Epigenetic and Drug Response Modulation of Epigalocaten-In-3-Gallate in Staphylococcus aureus with Divergent Resistance Phenotypes
Abstract
:1. Introduction
2. Results
2.1. Exposure to EGCG Induced a Synergistic Effect with Amoxicillin in MSSA Resistant Phenotypes
2.2. EGCG Reverted the Resistant Phenotype to Gentamicin in MSSA and MRSA Strains
2.3. No Alterations in Imipenem Resistance Were Observed Associated with EGCG Exposure
2.4. EGCG Affects Zone of Inhibition Values after Co-Exposure with Imipenem, Tetracycline, Gentamycin and Amoxicillin
2.5. Exposure to EGCG Alters S. aureus Molecular Profile
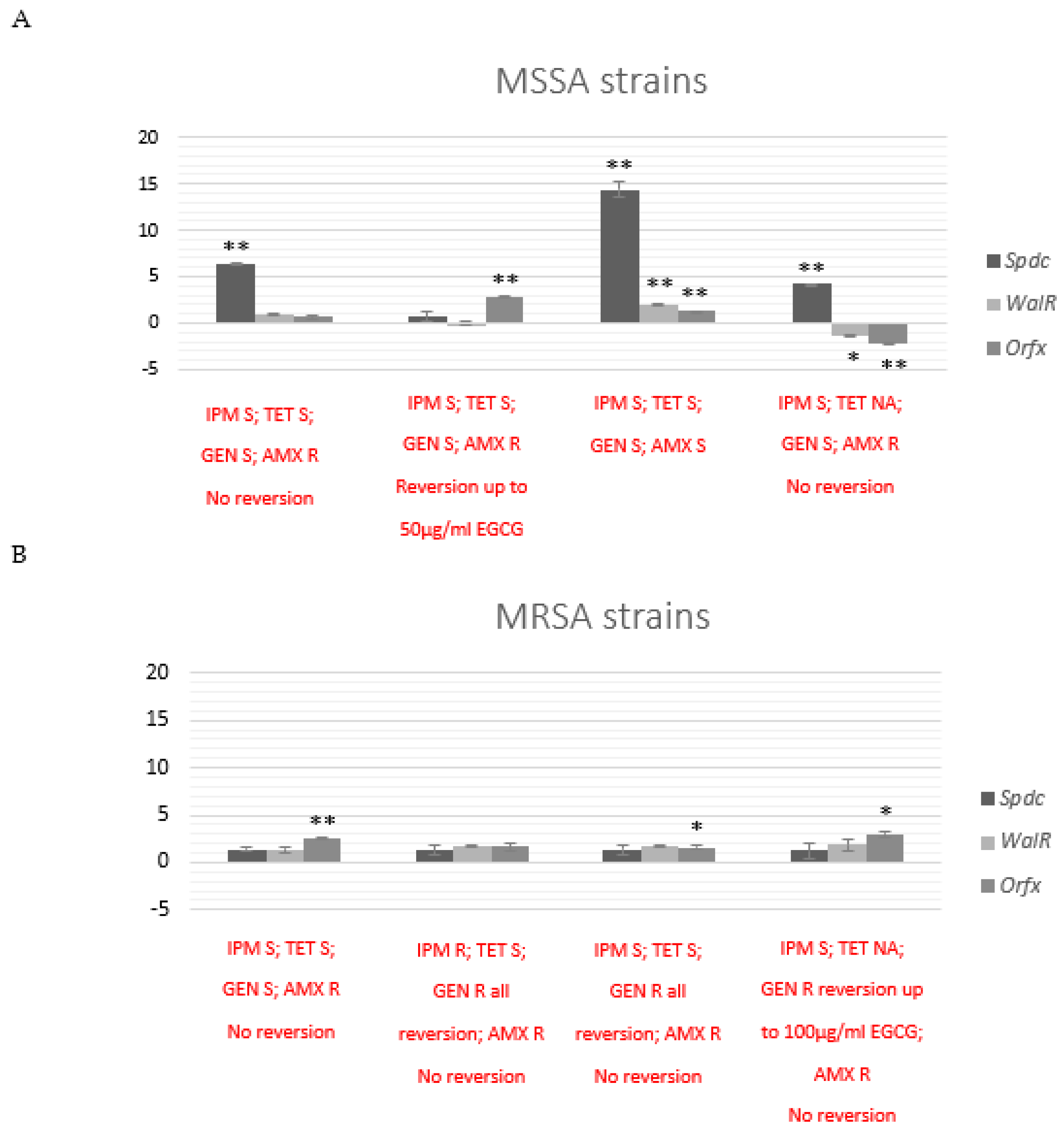
2.6. Exposure to EGCG Affects Transcriptional Patterns of Epigenetic and Drug Resistance Modulators Genes
2.6.1. Transcriptional Effects after Co-Exposure with EGCG in Selected MSSA and MRSA Strains
2.6.2. Transcriptional Effects after 24 h Subculture of MSSA and MRSA Strains with Previous Co-Exposure with EGCG
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains Identification and Antibiotic-Resistant Profile Assessment
4.2. EGCG Synergism Evaluation
4.3. FTIR-Spectroscopic Analysis
4.4. Quantitative Real-Time PCR Gene Expression Analysis
4.5. Ethics Statement
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Morris, A.K.; Masterton, R.G. Antibiotic resistance surveillance: Action for international studies. J. Antimicrob. Chemother. 2002, 49, 7–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization (WHO). Antimicrobial Resistance: Fact Sheet No 194. Genebra: WHO. 2016. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 2 January 2023).
- Foster, T.J. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 2017, 41, 430–449. [Google Scholar] [CrossRef]
- Lakhundi, S.; Zhang, K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 2018, 31, e00020-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peacock, S.J.; Paterson, G.K. Mechanisms of Methicillin Resistance in Staphylococcus aureus. Annu. Rev. Biochem. 2015, 84, 577–601. [Google Scholar] [CrossRef]
- Rolo, J.; Miragaia, M.; Turlej-Rogacka, A.; Empel, J.; Bouchami, O.; Faria, N.A.; Tavares, A.; Hryniewicz, W.; Fluit, A.C.; de Lencastre, H.; et al. High genetic diversity among community-associated staphylococcus aureus in europe: Results from a multicenter study. PLoS ONE 2012, 7, e34768. [Google Scholar] [CrossRef] [Green Version]
- Klevens, R.M.; Morrison, M.A.; Nadle, J.; Petit, S.; Gershman, K.; Ray, S.; Harrison, L.H.; Lynfield, R.; Dumyati, G.; Townes, J.M.; et al. FSABC surveillance (ABCs) M.I. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 2007, 298, 1763–1771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shafer, C.; Allison, J.; Hogue, A.; Huntington, M. Infectious Disease: Health Care-Associated Infections. FP Essent. 2019, 476, 30–42. [Google Scholar] [PubMed]
- Steinmann, J.; Buer, J.; Pietschmann, T.; Steinmann, E. Anti-infective properties of (EGCG), a component of green tea. Br. J. Pharmacol. 2013, 168, 1059–1073. [Google Scholar] [CrossRef] [Green Version]
- Novy, P.; Rondevaldova, J.; Kourimska, L.; Kokoska, L. Synergistic interactions of epigallocatechin gallate and oxytetracycline against various drug resistant Staphylococcus aureus strains in vitro. Phytomedicine 2013, 20, 432–435. [Google Scholar] [CrossRef]
- Hu, Z.; Zhao, W.; Asano, N.; Yoda, Y.; Hara, Y.; Shimamura, T. Epigallocatechin Gallate Synergistically Enhances the Activity of Carbapenems against Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2002, 46, 558–560. [Google Scholar] [CrossRef] [Green Version]
- Parvez, M.A.K.; Saha, K.; Rahman, J.; Munmun, R.A.; Rahman, M.A.; Dey, S.K.; Rahman, M.S.; Islam, S.; Shariare, M. Antibacterial activities of green tea crude extracts and synergistic effects of epigallocatechingallate ( EGCG ) with gentamicin against MDR pathogens. Heliyon 2019, 5, e02126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Z.; Zhao, W.; Hara, Y.; Shimamura, T. Epigallocatechin gallate synergy with ampicilin/sulfabactam against 28 clinical isolates of methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 2001, 48, 361–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araújo, R.; Ramalhete, L.; Da Paz, H.; Ribeiro, E.; Calado, C.R.C. A Simple, Label-Free, and High-Throughput Method to Evaluate the Epigallocatechin-3-Gallate Impact in Plasma Molecular Profile. High-Throughput 2020, 9, 9. [Google Scholar] [CrossRef] [Green Version]
- Chow, H.S.; Cai, Y.; Hakim, I.A.; Crowell, J.A.; Shahi, F.; Brooks, C.A.; Dorr, R.T.; Hara, Y.; Alberts, D.S. Pharmacokinetics and Safety of Green Tea Polyphenols after Multiple-Dose Administration of Epigallocatechin Gallate and Polyphenon E in Healthy Individuals. Clin. Cancer Res. 2003, 9, 3312–3319. [Google Scholar]
- Negri, A.; Naponelli, V.; Rizzi, F.; Bettuzzi, S. Molecular Targets of Epigallocatechin-Gallate (EGCG): A Special Focus on Signal Transduction and Cancer. Nutrients 2018, 10, 1936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boundy, S.; Safo, M.K.; Wang, L.; Musayev, F.N.; Farrell, H.C.O.; Rife, J.P.; Archer, G.L. Characterization of the Staphylococcus aureus rRNA Methyltransferase Encoded by orfX, the Gene Containing the Staphylococcal Chromosome Cassette mec ( SCC mec ) Insertion Site. J. Biol. Chem. 2013, 288, 132–140. [Google Scholar] [CrossRef] [Green Version]
- Ji, Q.; Chen, P.J.; Qin, G.; Deng, X.; Hao, Z.; Wawrzak, Z.; Yeo, W.S.; Quang, J.W.; Cho, H.; Luo, G.Z.; et al. Structure and mechanism of the essential two-component signal-transduction system WalKR in Staphylococcus aureus. Nat. Commun. 2016, 7, 11000, Retraction in Nat Commun. 2017, 8, 14331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delauné, A.; Dubrac, S.; Blanchet, C.; Poupel, O.; Mäder, U.; Hiron, A.; Leduc, A.; Fitting, C.; Nicolas, P.; Cavaillon, J.M.; et al. The WalKR System Controls Major Staphylococcal Virulence Genes and Is Involved in Triggering the Host Inflammatory Response. Infect. Immun. 2012, 80, 3438–3453. [Google Scholar] [CrossRef] [Green Version]
- Poupel, O.; Proux, C.; Jagla, B.; Msadek, T.; Dubrac, S. SpdC, a novel virulence factor, controls histidine kinase activity in Staphylococcus aureus. PLoS Pathog. 2018, 14, e1006917. [Google Scholar] [CrossRef] [Green Version]
- EUCAST. EUCAST Clinical Breakpoint Tables for Interpretation of MICs and Zone Diameters. 2022. Available online: https://www.eucast.org/clinical_breakpoints (accessed on 2 January 2023).
- Zarnowiec, P.; Lechowicz, Ł.; Czerwonka, G.; Kaca, W. Fourier Transform Infrared Spectroscopy (FTIR) as a Tool for the Identification and Differentiation of Pathogenic Bacteria. Curr. Med. Chem. 2015, 22, 1710–1718. [Google Scholar] [CrossRef]
- Shi, H.; Sun, J.; Han, R.; Ding, C.; Hu, F.; Yu, S. The strategy for correcting interference from water in Fourier transform infrared spectrum based bacterial typing. Talanta 2020, 208, 120347. [Google Scholar] [CrossRef]
- Wongthong, S.; Tippayawat, P.; Wongwattanakul, M.; Poung-Ngern, P.; Wonglakorn, L.; Chanawong, A.; Heraud, P.; Lulitanond, A. Attenuated total reflection: Fourier transform infrared spectroscopy for detection of heterogeneous vancomycin—Intermediate Staphylococcus aureus. World J. Microbiol. Biotechnol. 2020, 36, 22. [Google Scholar] [CrossRef]
- Cunha, B.R.; Aleixo, S.M.; Fonseca, L.P.; Calado, C.R.C. Fast identification of off-target liabilities in early antibiotic discovery with Fourier-transform infrared spectroscopy. Biotechnol. Bioeng. 2021, 118, 4465–4476. [Google Scholar] [CrossRef] [PubMed]
- Cunha, B.R.; Fonseca, L.P.; Calado, C.R.C. Simultaneous elucidation of antibiotics mechanism-of-action and potency with high-throughput fourier-transform Infrared spectroscopy and machine-learning. App. Microb. Biot. 2021, 105, 1269–1286. [Google Scholar] [CrossRef]
- Sampaio, P.N.; Calado, C.R.C. Potential of FTIR spectroscopy for drugs screening against Helicobacter pylori. Antibiotics 2020, 9, 897. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Chen, L.; Wang, J.; Wang, W.; Chen, D.; Li, L. Current methodologies on genotyping for nosocomial pathogen methicillin-resistant Staphylococcus aureus (MRSA). Microb. Pathog. 2017, 107, 17–28. [Google Scholar] [CrossRef]
- Pathare, N.A.; Tejani, S.; Asogan, H.; Al Mahruqi, G.; Al Fakhri, S.; Zafarulla, R. Comparison of Methicillin Resistant Staphylococcus Aureus in Healthy Community. Mediterr. J. Hematol. Infect. Dis. 2015, 7, e2015053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, H.; Liu, D.; Ma, Y.; Gao, W. Comparison of community- and healthcare-associated methicillin-resistant Staphylococcus aureus isolates at a Chinese tertiary hospital, 2012–2017. Sci. Rep. 2018, 8, 17916. [Google Scholar] [CrossRef] [Green Version]
- Kluytmans-VandenBergh, M.F.Q.; Kluytmans, J.A.J.W. Community-acquired methicillin-resistant Staphylococcus aureus: Current perspectives. Clin. Microbiol. Infect. 2006, 12 (Suppl. 1), 9–15. [Google Scholar] [CrossRef] [Green Version]
- Hsiao, C.; Ong, S.J.; Chuang, C.; Ma, D.H.K.; Huang, Y. A Comparison of Clinical Features between Community-Associated and Healthcare-Associated Methicillin-Resistant Staphylococcus aureus Keratitis. J. Ophthalmol. 2015, 2015, 923941. [Google Scholar] [CrossRef] [Green Version]
- Bonesso, M.F.; Marques, S.A.; Cunha, M.L.R.S.C. Community-acquired methicillin-resistant Staphylococcus aureus (CA- MRSA): Molecular background, virulence, and relevance for public health. J. Venom. Anim. Toxins. Incl. Trop. Dis. 2011, 17, 378–386. [Google Scholar] [CrossRef]
- Rayner, C.; Munckhof, W.J. Antibiotics currently used in the treatment of infections caused by Staphylococcus aureus. Intern. Med. J. 2005, 35, S3–S16. [Google Scholar] [CrossRef] [PubMed]
- Betts, J.W.; Sharili, A.S.; Phee, L.M.; Wareham, D.W. In Vitro Activity of Epigallocatechin Gallate and Quercetin Alone and in Combination versus Clinical Isolates of Methicillin-Resistant Staphylococcus aureus. J. Nat. Prod. 2015, 78, 2145–2148. [Google Scholar] [CrossRef]
- Haghjoo, B.; Lee, L.H.; Habiba, U.; Tahir, H.; Olabi, M.; Chu, T. The synergistic effects of green tea polyphenols and antibiotics against potential pathogens. Adv. Biosci. Biotechnol. 2013, 4, 959–967. [Google Scholar] [CrossRef] [Green Version]
- Reygaert, W.C. Green Tea Catechins: Their Use in Treating and Preventing Infectious Diseases. Biomed. Res. Int. 2018, 2018, 9105261. [Google Scholar] [CrossRef]
- Bakkir, L.K.; Yassen, R.T.; Mustaffa, R.M. In vitro and in vivo study of green and black tea antimicrobial activity on methicillin resistant Staphylococcus aureus. Bas. J. Vet. Res. 2011, 10, 1–12. Available online: https://bjvr.uobasrah.edu.iq/article_55014_555285214990f32d4e6f813cff7e1093.pdf (accessed on 2 January 2023).
- Cunha, B.R.; Ramalhete, L.; Fonseca, L.P.; Calado, C.R.C. Fourier-Transform Mid-Infrared (FT-MIR) Spectroscopy in Biomedicine. In Essential Techniques for Medical and Life Scientists: A Guide to Contemporary Methods and Current Applications-Part II; Bentham Science: Hilversum, The Netherlands, 2020; Volume 1, p. 1. [Google Scholar] [CrossRef]
- Amiali, N.M.; Golding, G.R.; Sedman, J.; Simor, A.E.; Ismail, A.A. Rapid identification of community-associated methicillin-resistant Staphylococcus aureus by Fourier transform infrared spectroscopy. Diagn. Microbiol. Infect Dis. 2011, 70, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Amiali, N.M.; Mulvey, M.R.; Sedman, J.; Simor, A.E.; Ismail, A.A. Epidemiological typing of methicillin-resistant Staphylococcus aureus strains by Fourier transform infrared spectroscopy. J. Microbiol. Methods. 2007, 69, 146–153. [Google Scholar] [CrossRef]
- Stapleton, P.D.; Taylor, P.W. Methicillin resistance in Staphylococcus aureus: Mechanisms and modulation. Sci. Prog. 2002, 85 Pt 1, 57–72. [Google Scholar] [CrossRef]
- Zhao, W.; Hu, Z.; Okubo, S.; Hara, Y. Mechanism of Synergy between Epigallocatechin Gallate and β-Lactams against Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2001, 45, 1737–1742. [Google Scholar] [CrossRef] [Green Version]
- Roccaro, A.S.; Blanco, A.R.; Giuliano, F.; Rusciano, D.; Enea, V. Epigallocatechin-Gallate Enhances the Activity of Tetracycline in Staphylococci by Inhibiting Its Efflux from Bacterial Cells. Antimicrob. Agents Chemother. 2004, 48, 1968–1973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanco, A.R.; Sudano-roccaro, A.; Spoto, G.C.; Nostro, A.; Rusciano, D. Epigallocatechin Gallate Inhibits Biofilm Formation by Ocular Staphylococcal Isolates. Antimicrob. Agents Chemother. 2005, 49, 4339–4343. [Google Scholar] [CrossRef] [Green Version]
- Yam, T.S.; Hamilton-Miller, J.M.T.; Shah, S. The effect of a component of tea (Camelia sinensis) on methicilin resistance, PBP2’ synthesis and b-lactamase production in Staphylococcus aureus. J. Antimicrob. Chemother. 1998, 42, 211–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jenkins, A.; An Diep, B.; Mai, T.T.; Vo, N.H.; Warrener, P.; Suzich, J.; Stover, C.K.; Sellman, B.R. Differential expression and roles of Staphylococcus aureus virulence determinants during colonization and disease. MBio 2015, 6, e02272-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bitrus, A.A.; Zunita, Z.; Khairani-Bejo, S.; Othman, S.; Ahmad Nadzir, N.A. Staphylococcal cassette chromosome mec (SCCmec) and characterization of the attachment site (attB) of methicillin resistant Staphylococcus aureus (MRSA) and methicillin susceptible Staphylococcus aureus (MSSA) isolates. Microb. Pathog. 2018, 123, 323–329. [Google Scholar] [CrossRef]
- Osterman, I.A.; Dontsova, O.A.; Sergiev, P.V. rRNA Methylation and Antibiotic Resistance. Biochemistry 2020, 85, 1335–1349. [Google Scholar] [CrossRef]
- Kitichalermkiat, A.; Katsuki, M.; Sato, J.; Sonoda, T.; Masuda, Y.; Honjoh, K.; Miyamoto, T. Effect of epigallocatechin gallate on gene expression of Staphylococcus aureus. J. Glob. Antimicrob. Resist. 2020, 22, 854–859. [Google Scholar] [CrossRef]
- Zeferino, A.S.; Mira, A.R.; Delgadinho, M.; Brito, M.; Ponte, T.; Ribeiro, E. Drug Resistance and Epigenetic Modulatory Potential of Epigallocatechin-3-Gallate Against Staphylococcus aureus. Curr. Microbiol. 2022, 79, 149. [Google Scholar] [CrossRef]
- Ghosh, D.; Veeraraghavan, B.; Elangovan, R.; Vivekanandan, P. Antibiotic Resistance and Epigenetics: More to It than Meets the Eye. Antimicrob. Agents Chemother. 2020, 64, e02225-19. [Google Scholar] [CrossRef]
- Oliveira, P.H.; Fang, G. Conserved DNA Methyltransferases: A Window into Fundamental Mechanisms of Epigenetic Regulation in Bacteria. Trends Microbiol. 2021, 29, 28–40. [Google Scholar] [CrossRef]
- Araújo, R.; Ramalhete, L.; Ribeiro, E.; Calado, C. Plasma versus Serum Analysis by FTIR Spectroscopy to Capture the Human Physiological State. BioTech 2022, 11, 56. [Google Scholar] [CrossRef] [PubMed]



| GEN | GEN + 250 µg/mL EGCG | GEN + 100 µg/mL EGCG | GEN + 50 µg/mL EGCG | GEN + 25 µg/mL EGCG | |
|---|---|---|---|---|---|
| MS13 | R | R | R | R | R |
| MR3 | R | S | S | S | S |
| MR4 | R | S | S | S | S |
| MR15 | R | S | S | R | R |
| Code | Resistance Phenotype |
|---|---|
| MS3 | IPM S; TET S; GEN S; AMX R no reversion |
| MS5 | IPM S; TET S; GEN S; AMX R reversion up to 50 µg/mL EGCG |
| MS13 | IPM S; TET S; GEN S; AMX S |
| MS15 | IPM S; TET NA; GEN S; AMX R no reversion |
| MS16 | IPM S; TET S; GEN R no reversion; AMX R no reversion |
| MR7 | IPM S; TET S; GEN S; AMX All R no reversion |
| MR9 | IPM R no reversion; TET S; GEN R all reversion; AMX all R no reversion |
| MR18 | IPM S; TET S; GEN R all reversion; AMX all R no reversion |
| MR31 | IPM S; TET NA; GEN R reversion up to 100 ugEGCG; AMX all R no reversion |
| MR22 | IPM R no reversion; TET S; GEN S; AMX R no reversion |
| Biological Sample | Sensible | Resistent | |
|---|---|---|---|
| MS1: Purulent exudate right foot ulcer | OXA;GEN;ERY;CLI;TET;SXT | PEN;TEC | |
| MS2: Sputum | OXA;GEN;CLI;LVX;TET;SXT | PEN | |
| MS3: Purulent exudate left hip | OXA;GEN;ERY;CLI;TET;SXT | PEN;TEC | |
| MS4: Sputum | OXA;GEN;CLI;LVX;TET;SXT | PEN | |
| MS5: Blood culture | OXA;GEN;CLI;SXT | ||
| MS6: Purulent exudate left foot ulcer | OXA;GEN;ERY;CLI;TET;SXT | ||
| M | MS7: Sputum | OXA;GEN;LVX;TET;SXT | ERY;CLI |
| S | MS8: Superficial exudate aspirated Forearm | OXA;GEN;ERY;CLI;TET;SXT | PEN |
| S | MS9: Urine | OXA;GEN;CLI;SXT | PEN;MXF;LVX |
| A | MS10: Superficial exudate aspirated Forearm | PEN;OXA;GEN;CLI;TET; SXT | |
| MS11: Sputum | OXA;GEN;CLI;SXT | PEN; ERY;TEC | |
| MS12: Sputum | OXA;GEN;ERY;CLI;LVX;TET;SXT | PEN | |
| MS13: Sputum | OXA;CLI;LVX;TET;SXT | GEN; ERY | |
| MS14: Dialysis Fluid | OXA;GEN;ERY;CLI;TET; SXT | PEN; FA | |
| MS15: Urine | OXA;GEN;SXT | PEN | |
| MS16: Exudate surgical wound- Lumbar | OXA;GEN;CLI;TET;SXT | PEN | |
| MR1: Purulent exudate from wound on back of hand | GEN; SXT;VAN | PEN;OXA;ERY;CLI;MXF;LVX | |
| MR2: Purulent exudate from right foot ulcer | GEN;ERY;CLI;TET;SXT;VAN | PEN; OXA; FA | |
| MR3: Sputum | TET; LVX; VAN | PEN; OXA; GEN; ERY;CLI;SXT | |
| MR4: Bronchoalveolar lavage | TET; VAN | PEN; OXA; GEN; ERY;CLI;SXT | |
| MR5: Blood culture | GEN;CLI;VAN | PEN;OXA | |
| M | MR6: Purulent swab exudate—schematic foot | GEN;TET;VAN;SXT | PEN;OXA;ERY;CLIMXFLVX |
| R | MR7: Pressure ulcer biopsy | GEN;TET;SXTVAN | PEN;OXA;ERY;CLI;MXFLVX |
| S | MR8: Purulent exudate swab—leg | GEN;TET;SXTVAN | PEN;OXA;ERY;CLI;MXF;LVX; FA |
| A | MR9: Blood culture | GEN;VAN | PEN;OXA;ERY;CLI;MXF;LVX |
| MR10: Biopsy/tissue fragment—amputation | GEN;TET;SXT;VAN | PEN;OXA;ERY;CLI;MXF;LVX | |
| MR11: Blood culture—catéter | GEN;CLI;VAN | PEN;OXA;MXF;LVX | |
| MR12: Aspirated purulent exudate—inguino-scrotal hernia | GEN; TET; SXT; VAN | PEN;OXA;ERY;CLI;MXF;LVX | |
| MR13: Purulent swab exudate—burnt skin | GEN;CLI;TET;SXT; VAN | PEN;OXA; FA | |
| MR14: Exudate Operative wound partial swab-toe matricectomy | GEN;CLI;TET;VAN | PEN;OXA | |
| MR15: Blood culture | VAN | PEN;OXA;GEN;ERY;CLI; MXF;LVX | |
| MR16: Blood culture | GEN;VAN | PEN;OXA;ERY;CLI;MXF; LVX |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mira, A.R.; Zeferino, A.S.; Inácio, R.; Delgadinho, M.; Brito, M.; Calado, C.R.C.; Ribeiro, E. Epigenetic and Drug Response Modulation of Epigalocaten-In-3-Gallate in Staphylococcus aureus with Divergent Resistance Phenotypes. Antibiotics 2023, 12, 519. https://doi.org/10.3390/antibiotics12030519
Mira AR, Zeferino AS, Inácio R, Delgadinho M, Brito M, Calado CRC, Ribeiro E. Epigenetic and Drug Response Modulation of Epigalocaten-In-3-Gallate in Staphylococcus aureus with Divergent Resistance Phenotypes. Antibiotics. 2023; 12(3):519. https://doi.org/10.3390/antibiotics12030519
Chicago/Turabian StyleMira, Ana Rita, Ana Sofia Zeferino, Raquel Inácio, Mariana Delgadinho, Miguel Brito, Cecília R. C. Calado, and Edna Ribeiro. 2023. "Epigenetic and Drug Response Modulation of Epigalocaten-In-3-Gallate in Staphylococcus aureus with Divergent Resistance Phenotypes" Antibiotics 12, no. 3: 519. https://doi.org/10.3390/antibiotics12030519





