. [Google Scholar] [CrossRef]
- O’Neill, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. Available online: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf (accessed on 12 November 2021).
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-spectrum β-lactamases: An update on their characteristics, epidemiology and detection. JAC-Antimicrob. Resist. 2021, 3, 3. [Google Scholar] [CrossRef]
- Madec, J.-Y.; Haenni, M.; Nordmann, P.; Poirel, L. Extended-spectrum β-lactamase/AmpC- and carbapenemase-producing Enterobacteriaceae in animals: A threat for humans? Clin. Microbiol. Infect. 2017, 23, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.; Bradford, P.A. Epidemiology of β-lactamase-producing pathogens. Clin. Microbiol. Rev. 2020, 33, 2. [Google Scholar] [CrossRef]
- Nhung, N.T.; Chansiripornchai, N.; Carrique-Mas, J.J. Antimicrobial resistance in bacterial poultry pathogens: A review. Front. Vet. Sci. 2017, 4, 126. [Google Scholar] [CrossRef] [PubMed]
- Hedman, H.D.; Vasco, K.A.; Zhang, L. A review of antimicrobial resistance in poultry farming within low-resource settings. Animals 2020, 10, 1264. [Google Scholar] [CrossRef]
- Roth, N.; Käsbohrer, A.; Mayrhofer, S.; Zitz, U.; Hofacre, C.; Domig, K.J. The application of antibiotics in broiler production and the resulting antibiotic resistance in Escherichia coli: A global overview. Poult. Sci. 2019, 98, 1791–1804. [Google Scholar] [CrossRef] [PubMed]
- Rabello, R.F.; Bonelli, R.R.; Penna, B.A.; Albuquerque, J.P.; Souza, R.M.; Cerqueira, A.M.F. Antimicrobial resistance in farm animals in Brazil: An update overview. Animals 2020, 10, 552. [Google Scholar] [CrossRef] [PubMed]
- Faccone, D.; Moredo, F.A.; Giacoboni, G.I.; Albornoz, E.; Alarcón, L.; Nievas, V.F.; Corso, A. Multidrug-resistant Escherichia coli harbouring mcr-1 and blaCTX-M genes isolated from swine in Argentina. J. Glob. Antimicrob. Resist. 2019, 18, 160–162. [Google Scholar] [CrossRef]
- Abdalla, S.E.; Abia, A.L.K.; Amoako, D.G.; Perrett, K.; Bester, L.A.; Essack, S.Y. From farm-to-fork: E. coli from an intensive pig production system in South Africa shows high resistance to critically important antibiotics for human and animal use. Antibiotics 2021, 10, 178. [Google Scholar] [CrossRef] [PubMed]
- Viana, C.; Grossi, J.L.; Sereno, M.J.; Yamatogi, R.S.; Bersot, L.D.S.; Call, D.R.; Nero, L.A. Phenotypic and genotypic characterization of non-typhoidal Salmonella isolated from a brazilian pork production chain. Food Res. Int. 2020, 137, 109406. [Google Scholar] [CrossRef] [PubMed]
- Cyoia, P.S.; Koga, V.L.; Nishio, E.K.; Houle, S.; Dozois, C.M.; de Brito, K.C.T.; de Brito, B.G.; Nakazato, G.; Kobayashi, R.K.T. Distribution of ExPEC virulence factors, blaCTX-M, fosA3, and mcr-1 in Escherichia coli isolated from commercialized chicken carcasses. Front. Microbiol. 2018, 9, 3254. [Google Scholar] [CrossRef] [PubMed]
- de Souza Gazal, L.E.; Medeiros, L.P.; Dibo, M.; Nishio, E.K.; Koga, V.L.; Gonçalves, B.C.; Grassotti, T.T.; de Camargo, T.C.L.; Pinheiro, J.J.; Vespero, E.C.; et al. Detection of ESBL/AmpC-producing and fosfomycin-resistant Escherichia coli from different sources in poultry production in southern Brazil. Front. Microbiol. 2020, 11, 604544. [Google Scholar] [CrossRef]
- Oliveira, W.D.; Barboza, M.G.L.; Faustino, G.; Inagaki, W.T.Y.; Sanches, M.S.; Kobayashi, R.K.T.; Vespero, E.C.; Rocha, S.P.D. Virulence, resistance and clonality of Proteus mirabilis isolated from patients with community-acquired urinary tract infection (CA-UTI) in Brazil. Microb. Pathog. 2021, 152, 104642. [Google Scholar] [CrossRef]
- Rodríguez-Baño, J.; Gutiérrez-Gutiérrez, B.; Machuca, I.; Pascual, A. Treatment of infections caused by extended-spectrum-beta-lactamase-, AmpC-, and carbapenemase-producing Enterobacteriaceae. Clin. Microbiol. Rev. 2018, 31, 2. [Google Scholar] [CrossRef]
- Dhanji, H.; Murphy, N.M.; Doumith, M.; Durmus, S.; Lee, S.S.; Hope, R.; Woodford, N.; Livermore, D.M. Cephalosporin resistance mechanisms in Escherichia coli isolated from raw chicken imported into the UK. J. Antimicrob. Chemother. 2010, 65, 2534–2537. [Google Scholar] [CrossRef]
- Faife, S.L.; Zimba, T.; Sekyere, J.O.; Govinden, U.; Chenia, H.Y.; Simonsen, G.S.; Sundsfjord, A.; Essack, S.Y. β-lactam and fluoroquinolone resistance in Enterobacteriaceae from imported and locally-produced chicken in Mozambique. J. Infect. Dev. Ctries. 2020, 14, 471–478. [Google Scholar] [CrossRef]
- Park, H.; Kim, J.; Ryu, S.; Jeon, B. Predominance of blaCTX-M-65 and blaCTX-M-55 in extended-spectrum β-lactamase-producing Escherichia coli from raw retail chicken in South Korea. J. Glob. Antimicrob. Resist. 2019, 17, 216–220. [Google Scholar] [CrossRef]
- Leão, C.; Clemente, L.; Moura, L.; Seyfarth, A.M.; Hansen, I.M.; Hendriksen, R.S.; Amaro, A. Emergence and clonal spread of CTX-M-65-Producing Escherichia coli from retail meat in Portugal. Front. Microbiol. 2021, 12, 653595. [Google Scholar] [CrossRef]
- Hounmanou, Y.M.G.; Bortolaia, V.; Dang, S.T.T.; Truong, D.; Olsen, J.E.; Dalsgaard, A. ESBL and AmpC β-lactamase encoding genes in E. coli from pig and pig farm workers in Vietnam and their Association with mobile genetic elements. Front. Microbiol. 2021, 12, 629139. [Google Scholar] [CrossRef] [PubMed]
- Tate, H.; Folster, J.P.; Hsu, C.-H.; Chen, J.; Hoffmann, M.; Li, C.; Morales, C.; Tyson, G.H.; Mukherjee, S.; Brown, A.C.; et al. Comparative analysis of extended-spectrum-β-lactamase CTX-M-65-producing Salmonella enterica Serovar Infantis isolates from humans, food animals, and retail chickens in the United States. Antimicrob. Agents Chemother. 2017, 61, 7. [Google Scholar] [CrossRef] [PubMed]
- Vinueza-Burgos, C.; Ortega-Paredes, D.; Narváez, C.; De Zutter, L.; Zurita, J. Characterization of cefotaxime resistant Escherichia coli isolated from broiler farms in Ecuador. PLoS ONE 2019, 14, 4. [Google Scholar] [CrossRef]
- Martínez-Puchol, S.; Riveros, M.; Ruidias, K.; Granda, A.; Ruiz-Roldán, L.; Zapata-Cachay, C.; Ochoa, T.J.; Pons, M.J.; Ruiz, J. Dissemination of a multidrug resistant CTX-M-65 producer Salmonella enterica Serovar Infantis clone between marketed chicken meat and children. Int. J. Food Microbiol. 2021, 344, 109109. [Google Scholar] [CrossRef] [PubMed]
- Furlan, J.P.R.; Lopes, R.; Gonzalez, I.H.L.; Ramos, P.L.; Stehling, E.G. Comparative analysis of multidrug resistance plasmids and genetic background of CTX-M-producing Escherichia coli recovered from captive wild animals. Appl. Microbiol. Biotechnol. 2020, 104, 6707–6717. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zeng, Z.-L.; Huang, X.-Y.; Ma, Z.-B.; Guo, Z.-W.; Lv, L.-C.; Xia, Y.-B.; Zeng, L.; Song, Q.-H.; Liu, J.-H. Evolution and comparative genomics of F33:A-:B- plasmids carrying blaCTX-M-55 or blaCTX-M-65 in Escherichia coli and Klebsiella pneumoniae isolated from animals, food products, and humans in China. mSphere 2018, 3, 4. [Google Scholar] [CrossRef]
- Wu, C.; Wang, Y.; Shi, X.; Wang, S.; Ren, H.; Shen, Z.; Wang, Y.; Lin, J.; Wang, S. Rapid rise of the ESBL and mcr-1 Genes in Escherichia coli of chicken origin in China, 2008–2014. Emerg. Microbes Infect. 2018, 7, 1. [Google Scholar] [CrossRef]
- Rizi, K.S.; Mosavat, A.; Youssefi, M.; Jamehdar, S.A.; Ghazvini, K.; Safdari, H.; Amini, Y.; Farsiani, H. High prevalence of blaCMY AmpC beta-lactamase in ESBL co-producing Escherichia coli and Klebsiella spp. clinical isolates in the northeast of Iran. J. Glob. Antimicrob. Resist. 2020, 22, 477–482. [Google Scholar] [CrossRef]
- Kumar, N.; Chatterjee, K.; Deka, S.; Shankar, R.; Kalita, D. Increased isolation of extended-spectrum beta-lactamase-producing Escherichia coli from community-onset urinary tract infection cases in Uttarakhand, India. Cureus 2021, 13, 3. [Google Scholar] [CrossRef]
- Kimera, Z.I.; Mgaya, F.X.; Misinzo, G.; Mshana, S.E.; Moremi, N.; Matee, M.I.N. Multidrug-resistant, including extended-spectrum beta lactamase-producing and quinolone-Resistant, Escherichia coli isolated from poultry and domestic pigs in Dar Es Salaam, Tanzania. Antibiotics 2021, 10, 406. [Google Scholar] [CrossRef] [PubMed]
- Koga, L.; Maluta, R.P.; da Silveira, W.D.; Ribeiro, R.A.; Hungria, M.; Vespero, E.C.; Nakazato, G.; Kobayashi, R.K.T. Characterization of CMY-2-type beta-lactamase-producing Escherichia coli isolated from chicken carcasses and human infection in a city of south Brazil. BMC Microbiol. 2019, 19, 174. [Google Scholar] [CrossRef] [PubMed]
- Kraychete, G.B.; Campana, E.H.; Picão, R.C.; Bonelli, R.R. qnrD-Harboring plasmids in Providencia spp. recovered from food and environmental brazilian sources. Sci. Total Environ. 2019, 646, 1290–1292. [Google Scholar] [CrossRef] [PubMed]
- Cai, T.; Palagin, I.; Brunelli, R.; Cipelli, R.; Pellini, E.; Truzzi, J.C.; Van Bruwaene, S. Office-Based approach to urinary tract infections in 50.000 patients: Results from the REWIND study. Int. J. Antimicrob. Agents 2020, 56, 105966. [Google Scholar] [CrossRef] [PubMed]
- Michelim, L.; Muller, G.; Zacaria, J.; Delamare, A.P.L.; da Costa, S.O.P.; Echeverrigaray, S. Comparison of PCR-based molecular markers for the characterization of Proteus mirabilis clinical isolates. Braz. J. Infect. Dis. 2008, 12, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Approved Standard; thirty-first Informational Supplement; CLSI Document M100-S31; CLSI: Wayne, PA, USA, 2021. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; Approved Standard; Third Informational Supplement; CLSI Document M31-A3; CLSI: Wayne, PA, USA, 2008. [Google Scholar]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Standard Operating Procedure for PulseNet PFGE of Escherichia coli O157:H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri. Available online: https://www.cdc.gov/pulsenet/pathogens/pfge.html (accessed on 12 July 2022).
- Basic Local Alignment Search Tool (BLAST). Available online: http://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 12 November 2021).
- Dallenne, C.; Da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef]
- Arlet, G.; Philippon, A. Construction by polymerase chain reaction and intragenic DNA probes for three main types of transferable β-lactamases (TEM, SHV, CARB). FEMS Microbiol. Lett. 1991, 82, 19–25. [Google Scholar] [CrossRef]
- Saladin, M.; Cao, V.T.B.; Lambert, T.; Donay, J.-L.; Herrmann, J.-L.; Ould-Hocine, Z.; Verdet, C.; Delisle, F.; Philippon, A.; Arlet, G. Diversity of CTX-M β-lactamases and their promoter regions fromEnterobacteriaceaeisolated in three Parisian hospitals. FEMS Microbiol. Lett. 2002, 209, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pérez, F.J.; Hanson, N.D. Detection of Plasmid-Mediated AmpC β-Lactamase Genes in Clinical Isolates by Using Multiplex PCR. J. Clin. Microbiol. 2002, 40, 2153–2162. [Google Scholar] [CrossRef]
- Cattoir, V.; Poirel, L.; Rotimi, V.; Soussy, C.-J.; Nordmann, P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 2007, 60, 394–397. [Google Scholar] [CrossRef]
- Wang, M.; Guo, Q.; Xu, X.; Wang, X.; Ye, X.; Wu, S.; Hooper, D.C.; Wang, M. New Plasmid-Mediated Quinolone Resistance Gene, qnrC, Found in a Clinical Isolate of Proteus mirabilis. Antimicrob. Agents Chemother. 2009, 53, 1892–1897. [Google Scholar] [CrossRef] [PubMed]
- Cavaco, L.M.; Hasman, H.; Xia, S.; Aarestrup, F.M. qnrD, a Novel Gene Conferring Transferable Quinolone Resistance in Salmonella enterica Serovar Kentucky and Bovismorbificans Strains of Human Origin. Antimicrob. Agents Chemother. 2009, 53, 603–608. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, W.; Pan, W.; Yin, J.; Pan, Z.; Gao, S.; Jiao, X. Prevalence of qnr, aac(6 ′ )-Ib-cr, qepA, and oqxAB in Escherichia coli Isolates from Humans, Animals, and the Environment. Antimicrob. Agents Chemother. 2012, 56, 3423–3427. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Sherwood, J.; Logue, C. Characterization of antimicrobial resistant Escherichia coli isolated from processed bison carcasses. J. Appl. Microbiol. 2007, 103, 2361–2369. [Google Scholar] [CrossRef]
- Guerra, B.; Soto, S.M.; Argüelles, J.M.; Mendoza, M.C. Multidrug Resistance Is Mediated by Large Plasmids Carrying a Class 1 Integron in the Emergent Salmonella enterica Serotype [4,5,12:i:−]. Antimicrob. Agents Chemother. 2001, 45, 1305–1308. [Google Scholar] [CrossRef]
- Sato, N.; Kawamura, K.; Nakane, K.; Wachino, J.-I.; Arakawa, Y. First Detection of Fosfomycin Resistance Gene fosA3 in CTX-M-Producing Escherichia coli Isolates from Healthy Individuals in Japan. Microb. Drug Resist. 2013, 19, 477–482. [Google Scholar] [CrossRef]



| Antimicrobials | Genes | Chicken Meat | Pork | Beef | Human |
|---|---|---|---|---|---|
| β-lactams | blaCTX-M-2 | 14 (7.0%) | 0 (0.0%) | 0 (0.0%) | 5 (2.5%) |
| blaCTX-M-65 | 33 (16.5%) | 6 (7.2%) | 3 (3.0%) | 14 (7.0%) | |
| blaCMY-2 | 26 (13.0%) | 0 (0.0%) | 0 (0.0%) | 9 (4.5%) | |
| Fosfomycin | fosA3 | 30 (15.0%) | 0 (0.0%) | 0 (0.0%) | 11 (5.5%) |
| Sulfonamides | sul1 | 57 (28.5%) | 9 (10.8%) | 2 (2.0%) | 19 (9.5%) |
| sul2 | 124 (62.0%) | 31 (37.3%) | 13 (13.0%) | 46 (23.0%) | |
| Amphenicols | cmlA | 36 (18.0%) | 6 (7.2%) | 0 (0.0%) | 21 (10.5%) |
| floR | 6 (3.0%) | 50 (60.2%) | 0 (0.0%) | 14 (7.0%) | |
| Quinolones | qnrD | 125 (62.5%) | 26 (31.3%) | 10 (10.0%) | 34 (17.0%) |
| Aminoglycosides/quinolones | aac(6′)-Ib-cr | 24 (12.0%) | 0 (0.0%) | 0 (0.0%) | 11 (5.5%) |
| Sources | Resistance Genes | |||||||
|---|---|---|---|---|---|---|---|---|
| ESBL/AmpC | fosA3 | sul1 | sul2 | cmlA | floR | qnrD | aac(6′)-Ib-cr | |
| Chicken meat | ESBL | 30 (64%) * | 26 (55%) * | 43 (91%) * | 31 (66%) * | (-) | 45 (96%) * | 17 (36%) * |
| Non-ESBL | (-) | 31 (20%) | 81 (53%) | 5 (3%) | 6 (4%) | 80 (52%) | 7 (5%) | |
| AmpC | 13 (50%) * | 14 (54%) * | 23 (88%) * | 14 (54%) * | (-) | 24 (92%) * | 7 (27%) * | |
| Non-AmpC | 17 (10%) | 43 (25%) | 101 (58%) | 22 (13%) | 6 (3%) | 101 (58%) | 17 (10%) | |
| Pork | ESBL | (-) | (-) | 5 (83%) * | 1 (17%) | 5 (83%) | 5 (83%) * | (-) |
| Non-ESBL | (-) | 9 (12%) | 26 (34%) | 5 (6%) | 45 (58 %) | 21 (27%) | (-) | |
| AmpC | (-) | (-) | (-) | (-) | (-) | (-) | (-) | |
| Non-AmpC | (-) | 9 (11%) | 31 (37%) | 6 (7%) | 50 (60%) * | 26 (31%) | (-) | |
| Beef | ESBL | 0 (0%) | (-) | 3 (100%) * | (-) | (-) | 3 (100%) * | (-) |
| Non-ESBL | 0 (0) | 2 (2%) | 10 (10%) | (-) | (-) | 7 (7%) | (-) | |
| AmpC | (-) | (-) | (-) | (-) | (-) | (-) | (-) | |
| Non-AmpC | (-) | 2 (2%) | 13 (13%) | (-) | (-) | 10 (10%) | (-) | |
| Human | ESBL | 11 (58%) * | 4 (21%) | 19 (100%) * | 9 (47%) * | (-) | 19 (100%) * | 6 (32%) * |
| Non-ESBL | (-) | 15 (8%) | 27 (15%) | 12 (7%) | 14 (8%) | 15 (8%) | 5 (3%) | |
| AmpC | 1 (10%) | 4 (40%) | 8 (80%) * | 4 (40%) | (-) | 7 (70%) * | 3 (30%) | |
| Non-AmpC | 10 (5%) | 15 (8%) | 38 (20%) | 17 (9%) | 14 (7%) | 27 (14%) | 8 (4%) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanches, M.S.; Silva, L.C.; Silva, C.R.d.; Montini, V.H.; Oliva, B.H.D.d.; Guidone, G.H.M.; Nogueira, M.C.L.; Menck-Costa, M.F.; Kobayashi, R.K.T.; Vespero, E.C.; et al. Prevalence of Antimicrobial Resistance and Clonal Relationship in ESBL/AmpC-Producing Proteus mirabilis Isolated from Meat Products and Community-Acquired Urinary Tract Infection (UTI-CA) in Southern Brazil. Antibiotics 2023, 12, 370. https://doi.org/10.3390/antibiotics12020370
Sanches MS, Silva LC, Silva CRd, Montini VH, Oliva BHDd, Guidone GHM, Nogueira MCL, Menck-Costa MF, Kobayashi RKT, Vespero EC, et al. Prevalence of Antimicrobial Resistance and Clonal Relationship in ESBL/AmpC-Producing Proteus mirabilis Isolated from Meat Products and Community-Acquired Urinary Tract Infection (UTI-CA) in Southern Brazil. Antibiotics. 2023; 12(2):370. https://doi.org/10.3390/antibiotics12020370
Chicago/Turabian StyleSanches, Matheus Silva, Luana Carvalho Silva, Caroline Rodrigues da Silva, Victor Hugo Montini, Bruno Henrique Dias de Oliva, Gustavo Henrique Migliorini Guidone, Mara Corrêa Lelles Nogueira, Maísa Fabiana Menck-Costa, Renata Katsuko Takayama Kobayashi, Eliana Carolina Vespero, and et al. 2023. "Prevalence of Antimicrobial Resistance and Clonal Relationship in ESBL/AmpC-Producing Proteus mirabilis Isolated from Meat Products and Community-Acquired Urinary Tract Infection (UTI-CA) in Southern Brazil" Antibiotics 12, no. 2: 370. https://doi.org/10.3390/antibiotics12020370
APA StyleSanches, M. S., Silva, L. C., Silva, C. R. d., Montini, V. H., Oliva, B. H. D. d., Guidone, G. H. M., Nogueira, M. C. L., Menck-Costa, M. F., Kobayashi, R. K. T., Vespero, E. C., & Rocha, S. P. D. (2023). Prevalence of Antimicrobial Resistance and Clonal Relationship in ESBL/AmpC-Producing Proteus mirabilis Isolated from Meat Products and Community-Acquired Urinary Tract Infection (UTI-CA) in Southern Brazil. Antibiotics, 12(2), 370. https://doi.org/10.3390/antibiotics12020370








