Genotypic Diversity, Antibiotic Resistance, and Virulence Phenotypes of Stenotrophomonas maltophilia Clinical Isolates from a Thai University Hospital Setting
Abstract
:1. Introduction
2. Results
2.1. Bacterial Collection and Clinical Characteristics
2.2. Antibiotic Susceptibility and Antibiotic Resistances
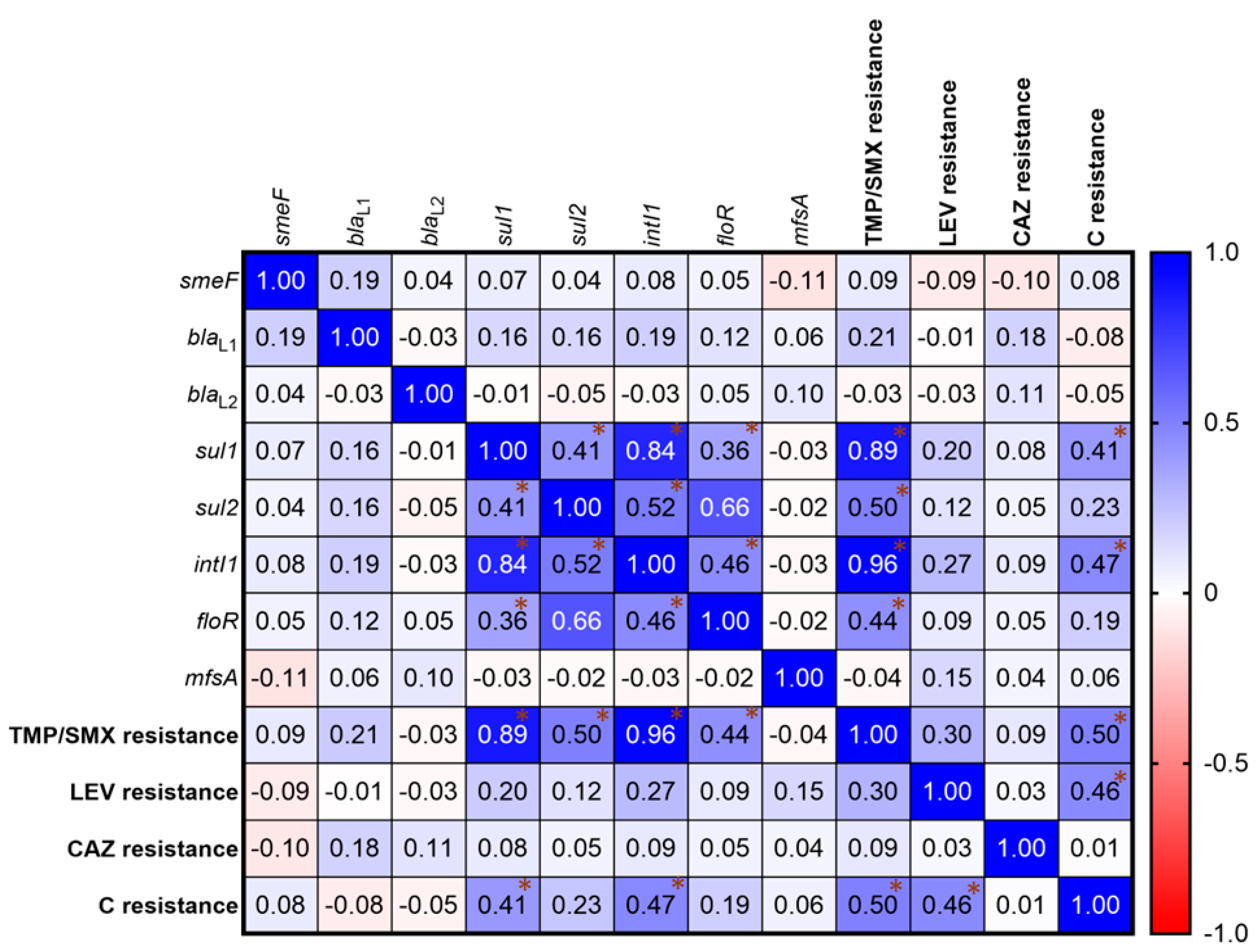
2.3. Detection of Antibiotic Resistance Genes
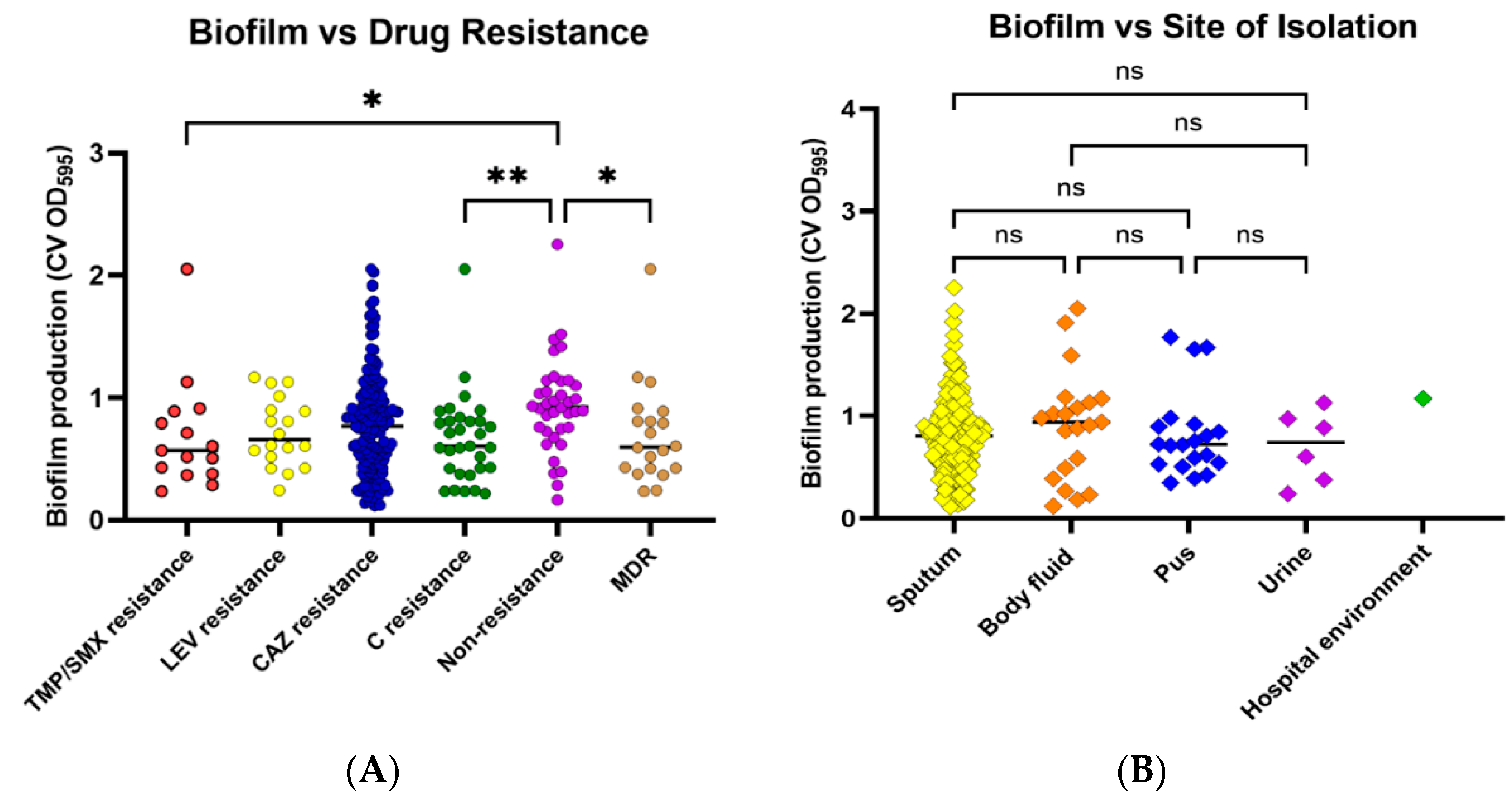
2.4. Biofilm Formation
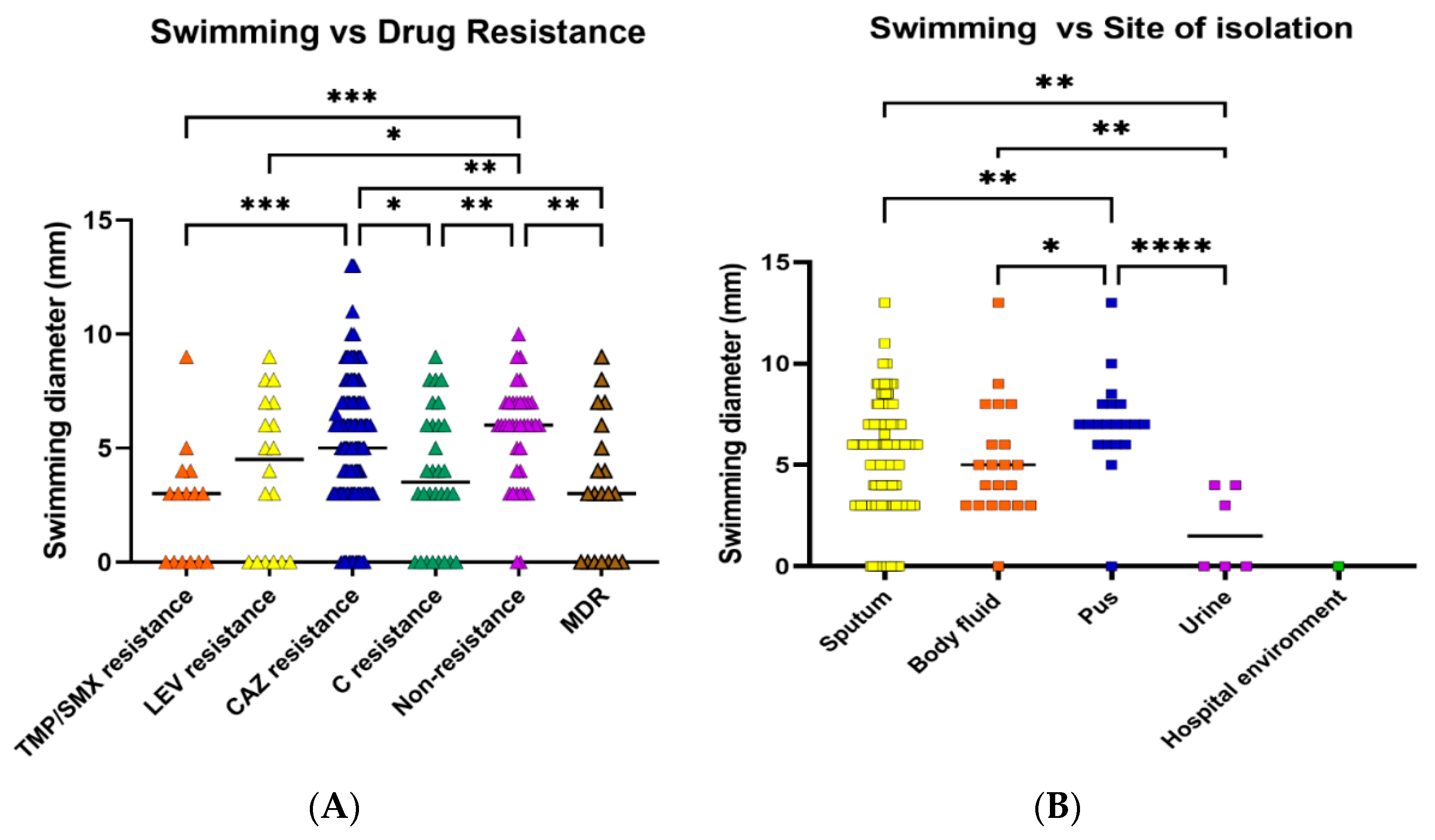
2.5. Motility
2.6. Toxin and Enzyme Production
2.7. Correlation of Antibiotic Resistance
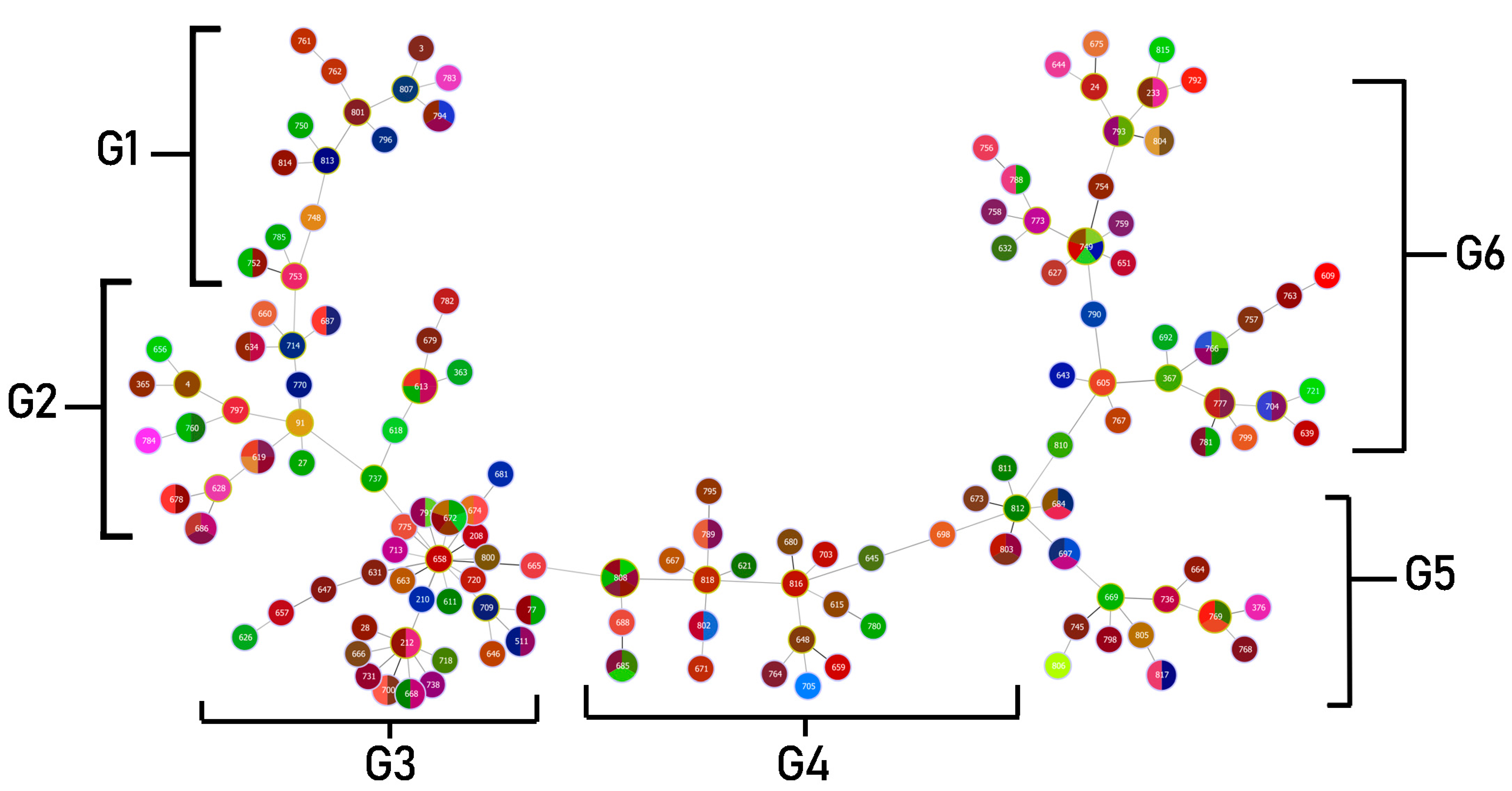
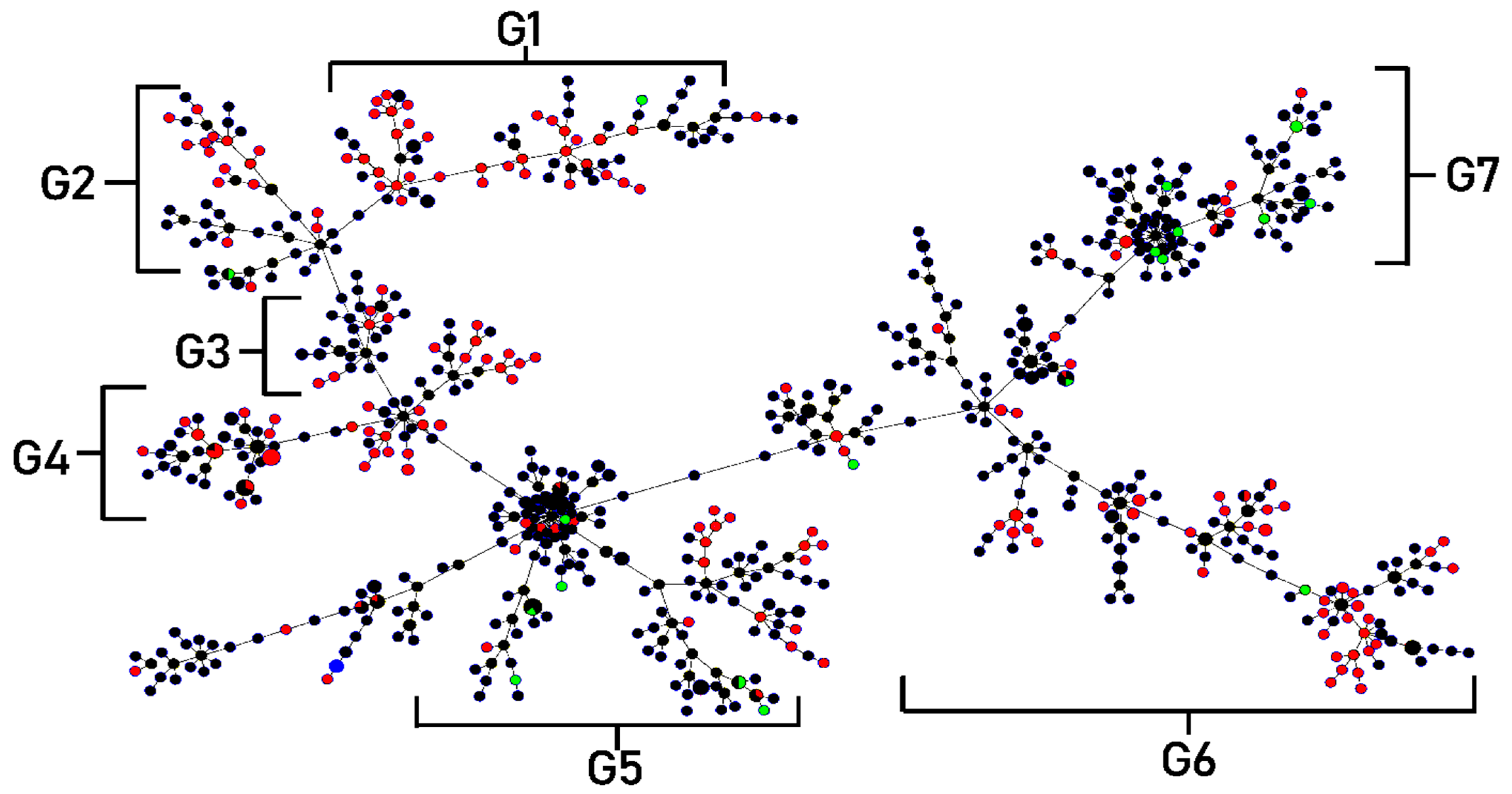
2.8. MLST Analysis and Clonal Complexes
3. Discussion
4. Materials and Methods
4.1. Bacterial Collection, Culture, and Clinical Information
4.2. Species Confirmation by 23S rRNA PCR
4.3. Antibiotic Susceptibility
4.4. Detection of Drug Resistance Genes
4.5. Biofilm Formation Assay
4.6. Motility Test
4.7. Screening of Toxin and Enzymes Production
4.8. Multi-Locus Sequence Typing (MLST) Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adegoke, A.A.; Stenström, T.A.; Okoh, A.I. Stenotrophomonas maltophilia as an emerging ubiquitous pathogen: Looking beyond contemporary antibiotic therapy. Front. Microbiol. 2017, 8, 2276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singhal, L.; Kaur, P.; Gautam, V. Stenotrophomonas maltophilia: From trivial to grievous. Indian J. Med. Microbiol. 2017, 35, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Looney, W.J.; Narita, M.; Mühlemann, K. Stenotrophomonas maltophilia: An emerging opportunist human pathogen. Lancet Infect. Dis. 2009, 9, 312–323. [Google Scholar] [CrossRef]
- Caylan, R.; Kaklikkaya, N.; Aydin, K.; Aydin, F.; Yilmaz, G.; Ozgumus, B.; Koksal, I. An epidemiological analysis of Stenotrophomonas maltophilia strains in a university hospital. Jpn. J. Infect. Dis. 2004, 57, 37–40. [Google Scholar] [PubMed]
- Chang, Y.T.; Lin, C.Y.; Chen, Y.H.; Hsueh, P.R. Update on infections caused by Stenotrophomonas maltophilia with particular attention to resistance mechanisms and therapeutic options. Front. Microbiol. 2015, 6, 893. [Google Scholar] [CrossRef]
- Gajdács, M.; Urbán, E. Prevalence and Antibiotic Resistance of Stenotrophomonas maltophilia in Respiratory Tract Samples: A 10-Year Epidemiological Snapshot. Health Serv. Res. Manag. Epidemiol. 2019, 6, 2333392819870774. [Google Scholar] [CrossRef] [Green Version]
- Mojica, M.F.; Humphries, R.; Lipuma, J.J.; Mathers, A.J.; Rao, G.G.; Shelburne, S.A.; Fouts, D.E.; Van Duin, D.; Bonomo, R.A. Clinical challenges treating Stenotrophomonas maltophilia infections: An update. JAC Antimicrob. Resist. 2022, 4, dlac040. [Google Scholar] [CrossRef]
- Looney, W.J. Role of Stenotrophomonas maltophilia in hospital-acquired infection. Br. J. Biomed. Sci. 2005, 62, 145–154. [Google Scholar] [CrossRef]
- Guyot, A.; Turton, J.F.; Garner, D. Outbreak of Stenotrophomonas maltophilia on an intensive care unit. J. Hosp. Infect. 2013, 85, 303–307. [Google Scholar] [CrossRef]
- Cervia, S.J.; Ortolano, A.G.; Canonica, P.F. Hospital tap water as a source of Stenotrophomonas maltophilia infection. Clin. Infect. Dis. 2008, 46, 1485–1487. [Google Scholar] [CrossRef]
- Trifonova, A.; Strateva, T. Stenotrophomonas maltophilia—A low-grade pathogen with numerous virulence factors. Infect. Dis. 2019, 51, 168–178. [Google Scholar] [CrossRef]
- de Oliveira-Garcia, D.; Dall’Agnol, M.; Rosales, M.; Azzuz, A.C.; Alcántara, N.; Martinez, M.B.; Girón, J.A. Fimbriae and adherence of Stenotrophomonas maltophilia to epithelial cells and to abiotic surfaces. Cell Microbiol. 2003, 5, 625–636. [Google Scholar] [CrossRef]
- Brooke, J.S. Advances in the Microbiology of Stenotrophomonas maltophilia. Clin. Microbiol. Rev. 2021, 34, e0003019. [Google Scholar] [CrossRef] [PubMed]
- Igbinosa, E.O.; Oviasogie, F.E. Multiple antibiotics resistant among environmental isolates of Stenotrophomonas maltophilia. J. Appl. Sci. Environ. Manag. 2014, 18, 255–261. [Google Scholar] [CrossRef] [Green Version]
- Kwa, A.L.; Low, J.G.; Lim, T.P.; Leow, P.C.; Kurup, A.; Tam, V.H. Independent predictors for mortality in patients with positive Stenotrophomonas maltophilia cultures. Ann. Acad. Med. Singap. 2008, 37, 826–830. [Google Scholar] [CrossRef]
- Gibb, J.; Wong, D.W. Antimicrobial Treatment Strategies for Stenotrophomonas maltophilia: A Focus on Novel Therapies. Antibiotics 2021, 10, 1226. [Google Scholar] [CrossRef] [PubMed]
- Gherardi, G.; Creti, R.; Pompilio, A.; Di Bonaventura, G. An overview of various typing methods for clinical epidemiology of the emerging pathogen Stenothophomonas maltophilia. Diagn. Microbiol. Infect. Dis. 2015, 81, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Maiden, M.C.; Bygraves, J.A.; Feil, E.; Morelli, G.; Russell, J.E.; Urwin, R.; Zhang, Q.; Zhou, J.; Zurth, K.; Caugant, D.A.; et al. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 1998, 95, 3140–3145. [Google Scholar] [CrossRef] [Green Version]
- Kaiser, S.; Biehler, K.; Jonas, D.A. A Stenotrophomonas maltophilia multilocus sequence typing scheme for inferring population structure. J. Bacteriol. 2009, 191, 2934–2943. [Google Scholar] [CrossRef] [Green Version]
- Zając, O.M.; Tyski, S.; Laudy, A.E. Phenotypic and Molecular Characteristics of the MDR Efflux Pump Gene-Carrying Stenotrophomonas maltophilia Strains Isolated in Warsaw, Poland. Biology 2022, 11, 105. [Google Scholar] [CrossRef]
- Crossman, L.C.; Gould, V.C.; Dow, J.M.; Vernikos, G.S.; Okazaki, A.; Sebaihia, M.; Saunders, D.; Arrowsmith, C.; Carver, T.; Peters, N.; et al. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 2008, 9, R74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, R.; Monchy, S.; Cardinale, M.; Taghavi, S.; Crossman, L.; Avison, B.M.; Berg, G.; van der Lelie, D.; Dow, J.M. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 2009, 7, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Gil-Gil, T.; Martínez, J.L.; Blanco, P. Mechanisms of antimicrobial resistance in Stenotrophomonas maltophilia: A review of current knowledge. Expert Rev. Anti. Infect. Ther. 2020, 18, 335–347. [Google Scholar] [CrossRef]
- Diagnostic Laboratory, Maharaj Nakorn Chiang Mai Hospital. Antibiotic Susceptibility Testing Summary Report of Important Organisms from Wards. 2016. Available online: https://www.med.cmu.ac.th/hospital/lab/2011/index.php?option=com_content&view=article&layout=edit&id=87 (accessed on 3 June 2017).
- Paopradit, P.; Srinitiwarawong, K.; Ingviya, N.; Singkhamanan, K.; Vuddhakul, V. Distribution and characterization of Stenotrophomonas maltophilia isolates from environmental and clinical samples in Thailand. J. Hosp. Infect. 2017, 97, 185–191. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 29th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]
- Neela, V.; Rankouhi, S.Z.; van Belkum, A.; Goering, R.V.; Awang, R. Stenotrophomonas maltophilia in Malaysia: Molecular epidemiology and trimethoprim-sulfamethoxazole resistance. Int. J. Infect. Dis. 2012, 16, 603–607. [Google Scholar] [CrossRef] [Green Version]
- Jia, W.; Wang, J.; Xu, H.; Li, G. Resistance of Stenotrophomonas maltophilia to fluoroquinolones: Prevalence in a university hospital and possible mechanisms. Int. J. Environ. Res. Public Health 2015, 12, 5177–5195. [Google Scholar] [CrossRef] [Green Version]
- Gallo, S.W.; Figueiredo, T.P.; Bessa, M.C.; Pagnussatti, V.E.; Ferreira, C.A.; Oliveira, S.D. Isolation and characterization of Stenotrophomonas maltophilia isolates from a brazilian hospital. Microb. Drug Resist. 2016, 22, 688–695. [Google Scholar] [CrossRef]
- Waters, V.; Atenafu, E.G.; Lu, A.; Yau, Y.; Tullis, E.; Ratjen, F. Chronic Stenotrophomonas maltophilia infection and mortality or lung transplantation in cystic fibrosis patients. J. Cyst. Fibros. 2013, 12, 482–486. [Google Scholar] [CrossRef] [Green Version]
- Herrera-Heredia, S.A.; Pezina-Cantú, C.; Garza-González, E.; Bocanegra-Ibarias, P.; Mendoza-Olazarán, S.; Morfín-Otero, R.; Camacho-Ortiz, A.; Villarreal-Treviño, L.; Rodríguez-Noriega, E.; Paláu-Davila, L.; et al. Risk factors and molecular mechanisms associated with trimethoprim-sulfamethoxazole resistance in Stenotrophomonas maltophilia in Mexico. J. Med. Microbiol. 2017, 66, 1102–1109. [Google Scholar] [CrossRef]
- Gülmez, D.; Hasçelik, G. Stenotrophomonas maltophilia: Antimicrobial resistance and molecular typing of an emerging pathogen in a Turkish university hospital. Clin. Microbiol. Infect. 2005, 11, 880–886. [Google Scholar] [CrossRef] [Green Version]
- Duan, Z.; Qin, J.; Liu, Y.; Li, C.; Ying, C. Molecular epidemiology and risk factors of Stenotrophomonas maltophilia infections in a Chinese teaching hospital. BMC Microbiol. 2020, 20, 294. [Google Scholar] [CrossRef]
- Samonis, G.; Karageorgopoulos, D.E.; Maraki, S.; Levis, P.; Dimopoulou, D.; Spernovasilis, N.A.; Kofteridis, D.P.; Falagas, M.E. Stenotrophomonas maltophilia infections in a general hospital: Patient characteristics, antimicrobial susceptibility, and treatment outcome. PLoS ONE 2012, 7, e37375. [Google Scholar] [CrossRef]
- Farrell, D.J.; Sader, H.S.; Jones, R.N. Antimicrobial susceptibilities of a worldwide collection of Stenotrophomonas maltophilia isolates tested against tigecycline and agents commonly used for S. maltophilia infections. Antimicrob. Agents Chemother. 2010, 54, 2735–2737. [Google Scholar] [CrossRef] [Green Version]
- Rhee, J.Y.; Choi, J.Y.; Choi, M.J.; Song, J.H.; Peck, K.R.; Ko, K.S. Distinct groups and antimicrobial resistance of clinical Stenotrophomonas maltophilia complex isolates from Korea. J. Med. Microbiol. 2013, 62, 748–753. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Wang, J.T.; Shiau, Y.R.; Wang, H.Y.; Lauderdale, T.L.; Chang, S.C.; TSAR Hospitals. A multicenter surveillance of antimicrobial resistance on Stenotrophomonas maltophilia in Taiwan. J. Microbiol. Immunol. Infect. 2012, 45, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.F.; Gao, L.P.; Ye, Y.; Chen, X.; Zhou, X.T.; Yang, H.F.; Liiu, Y.Y.; Mei, Q.; Li, J.B. Susceptibility of Stenotrophomonas maltophilia clinical strains in China to antimicrobial combinations. J. Chemother. 2014, 26, 282–286. [Google Scholar] [CrossRef]
- Çalışkan, A.; Çopur Çicek, A.; Aydogan Ejder, N.; Karagöz, A.; Kirişc, i.Ö.; Kılıç, S. The Antibiotic sensitivity of Stenotrophomonas maltophilia in a 5-year period and investigation of clonal outbreak with PFGE. J. Infect. Dev. Ctries. 2019, 13, 634–639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Insuwanno, W.; Kiratisin, P.; Jitmuang, A. Stenotrophomonas maltophilia infections: Clinical characteristics and factors associated with mortality of hospitalized patients. Infect. Drug Resist. 2020, 13, 1559–1566. [Google Scholar] [CrossRef]
- Hamdi, A.M.; Fida, M.; Abu Saleh, O.M.; Beam, E. Stenotrophomonas bacteremia antibiotic susceptibility and prognostic determinants: Mayo Clinic 10-year experience. Open Forum Infect. Dis. 2020, 7, ofaa008. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Yang, L.; Liu, H.; Gao, F. Stenotrophomonas maltophilia in a university hospital of traditional Chinese medicine: Molecular epidemiology and antimicrobial resistance. J. Hosp. Infect. 2017, 96, 286–289. [Google Scholar] [CrossRef]
- Avison, M.B.; Higgins, C.S.; von Heldreich, C.J.; Bennett, P.M.; Walsh, T.R. Plasmid location and molecular heterogeneity of the L1 and L2 β-lactamase genes of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2001, 45, 413–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bostanghadiri, N.; Ghalavand, Z.; Fallah, F.; Yadegar, A.; Ardebili, A.; Tarashi, S.; Pournajaf, A.; Mardaneh, J.; Shams, S.; Hashemi, A. Characterization of phenotypic and genotypic diversity of Stenotrophomonas maltophilia strains isolated from selected hospitals in Iran. Front. Microbiol. 2019, 29, 1191. [Google Scholar] [CrossRef] [PubMed]
- Kaur, P.; Gautam, V.; Tewari, R. Distribution of class 1 integrons, sul1 and sul2 genes among clinical isolates of Stenotrophomonas maltophilia from a tertiary care hospital in North India. Microb. Drug Resist. 2015, 21, 380–385. [Google Scholar] [CrossRef] [PubMed]
- Le, T.H.; Ng, C.; Chen, H.; Yi, X.Z.; Koh, T.H.; Barkham, T.M.; Zhou, Z.; Gin, K.Y. Occurrences and Characterization of Antibiotic-Resistant Bacteria and Genetic Determinants of Hospital Wastewater in a Tropical Country. Antimicrob. Agents Chemother. 2016, 60, 7449–7456. [Google Scholar] [CrossRef] [Green Version]
- Adelowo, O.O.; Osuntade, A.I. Class 1 Integron, Sulfonamide and Florfenicol Resistance Genes in Bacteria from Three Unsanitary Landfills, Ibadan, Nigeria. J. Microbiol. Infect. Dis. 2019, 9, 34–42. [Google Scholar] [CrossRef] [Green Version]
- Pompilio, A.; Ranalli, M.; Piccirilli, A.; Perilli, M.; Vukovic, D.; Savic, B.; Krutova, M.; Drevinek, P.; Jonas, D.; Fiscarelli, E.V.; et al. Biofilm formation among Stenotrophomonas maltophilia isolates has clinical relevance: The ANSELM prospective multicenter study. Microorganisms 2020, 9, 49. [Google Scholar] [CrossRef] [PubMed]
- Pompilio, A.; Crocetta, V.; Ghosh, D.; Chakrabarti, M.; Gherardi, G.; Vitali, L.A.; Fiscarelli, E.; Di Bonaventura, G. Stenotrophomonas maltophilia phenotypic and genotypic diversity during a 10-year colonization in the lungs of a cystic fibrosis patient. Front. Microbiol. 2016, 7, 1551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanimoto, K. Stenotrophomonas maltophilia strains isolated from a university hospital in Japan: Genomic variability and antibiotic resistance. J. Med. Microbiol. 2013, 62, 565–570. [Google Scholar] [CrossRef] [Green Version]
- Zöttl, A.; Yeomans, J.M. Enhanced bacterial swimming speeds in macromolecular polymer solutions. Nat. Phys. 2019, 15, 554–558. [Google Scholar] [CrossRef] [Green Version]
- Luterbach, C.L.; Mobley, H. Cross talk between MarR-like transcription factors coordinates the regulation of motility in uropathogenic Escherichia coli. Infect. Immun. 2018, 86, e00338-18. [Google Scholar] [CrossRef] [Green Version]
- Bender, J.; Flieger, A. Lipases as pathogenicity factors of bacterial pathogens of humans. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 3241–3258. [Google Scholar]
- Cho, H.H.; Sung, J.Y.; Kwon, K.C.; Koo, S.H. Expression of Sme efflux pumps and multilocus sequence typing in clinical isolates of Stenotrophomonas maltophilia. Ann. Lab. Med. 2012, 32, 38–43. [Google Scholar] [CrossRef]
- Emami, S.; Nowroozi, J.; Abiri, R.; Mohajeri, P. Multilocus Sequence Typing for Molecular Epidemiology of Stenotrophomonas maltophilia Clinical and Environmental Isolates from a Tertiary Hospital in West of Iran. Iran Biomed. J. 2022, 26, 142–152. [Google Scholar]
- Gideskog, M.; Welander, J.; Melhus, Å. Cluster of S. maltophilia among patients with respiratory tract infections at an intensive care unit. Infect. Prev. Pract. 2020, 2, 100097. [Google Scholar] [CrossRef] [PubMed]
- Rizek, C.F.; Jonas, D.; Garcia Paez, J.I.; Rosa, J.F.; Perdigão Neto, L.V.; Martins, R.R.; Moreno, L.Z.; Rossi Junior, A.; Levin, A.S.; Costa, S.F. Multidrug-resistant Stenotrophomonas maltophilia: Description of new MLST profiles and resistance and virulence genes using whole-genome sequencing. J. Glob. Antimicrob. Resist. 2018, 15, 212–214. [Google Scholar] [CrossRef]
- Gallo, S.W.; Ramos, P.L.; Ferreira, C.A.S.; de Oliveira, S.D. A specific polymerase chain reaction method to identify Stenotrophomonas maltophilia. Mem. Inst. Oswaldo Cruz. 2013, 108, 390–391. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria:An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Wei, H.; Zhao, Y.; Shang, L.; Di, L.; Lyu, C.; Liu, J. Analysis of multidrug-resistant bacteria in 3223 patients with hospital-acquired infections (HAI) from a tertiary general hospital in China. Bosn. J. Basic Med. Sci. 2019, 19, 86–93. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Liu, W.; Cui, Q.; Niu, W.; Li, H.; Zhao, X.; Wei, X.; Wang, X.; Huang, S.; Dong, D.; et al. Prevalence and detection of Stenotrophomonas maltophilia carrying metallo-β-lactamase blaL1 in Beijing, China. Front. Microbiol. 2014, 9, 692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azimi, A.; Aslanimehr, M.; Yaseri, M.; Shadkam, M.; Douraghi, M. Distribution of smf-1, rmlA, spgM and rpfF genes among Stenotrophomonas maltophilia isolates in relation to biofilm-forming capacity. J. Glob. Antimicrob. Resist. 2020, 23, 321–326. [Google Scholar] [CrossRef]
- Toleman, M.A.; Bennett, P.M.; Walsh, T.R. Common regions e.g. orf513 and antibiotic resistance: IS91-like elements evolving complex class 1 integrons. J. Antimicrob. Chemother. 2006, 58, 1–6. [Google Scholar] [CrossRef]
- Toleman, M.A.; Bennett, P.M.; Bennett, D.M.; Jones, R.N.; Walsh, T.R. Global emergence of trimethoprim/sulfamethoxazole resistance in Stenotrophomonas maltophilia mediated by acquisition of sul genes. Emerg. Infect. Dis. 2007, 13, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Vattanaviboon, P.; Dulyayangkul, P.; Mongkolsuk, S.; Charoenlap, N. Overexpression of Stenotrophomonas maltophilia major facilitator superfamily protein MfsA increases resistance to fluoroquinolone antibiotics. J. Antimicrob. Chemother. 2018, 73, 1263–1266. [Google Scholar] [CrossRef] [PubMed]
- Madi, H.; Lukić, J.; Vasiljević, Z.; Biočanin, M.; Kojić, M.; Jovčić, B.; Lozo, J. Genotypic and phenotypic characterization of Stenotrophomonas maltophilia strains from a Pediatric Tertiary Care Hospital in Serbia. PLoS ONE 2016, 11, e0165660. [Google Scholar] [CrossRef]
- Cruz-Córdova, A.; Mancilla-Rojano, J.; Luna-Pineda, V.M.; Escalona-Venegas, G.; Cázares-Domínguez, V.; Ormsby, C.; Franco-Hernández, I.; Zavala-Vega, S.; Hernández, M.A.; Medina-Pelcastre, M. Molecular epidemiology, antibiotic resistance, and virulence traits of Stenotrophomonas maltophilia strains associated with an outbreak in a Mexican tertiary care hospital. Front. Cell Infect. Microbiol. 2020, 18, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, R.; Hamat, R.A.; Neela, V. Extracellular enzyme profiling of Stenotrophomonas maltophilia clinical isolates. Virulence 2014, 5, 326–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Characteristics | No. of Patients (%) (n = 192) | No. of Respiratory Infection (n = 148) | No. of Non-Respiratory Inection (n = 44) | p Value |
|---|---|---|---|---|
| Age | ||||
| Range | 1 M–99 Y | 1 M–99 Y | 6 M–89 Y | |
| Pediatrics (<15) | 32 (16.67) | 30 | 2 | 0.308 |
| Aged (≥65) | 79 (41.15) | 62 | 17 | 0.031 |
| Mean ± SD | 54.61Y ± 28.14 | 52.94 ± 29.78 | 55.82 ± 22.14 | |
| Gender: Male | 117 (60.94) | 91 | 26 | 0.398 |
| Underlying Diseases and Comorbidities | ||||
| Malignancy | 56 (29.17) | 40 | 16 | 0.041 |
| CNS and cerebrovascular diseases | 36 (18.75) | 31 | 5 | 0.040 |
| Urinary tract infection | 34 (17.71) | 28 | 6 | 0.044 |
| Hypertension | 33 (17.19) | 22 | 11 | 0.129 |
| Cardiovascular diseases | 28 (14.58) | 24 | 4 | 0.043 |
| Diabetes | 25 (13.02) | 20 | 5 | 0.281 |
| Chronic kidney disease | 20 (10.42) | 20 | 3 | 0.391 |
| Chronic pulmonary diseases | 18 (9.38) | 12 | 6 | 0.401 |
| Chronic viral infections | 11 (5.73) | 8 | 3 | 0.677 |
| Predisposing factors | ||||
| Invasive procedures | 190 (98.96) | 147 | 43 | 0.183 |
| Intravenous catheter | 116 (60.42) | 79 | 37 | 0.040 |
| Urinary catheter | 46 (23.96) | 41 | 5 | 0.129 |
| Suction catheter | 98 (51.04) | 80 | 18 | 0.258 |
| Endotracheal intubation | 57 (29.69) | 39 | 18 | 0.148 |
| Gastrostomy (feeding) intubation | 38 (19.79) | 32 | 6 | 0.529 |
| Surgery | 56 (29.17) | 40 | 16 | 0.046 |
| Chemotherapy, Radiotherapy | 19 (9.90) | 12 | 7 | 0.031 |
| Amputation | 5 (2.60) | 2 | 3 | |
| Organ transplant | 3 (1.56) | 2 | 1 | |
| Dialysis | 5 (2.60) | 5 | 0 | |
| Hospitalization | 190 (98.96) | 147 | 43 | |
| Medical wards | ||||
| General medicine | 53 (27.60) | 44 | 9 | |
| Pediatric | 33 (17.19) | 31 | 2 | |
| General surgery | 24 (12.5) | 14 | 10 | |
| Emergency surgery | 17 (8.85) | 11 | 6 | |
| Orthopedics | 9 (4.69) | 7 | 2 | |
| Neurosurgery | 3 (1.56) | 3 | 0 | |
| Others | 51 (26.56) | 37 | 14 | |
| Patient in ICU of each ward | 78 (40.63) | 71 | 7 | 0.001 |
| Antibiotic | MIC (µg/mL) | Susceptibility (%) | ||||
|---|---|---|---|---|---|---|
| MIC Range | MIC50 | MIC90 | S | I | R | |
| Trimethoprim/ Sulfamethoxazole * (TMP/SMX) | 0.047/0.893 → 32/608 | 0.19/3.61 | 0.5/9.5 | 185 (92.5) | 0 | 15 (7.5) |
| Levofloxacin ** (LEV) | 0.5 → 32 | 2 | 4 | 163 (81.5) | 19 (9.5) | 18 (9.0) |
| Ceftazidime ** (CAZ) | 2 → 128 | 128 | >128 | 21 (10.5) | 24 (12.0) | 150 (77.5) |
| Chloramphenicol ** (C) | 4 → 128 | 16 | 32 | 67 (33.5) | 97 (48.5) | 36 (18) |
| Minocycline ** (MH) | 0.5 → 4 | 0.5 | 2 | 200 (100) | 0 | 0 |
| Antibiotic Resistance | Antibiotic Resistance Genes | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Total of Isolates * | smeF | blaL1 | blaL2 | sul1 | sul2 | intI1 | floR | mfsA | |
| No. of Isolate (%) | |||||||||
| All isolates | 200 (100) | 183 (91.5) | 86 (43) | 20 (10) | 12 (6) | 4 (2) | 14 (7) | 8 (4) | 9 (4.5) |
| TMP/SMX resistance | 15 (7.5) | 15 (100) | 12 (80.0) | 1 (6.67) | 12 (80.0) | 4 (26.67) | 14 (93.33) | 5 (33.33) | 1 (6.67) |
| LEV resistance | 18 (9) | 16 (88.89) | 7 (38.89) | 1 (5.56) | 4 (22.22) | 2 (11.11) | 5 (27.78) | 2 (11.11) | 2 (11.11) |
| CAZ resistance | 157 (78.5) | 123 (78.34) | 80 (50.96) | 19 (12.1) | 12 (7.64) | 4 (5.09) | 13 (8.28) | 12 (7.64) | 7 (4.46) |
| C resistance | 31 (15.5) | 30 (96.77) | 11 (35.48) | 2 (6.45) | 10 (32.26) | 3 (9.68) | 11 (35.48) | 4 (12.90) | 2 (6.45) |
| Non-resistance | 37 (18.5) | 37 (100) | 6 (16.22) | 1 (2.70) | 0 | 0 | 0 | 5 (13.51) | 0 |
| MDR | 20 (10) | 20 (100) | 11 (55) | 1 (5) | 10 (50) | 3 (15) | 12 (60) | 4 (20) | 2 (10) |
| Antibiotic Resistance | Biofilm Formation | Swimming Motility | Toxin and Enzymes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Non | Weak | Moderate | Strong | Non | Weak | Moderate | Strong | α-Hemolys in | β-Hemolys in | Protease | Lipase | |
| Number of Isolate (%) | ||||||||||||
| All isolates | 1 (0.5) | 22 (11) | 31 (15.5) | 146 (73) | 21 (10.5) | 73 (36.5) | 81 (40.5) | 25 (12.5) | 106 (53) | 94(47) | 183 (91.5) | 45 (22.5) |
| TMP/SMX resistance | 0 | 2 (13.33) | 6 (40) | 7 (46.67) | 6 (40) | 8 (53.33) | 0 | 1 (6.67) | 11 (73.33) | 4 (26.67) | 11 (73.33) | 2 (13.33) |
| LEV resistance | 0 | 1 (5.56) | 4 (22.22) | 13 (72.22) | 6 (33.33) | 5 (27.78) | 6 (33.33) | 1 (5.55) | 9 (50) | 9 (50) | 14 (77.78) | 3 (16.67) |
| CAZ resistance | 1 (0.64) | 20 (12.74) | 28 (17.83) | 108 (68.79) | 18 (11.46) | 63 (40.13) | 55 (35.03) | 21 (13.38) | 89 (56.69) | 68 (43.31) | 144 (91.72) | 35 (22.29) |
| C resistance | 0 | 5 (16.13) | 7 (22.58) | 19 (61.29) | 8 (25.8) | 12 (38.71) | 9 (29.03) | 2 (6.45) | 15 (48.39) | 16 (51.61) | 25 (80.65) | 8 (5.10) |
| Non-resistance | 0 | 2 (5.4) | 3 (8.10) | 32 (86.49) | 2 (5.4) | 10 (27.02) | 22 (59.46) | 3 (8.11) | 21 (56.76) | 16 (43.24) | 34 (91.89) | 5 (13.51) |
| MDR | 0 | 2 (10) | 7 (35) | 11 (55) | 7 (35) | 8 (40) | 4 (20) | 1 (5) | 11 (55) | 9 (45) | 19 (95) | 4 (20) |
| Clonal * Complex | Sequence Types ** | Specimens (Sources of Isolates) | Genotypes and Phenotypes Exhibited by Most of the Isolates | ||||
|---|---|---|---|---|---|---|---|
| Drug a Resistance | Drug Resistance b Gene | Biofilm Formation | Swimming Motility | Toxin and c Enzymes Production | |||
| Group 1 | 3, 761, 762, 748, 750, 752, 753, 785, 796, 801, 807, 813, 814 | Sputum, Fluid | 1–2 drugs | 1–2 genes | Strong producer | Weak swimming | 2 types |
| Group 2 | 4, 27, 91, 363, 365, 613, 618, 619, 628, 634, 656, 660, 678, 686, 687, 697, 714, 737, 760, 770, 784 | Sputum, Fluid, Pus, Urine | 2–4 drugs | 2–4 genes | Moderate and strong producer | Weak to moderate swimming | 2 types |
| Group 3 | 28, 208, 212, 624, 626, 631, 647, 657, 658, 663, 665, 666, 668, 681, 700, 718, 720, 731, 738, 775, 791 | Sputum, Fluid, Pus, Urine | 1–3 drugs | 0–5 genes (most isolates contained 2 genes) | Strong producer | Weak swimming | 2 types |
| Group 4 | 621, 645, 648, 659, 667, 671, 673, 680, 685, 688, 697, 698, 705, 764, 789, 795, 802, 803, 808, 811, 812, 816, 818 | Sputum, Fluid, Pus, Urine | 0–1 drug | 1 gene | Strong producer (All isolates) | Moderate swimming | 2 types |
| Group 5 | 367, 605, 609, 627, 632, 643, 651, 692, 749, 754, 756, 757, 758, 759, 763, 766, 767, 773, 777, 781, 788, 790, 799, 810 | Sputum, Fluid, Pus | 1 drug | 2 genes | Strong producer | Moderate to strong swimming | 3 types |
| Group 6 | 376, 664, 669, 736, 745, 768, 769, 798, 805, 806, 817 | Sputum, Fluid, Pus | 1 drug | 1 gene | Strong producer (All isolates) | Weak, moderate and strong (found in similar rate) | 2–3 types |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yinsai, O.; Deeudom, M.; Duangsonk, K. Genotypic Diversity, Antibiotic Resistance, and Virulence Phenotypes of Stenotrophomonas maltophilia Clinical Isolates from a Thai University Hospital Setting. Antibiotics 2023, 12, 410. https://doi.org/10.3390/antibiotics12020410
Yinsai O, Deeudom M, Duangsonk K. Genotypic Diversity, Antibiotic Resistance, and Virulence Phenotypes of Stenotrophomonas maltophilia Clinical Isolates from a Thai University Hospital Setting. Antibiotics. 2023; 12(2):410. https://doi.org/10.3390/antibiotics12020410
Chicago/Turabian StyleYinsai, Orathai, Manu Deeudom, and Kwanjit Duangsonk. 2023. "Genotypic Diversity, Antibiotic Resistance, and Virulence Phenotypes of Stenotrophomonas maltophilia Clinical Isolates from a Thai University Hospital Setting" Antibiotics 12, no. 2: 410. https://doi.org/10.3390/antibiotics12020410






