The Genotypic and Phenotypic Characteristics Contributing to Flomoxef Sensitivity in Clinical Isolates of ESBL-Producing E. coli Strains from Urinary Tract Infections
Abstract
1. Introduction
2. Results
2.1. E. coli Isolates and Antibiotic Sensitivity
2.2. Multilocus Sequence Typing and β-Lactamase Gene PCR
2.3. The Clinical Efficacy of FMOX
3. Materials and Methods
3.1. ESBL-Producing E. coli
3.2. Flomoxef Efficacy
3.3. Statistical Analyses
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ramanath, K.V.; Shafiya, S.B. Prescription pattern of antibiotic usage for urinary tract infection treated in a rural tertiary care hospital. Indian J. Pharma. Pract. 2011, 4, 57–63. [Google Scholar]
- Jena, J.; Debata, N.K.; Subudhi, E. Prevalence of extended-spectrum-β-lactamase and metallo-β-lactamase producing multi drug rsistance gram negative bacteria from urinary isolates. Indian J. Med. Microbiol. 2013, 31, 420–421. [Google Scholar] [CrossRef] [PubMed]
- Wada, K.; Yokoyama, T.; Uno, S.; Araki, M.; Sadahira, T.; Maruyama, Y.; Acosta, H.; Nakajima, H.; Hiyama, Y.; Kunishima, Y.; et al. Nationwide surveillance of bacterial pathogens isolated from patients with acute uncomplicated cystitis in 2018: Conducted by the Japanese Research Group for Urinary Tract Infections (JRGU). J. Infect. Chemother. 2021, 27, 1169–1180. [Google Scholar] [CrossRef] [PubMed]
- Sadahira, T.; Wada, K.; Araki, M.; Ishii, A.; Takamoto, A.; Kobayashi, Y.; Watanabe, M.; Watanabe, T.; Nasu, Y.; Okayama Urological Research Group (OURG). Efficacy and safety of 3 day versus 7 day cefditoren pivoxil regimens for acute uncomplicated cystitis: Multicentre, randomized, open-label trial. J. Antimicrob. Chemother. 2017, 72, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Canton, R.; Coque, T.M. The CTX-M beta-lactamase pandemic. Curr. Opin. Microbiol. 2006, 9, 466–475. [Google Scholar] [CrossRef] [PubMed]
- Bradford, P.A. Extended-spectrum beta-lactamases in the 21st century: Characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 2001, 14, 933–951. [Google Scholar] [CrossRef]
- D’Andrea, M.M.; Arena, F.; Pallecchi, L.; Rossolini, G.M. CTX-M-type β-lactamases: A successful story of antibiotic resistance. Int. J. Med. Microbiol. 2013, 303, 305–317. [Google Scholar] [CrossRef]
- Pitout, J.D.; Gregson, D.B.; Campbell, L.; Laupland, K.B. Molecular characteristics of extended-spectrum-beta-lactamase-producing Escherichia coli isolates causing bacteremia in the Calgary Health Region from 2000 to 2007: Emergence of clone ST131 as a cause of community-acquired infections. Antimicrob. Agents Chemother. 2009, 53, 2846–2851. [Google Scholar] [CrossRef]
- Nicolas-Chanoie, M.H.; Bertrand, X.; Madec, J.Y. Escherichia coli ST131, an intriguing clonal group. Clin. Microbial. Rev. 2014, 27, 543–574. [Google Scholar] [CrossRef]
- Blanc, V.; Leflon-Guibout, V.; Blanco, J.; Haenni, M.; Madec, J.Y.; Rafignon, G.; Bruno, P.; Mora, A.; Lopez, C.; Dahbi, G.; et al. Prevalence of day-care centre children (France) with fecal CTX-M-producing Escherichia coli comprising O25b:H4 and O16:H5 ST131 strains. J. Antimicrob. Chemother. 2014, 69, 1231–1237. [Google Scholar] [CrossRef]
- Bevan, E.R.; Jones, A.M.; Hawkey, P.M. Global epidemiology of CTX-M β-lactamases: Temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 2017, 72, 2145–2155. [Google Scholar] [CrossRef]
- Kawamoto, I.; Miyauchi, M. β-Lactams and Other Antimicrobial Agents. In Antibiotics; Kawamoto, I., Ed.; CRC Press: Boca Raton, FL, USA, 1992. [Google Scholar]
- Sato, H.; Narita, A.; Suzuki, H.; Mastumoto, K.; Nakanishi, Y.; Nakazawa, S.; Niino, K.; Nakada, Y. The study of flomoxef in the pediatric field. Jpn. J. Antibiot. 1987, 40, 1349–1363. [Google Scholar]
- Yokoyama, T.; Kodama, T.; Takesue, Y.; Fujimoto, M.; Hiyama, E.; Ichikawa, T. Studies on antibacterial activity of flomoxef and its distribution to serum and intraperitoneal exudate in surgery. Jpn. J. Antibiot. 1987, 40, 1809–1819. [Google Scholar] [PubMed]
- Matsumura, Y.; Yamamoto, M.; Nagao, M.; Komori, T.; Fujita, N.; Hayashi, A.; Shimizu, T.; Watanabe, H.; Doi, S.; Tanaka, M.; et al. Multicenter retrospective study of cefmetazole and flomoxef for treatment of extended-spectrum-β-lactamase-producing Escherichia coli bacteremia. Antimicrob. Agents Chemother. 2015, 59, 5107–5113. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-H.; Su, L.-H.; Tang, Y.-F.; Liu, J.-W. Treatment of ESBL-producing Klebsiella pneumoniae bacteraemia with carbapenems or flomoxef: A retrospective study and laboratory analysis of the isolates. J. Antimicrob. Chemother. 2006, 58, 1074–1077. [Google Scholar] [CrossRef]
- Yang, Q.; Zhang, H.; Cheng, J.; Xu, Z.; Xu, Y.-C.; Cao, B.; Kong, H.; Ni, Y.; Yu, Y.; Sun, Z.; et al. In vitro activity of flomoxef and comparators against Escherichia coli, Klebsiella pneumoniae and Proteus mirabilis producing extended-spectrum β-lactamases in China. Int. J. Antimicrob. Agents. 2015, 45, 485–490. [Google Scholar] [CrossRef]
- Abe, Y.; Inan-Erdogan, I.; Fukuchi, K.; Wakabayashi, H.; Ogawa, Y.; Hibino, S.; Sakurai, S.; Matsuhashi, K.; Watanabe, Y.; Hashimoto, K.; et al. Efficacy of non-carbapenem antibiotics for pediatric patients with first febrile urinary tract infection due to extended-spectrum beta-lactamase-producing Escherichia coli. J. Infect. Chemother. 2017, 23, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Horie, A.; Nariai, A.; Katou, F.; Abe, Y.; Saito, Y.; Koike, D.; Hirade, T.; Ito, T.; Wakuri, M.; Fukuma, A. Increased community-acquired upper urinary tract infections caused by extended-spectrum beta-lactamase-producing Escherichia coli in children and the efficacy of flomoxef and cefmetazole. Clin. Exp. Nephrol. 2019, 23, 1306–1314. [Google Scholar] [CrossRef]
- Ngoi, S.T.; The, C.S.J.; Chong, C.W.; Abdul Jabar, K.; Tan, S.C.; Yu, L.H.; Leong, K.C.; Tee, L.H.; AbuBakar, S. In Vitro Efficacy of Flomoxef against Extended-Spectrum Beta-Lactamase-Producing Escherichia coli and Klebsiella pneumoniae Associated with Urinary Tract Infections in Malaysia. Antibiotics 2021, 10, 181. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; CLSI: Wayne, PA, USA, 2012. [Google Scholar]
- Dallenne, C.; Da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Kanda, Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013, 48, 452–458. [Google Scholar] [CrossRef]
- Knothe, H.; Shah, P.; Krcmery, V.; Antal, M.; Mitsuhashi, S. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection 1983, 11, 315–317. [Google Scholar] [CrossRef]
- Ishii, Y.; Ohno, A.; Taguchi, H.; Imajo, S.; Ishiguro, M.; Matsuzawa, H. Cloning and sequence of the gene encoding a cefotaxime-hydrolyzing class A beta-lactamase isolated from Escherichia coli. Antimicrob. Agents Chemother. 1995, 39, 2269–2275. [Google Scholar] [CrossRef]
- Chong, Y.; Yakushiji, H.; Ito, Y.; Kamimura, T. Clinical and molecular epidemiology of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in a long-term study from Japan. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 83–87. [Google Scholar] [CrossRef]
- Johnson, J.R.; Tchesnokova, V.; Johnston, B.; Clabots, C.; Roberts, P.L.; Billig, M.; Riddell, K.; Rogers, P.; Qin, X.; Butler-Wu, S.; et al. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J. Infect. Dis. 2013, 207, 919–928. [Google Scholar] [CrossRef]
- Demirci, M.; Unlu, O.; Tosun, A.I. Detection of O25b-ST131 clone, CTX-M-1 and CTX-M-15 genes via real-time PCR in Escherichia coli strains in patients with UTIs obtained from a university hospital in Istanbul. J. Infect. Public Health 2019, 12, 640–644. [Google Scholar] [CrossRef]
- Ismail, A.I.; Ali, S.S.; Altayeb, H.N.; Alla, A.A.; Moglad, E.H.; Elmissbah, T.E.; Elaskary, A.; Dahlawi, H. Detection of TEM, AmpC, SHV, CTX-M, and MCR-1 Genes in Gram Negative Isolates of Urinary Tract Infections. Clin. Lab. 2021, 67, 12. [Google Scholar] [CrossRef]
- Jena, J.; Sahoo, R.K.; Debata, N.K.; Subudhi, E. Prevalence of TEM, SHV, and CTX-M genes of extended-spectrum β-lactamase-producing Escherichia coli strains isolated from urinary tract infections in adults. 3 Biotech. 2017, 7, 244. [Google Scholar] [CrossRef]
- Sadahira, T.; Wada, K.; Araki, M.; Ishii, A.; Watanabe, T.; Nasu, Y.; Tsugawa, M.; Takenaka, T.; Nasu, Y.; Kumon, H. Impact of selective media for detecting fluoroquinolone-insusceptible/extended-spectrum beta-lactamase-producing Escherichia coli before transrectal prostate biopsy. Int. J. Urol. 2017, 24, 842–847. [Google Scholar] [CrossRef]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum beta-lactamases:a clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef]
- Nakagawa, S.; Hisada, H.; Nomura, N.; Mitsuyama, J.; Matsubara, S.; Yamaoka, K.; Watanabe, K.; Asano, Y.; Suematsu, H.; Sawamura, H.; et al. Antimicrobial activity of several drugs against extended-spectrum beta-lactamase positive Enterobacteriaceae isolates in Gifu and Aichi prefecture. Jpn. J. Antibiot. 2013, 66, 251–264. [Google Scholar]
- Hara, T.; Sato, T.; Horiyama, T.; Kanazawa, S.; Yamaguchi, T.; Maki, H. Prevalence and molecular characterization of CTX-M extended-spectrum β-lactamase-producing Escherichia coli from 2000 to 2010 in Japan. Jpn. J. Antibiot. 2015, 68, 75–84. [Google Scholar]
- Johnson, J.R.; Drawz, S.M.; Porter, S.; Kuskowski, M.A. Susceptibility to alternative oral antimicrobial agents in relation to sequence type ST131 status and Coresistance phenotype among recent Escherichia coli isolates from U.S. veterans. Antimicrob. Agents Chemother. 2013, 57, 4856–4860. [Google Scholar] [CrossRef]
- Popejoy, M.W.; Paterson, D.L.; Cloutier, D.; Huntington, J.A.; Miller, B.; Bliss, C.A.; Steenbergen, J.N.; Hershberger, E.; Umeh, O.; Kaye, K.S. Efficacy of ceftolozane/tazobactam against urinary tract and intra-abdominal infections caused by ESBL-producing Escherichia coli and Klebsiella pneumoniae: A pooled analysis of Phase 3 clinical trials. J. Antimicrob. Chemother. 2017, 72, 268–272. [Google Scholar] [CrossRef]
- Yang, C.C.; Li, S.H.; Chuang, F.R.; Chen, C.H.; Lee, C.H.; Chen, J.B.; Wu, C.H.; Lee, C.T. Discrepancy between effects of carbapenems and flomoxef in treating nosocomial hemodialysis access-related bacteremia secondary to extended spectrum beta-lactamase producing Klebsiella pneumoniae in patients on maintenance hemodialysis. BMC Infect Dis. 2012, 12, 206. [Google Scholar] [CrossRef]
- Tsujimoto, M.; Yokoyama, H.; Shimizu, K.; Yoneda, N.; Sano, H.; Ueyama, J.; Namba, N.; Tsuji, Y. Cases of Pediatric Pyelonephritis: A Single-Center Retrospective Study from an Extended-Spectrum β-Lactamase-Producing Escherichia coli Endemic Area in Japan. Yonago Acta Med. 2023, 66, 104–111. [Google Scholar] [CrossRef]
- Khonsari, M.; Behzadi, P.; Foroohi, F. The prevalence of type 3 fimbriae in Uropathogenic Escherichia coli isolated from clinical urine samples. Meta Gene 2021, 28, 100881. [Google Scholar] [CrossRef]
- Hozzari, A.; Behzadi, P.; Khiabani, K.P.; Sholeh, M.; Sabokroo, N. Clinical cases, drug resistance, and virulence genes profiling in Uropathogenic Escherichia coli. J. Appl. Genet. 2020, 61, 265–273. [Google Scholar] [CrossRef]


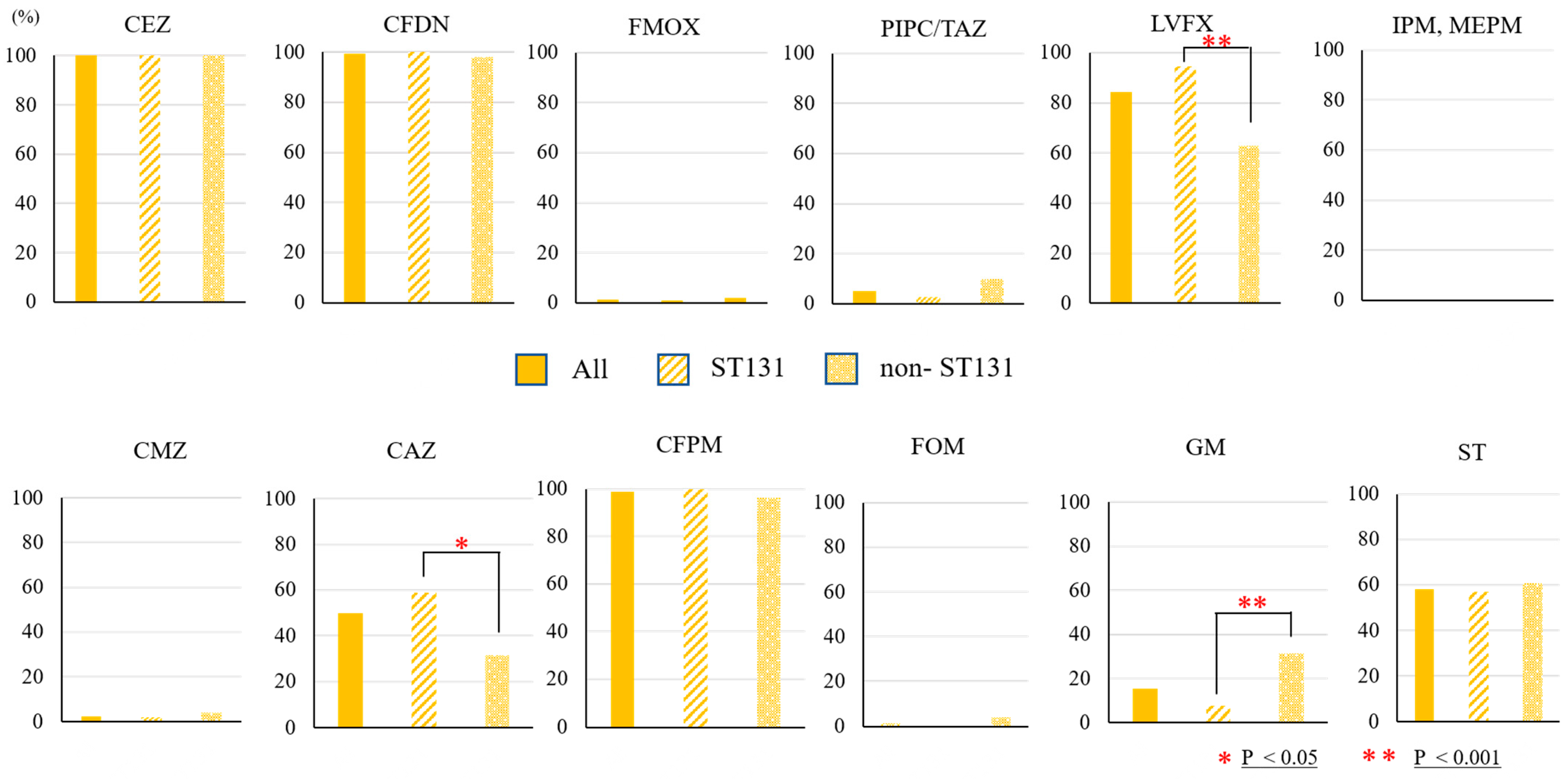

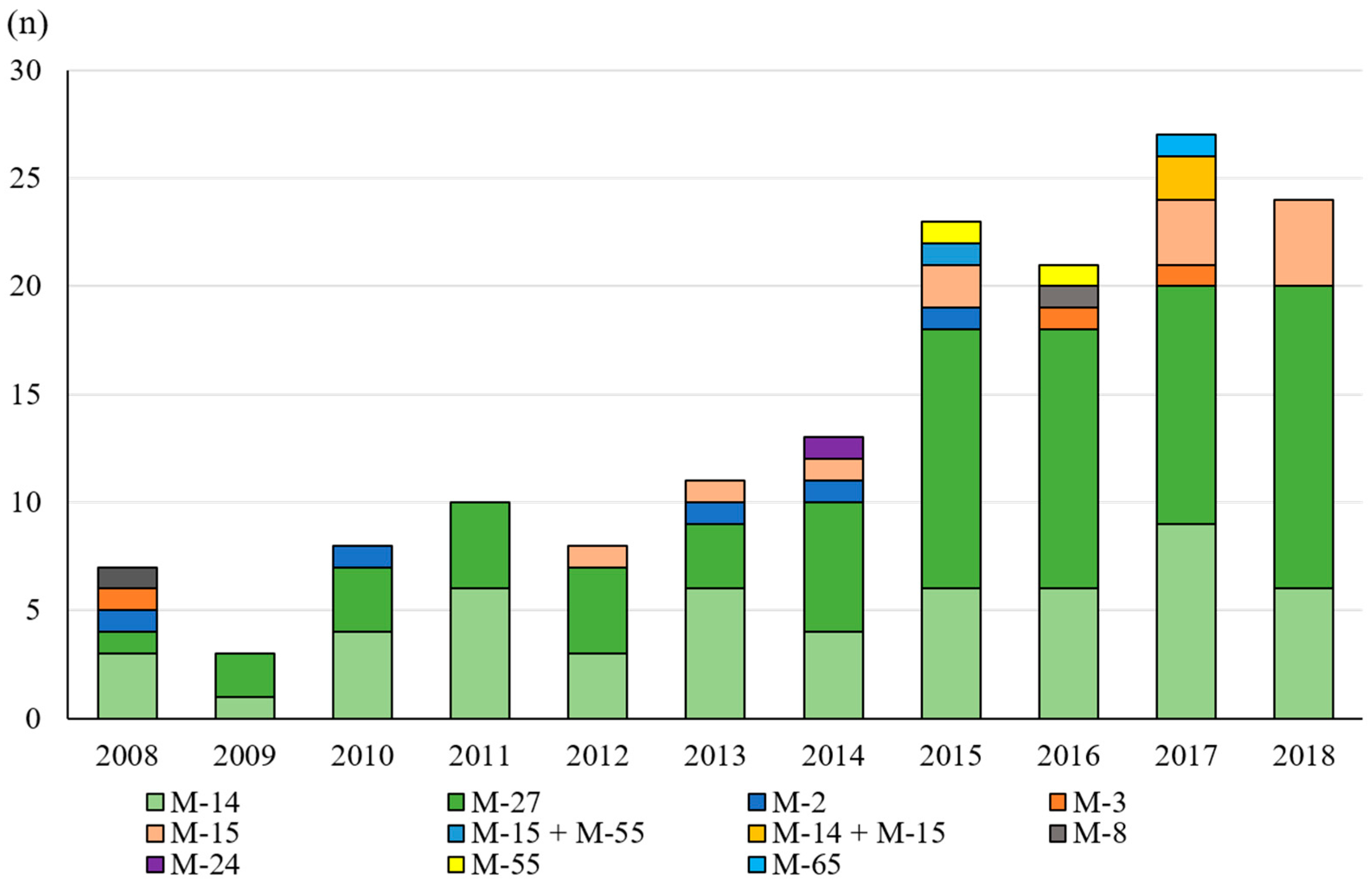


| Antibiotic Agent | MIC (μg/ml) for All Isolates (n = 158) | Susceptible Rate (%) | |||||
|---|---|---|---|---|---|---|---|
| Breakpoint * | MIC50 | MIC90 | 2008–2010 | 2011–2013 | 2014–2016 | 2017–2018 | |
| (n = 17) | (n = 31) | (n = 58) | (n = 52) | ||||
| cefazolin | 2 | >16 | >16 | 0 | 0 | 0 | 0 |
| cefdinir | 1 | >8 | >8 | 0 | 0 | 2 | 0 |
| flomoxef | 8 † | 0.125 | 0.5 | 94 | 100 | 100 | 98 |
| cefmetazole | 16 | 2 | 8 | 88 | 100 | 98 | 98 |
| ceftazidime | 4 | 4 | 32 | 59 | 65 | 45 | 44 |
| cefepime | 2 | >8 | >8 | 0 | 3 | 2 | 0 |
| piperacilin/tazobactam | 16/4 | 2/4 | 16/4 | 83 | 97 | 97 | 92 |
| imipenem | 1 | ≦0.5 | ≦0.5 | 100 | 100 | 100 | 100 |
| faropenem | - ‡ | 0.5 | 2 | - | - | - | - |
| meropenem | 1 | ≦0.5 | ≦0.5 | 100 | 100 | 100 | 100 |
| levofloxacin | 0.5 | 16 | 32 | 12 | 16 | 22 | 10 |
| sitafloxacin | -‡ | 1 | 2 | - | - | - | - |
| fosfomycin | 64 | 4 | 8 | 100 | 100 | 97 | 100 |
| gentamicin | 4 | ≦1 | >16 | 88 | 81 | 84 | 85 |
| sulfamethoxazole/trimethoprim | 2/38 | 8 | >64 | 24 | 42 | 50 | 38 |
| No. | ST | β-Lactamase Gene PCR | MIC50 (µg/mL) | Antibiotic Agent Sensitivity | |
|---|---|---|---|---|---|
| FMOX | CMZ | ||||
| 1 | ST131 | CTX-M-2 | 16 | 64 | PIPC/TAZ, IPM, MEPM, FOM, GM |
| 2 | ST354 | CTX-M-15,CMY161 | 64 | >64 | IPM, MEPM, FOM |
| Bacteria Types | n |
|---|---|
| Escherichiscoli | 16 |
| ESBL-producing | 5 |
| not ESBL-producing | 11 |
| Klebsiella oxytoca (ESBL-producing) | 1 |
| Proteus mirabillis | 3 |
| ESBL-producing | 1 |
| not ESBL-producing | 2 |
| Citrobacter koseri (ESBL-producing) | 1 |
| Pseudomonas aeruginosa | 1 |
| Other Gram-negative rods | 2 |
| Enterococcus | 2 |
| MRSA | 1 |
| Other Gram-positive cocci | 3 |
| Negative | 3 |
| Clinically Effective (Success) | Change to Other Antibiotic (Failure) | Other Antibiotic | |
|---|---|---|---|
| Surgical antibiotic prophylaxis | |||
| Escherichia coli ESBL-producing | 3 | - | |
| Other ESBL-producing bacteria | - | 1 | DRPM |
| Urinary tract infection treatment | |||
| Escherichia coli ESBL-producing | 1 | 1 | DRPM, STFX |
| Other ESBL-producing bacteria | 1 | 1 | DRPM |
| Primer | Target Gene | Nucleotide Sequence (5′---3′) | Denaturation | Annealing | Expansion | Number of Cycles | Product Length (b.p.) |
|---|---|---|---|---|---|---|---|
| adkF | adk | ATTCTGCTTGGCGCTCCGGG | 95 °C 1 min | 54 °C 1 min | 72 °C 2 min | 30 | 583 |
| adkR | CCGTCAACTTTCGCGTATTT | ||||||
| fumCF | fumC | TCACAGGTCGCCAGCGCTTC | 54 °C 1 min | 806 | |||
| fumCR | GTACGCAGCGAAAAAGATTC | ||||||
| gyrBF | gyrB | TCGGCGACACGGATGACGGC | 60 °C 1 min | 911 | |||
| gyrBR | ATCAGGCCTTCACGCGCATC | ||||||
| icdF | icd | ATGGAAAGTAAAGTAGTTGTTCCGGCACA | 54 °C 1 min | 878 | |||
| icdR | GGACGCAGCAGGATCTGTT | ||||||
| mdhF | mdh | ATGAAAGTCGCAGTCCTCGGCGCTGCTGGCGG | 60 °C 1 min | 932 | |||
| mdhR | TTAACGAACTCCTGCCCCAGAGCGATATCTTTCTT | ||||||
| purAF | purA | CGCGCTGATGAAAGAGATGA | 54 °C 1 min | 816 | |||
| purAR | CATACGGTAAGCCACGCAGA | ||||||
| recAF | recA | CGCATTCGCTTTACCCTGACC | 58 °C 1 min | 780 | |||
| recAR | TCGTCGAAATCTACGGACCGGA |
| PCR Name | β-Lactamase Targeted | Primer Name | Sequence(5′-3′) | Length (Bases) | Annealing Position | Amplicon Size(bp) | Primer Concentration |
|---|---|---|---|---|---|---|---|
| (pmol/μL) | |||||||
| Multiplex I TEM, SHV and OXA-1-like | TEM variants including TEM-1 and TEM-2 | MultiTSO-T_for | CATTTCCGTGTCGCCCTTATTC | 22 | 13–34 | 800 | 0.4 |
| MultiTSO-T_rev | CGTTCATCCATAGTTGCCTGAC | 22 | 812–791 | 0.4 | |||
| SHV variants including SHV-1 | MultiTSO-S_for | AGCCGCTTGAGCAAATTAAAC | 21 | 71–91 | 713 | 0.4 | |
| MultiTSO-S_rev | ATCCCGCAGATAAATCACCAC | 21 | 783–763 | 0.4 | |||
| OXA-1, OXA-4 and OXA-30 | MultiTSO-O_for | GGCACCAGATTCAACTTTCAAG | 22 | 201–222 | 564 | 0.4 | |
| MultiTSO-O_rev | GACCCCAAGTTTCCTGTAAGTG | 22 | 764–743 | 0.4 | |||
| Multiplex II CTX-M group 1, group 2 and group 9 | variants of CTX-M group 1 including CTX-M-1, CTX-M-3 and CTX-M-15 | MultiCTXMGp1_for | TTAGGAARTGTGCCGCTGYA b | 20 | 61–80 | 688 | 0.4 |
| MultiCTXMGp1-2_rev | CGATATCGTTGGTGGTRCCAT b | 21 | 748–728 | 0.2 | |||
| variants of CTX-M group 2 including CTX-M-2 | MultiCTXMGp2_for | CGTTAACGGCACGATGAC | 18 | 345–362 | 404 | 0.2 | |
| MultiCTXMGp1-2_rev | CGATATCGTTGGTGGTRCCAT b | 21 | 748–728 | 0.2 | |||
| variants of CTX-M group 9 including CTX-M-9 and CTX-M-14 | MultiCTXMGp9_for | TCAAGCCTGCCGATCTGGT | 19 | 299-317 | 561 | 0.4 | |
| MultiCTXMGp9_rev | TGATTCTCGCCGCTGAAG | 18 | 859–842 | 0.4 | |||
| CTX-M group 8/25 | CTX-M-8, CTX-M-25, CTX-M-26 and CTX-M-39 to CTX-M-41 | CTX-Mg8/25_for | AACRCRCAGACGCTCTAC b | 18 | 172–189 | 326 | 0.4 |
| CTX-Mg8/25_rev | TCGAGCCGGAASGTGTYAT b | 19 | 497–479 | 0.4 | |||
| Multiplex III ACC, FOX, MOX, DHA, CIT and EBC | ACC-1 and ACC-2 | MultiCaseACC_for | CACCTCCAGCGACTTGTTAC | 20 | 744–763 | 346 | 0.2 |
| MultiCaseACC_rev | GTTAGCCAGCATCACGATCC | 20 | 1089–1070 | 0.2 | |||
| FOX-1 to FOX-5 | MultiCaseFOX_for | CTACAGTGCGGGTGGTTT | 18 | 396–413 | 162 | 0.5 | |
| MultiCaseFOX_rev | CTATTTGCGGCCAGGTGA | 18 | 557–540 | 0.5 | |||
| MOX-1, MOX-2, CMY-1, CMY-8 to CMY-11 and CMY-19 | MultiCaseMOX_for | GCAACAACGACAATCCATCCT | 21 | 3–23 | 895 | 0.2 | |
| MultiCaseMOX_rev | GGGATAGGCGTAACTCTCCCAA | 22 | 900–879 | 0.2 | |||
| DHA-1 and DHA-2 | MultiCaseDHA_for | TGATGGCACAGCAGGATATTC | 21 | 113–133 | 997 | 0.5 | |
| MultiCaseDHA_rev | GCTTTGACTCTTTCGGTATTCG | 22 | 1109–1088 | 0.5 | |||
| LAT-1 to LAT-3, BIL-1, CMY-2 to CMY-7, CMY-12 to CMY-18 and CMY-21 to CMY-23 | MultiCaseCIT_for | CGAAGAGGCAATGACCAGAC | 20 | 570–589 | 538 | 0.2 | |
| MultiCaseCIT_rev | ACGGACAGGGTTAGGATAGY b | 20 | 1107–1088 | 0.2 | |||
| ACT-1 and MIR-1 | MultiCaseEBC_for | CGGTAAAGCCGATGTTGCG | 19 | 189–207 | 683 | 0.2 | |
| MultiCaseEBC_rev | AGCCTAACCCCTGATACA | 18 | 871–854 | 0.2 | |||
| Multiplex IV VEB, PER and GES | GES-1 to GES-9 and GES-11 | MultiGES_for | AGTCGGCTAGACCGGAAAG | 19 | 463–481 | 399 | 0.3 |
| MultiGES_rev | TTTGTCCGTGCTCAGGAT | 18 | 861–844 | 0.3 | |||
| PER-1 and PER-3 | MultiPER_for | GCTCCGATAATGAAAGCGT | 19 | 325–343 | 520 | 0.3 | |
| MultiPER_rev | TTCGGCTTGACTCGGCTGA | 19 | 844–826 | 0.3 | |||
| VEB-1 to VEB-6 | MultiVEB_for | CATTTCCCGATGCAAAGCGT | 20 | 187–206 | 648 | 0.3 | |
| MultiVEB_rev | CGAAGTTTCTTTGGACTCTG | 20 | 834–815 | 0.3 | |||
| Multiplex V GES and OXA-48-like | GES-1 to GES-9 and GES-11 | MultiGES_for | AGTCGGCTAGACCGGAAAG | 19 | 463–481 | 399 | 0.3 |
| MultiGES_rev | TTTGTCCGTGCTCAGGAT | 18 | 861–844 | 0.3 | |||
| OXA-48-like | MultiOXA-48_for | GCTTGATCGCCCTCGATT | 18 | 230–247 | 281 | 0.4 | |
| MultiOXA-48_rev | GATTTGCTCCGTGGCCGAAA | 20 | 490–510 | 0.4 | |||
| Multiplex VI IMP, VIM and KPC | IMP variants except IMP-9, IMP-16, IMP-18, IMP-22 and IMP-25 | MultiIMP_for | TTGACACTCCATTTACDG b | 18 | 194–211 | 139 | 0.5 |
| MultiIMP_rev | GATYGAGAATTAAGCCACYCT b | 21 | 332–313 | 0.5 | |||
| VIM variants including VIM-1 and VIM-2 | MultiVIM_for c | GATGGTGTTTGGTCGCATA | 19 | 151–169 | 390 | 0.5 | |
| MultiVIM_rev c | CGAATGCGCAGCACCAG | 17 | 540–524 | 0.5 | |||
| KPC-1 to KPC-5 | MultiKPC_for | CATTCAAGGGCTTTCTTGCTGC | 22 | 209–230 | 538 | 0.2 | |
| MultiKPC_rev | ACGACGGCATAGTCATTTGC | 20 | 746–727 | 0.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sakaeda, K.; Sadahira, T.; Maruyama, Y.; Iwata, T.; Watanabe, M.; Wada, K.; Araki, M. The Genotypic and Phenotypic Characteristics Contributing to Flomoxef Sensitivity in Clinical Isolates of ESBL-Producing E. coli Strains from Urinary Tract Infections. Antibiotics 2023, 12, 522. https://doi.org/10.3390/antibiotics12030522
Sakaeda K, Sadahira T, Maruyama Y, Iwata T, Watanabe M, Wada K, Araki M. The Genotypic and Phenotypic Characteristics Contributing to Flomoxef Sensitivity in Clinical Isolates of ESBL-Producing E. coli Strains from Urinary Tract Infections. Antibiotics. 2023; 12(3):522. https://doi.org/10.3390/antibiotics12030522
Chicago/Turabian StyleSakaeda, Kazuma, Takuya Sadahira, Yuki Maruyama, Takehiro Iwata, Masami Watanabe, Koichiro Wada, and Motoo Araki. 2023. "The Genotypic and Phenotypic Characteristics Contributing to Flomoxef Sensitivity in Clinical Isolates of ESBL-Producing E. coli Strains from Urinary Tract Infections" Antibiotics 12, no. 3: 522. https://doi.org/10.3390/antibiotics12030522
APA StyleSakaeda, K., Sadahira, T., Maruyama, Y., Iwata, T., Watanabe, M., Wada, K., & Araki, M. (2023). The Genotypic and Phenotypic Characteristics Contributing to Flomoxef Sensitivity in Clinical Isolates of ESBL-Producing E. coli Strains from Urinary Tract Infections. Antibiotics, 12(3), 522. https://doi.org/10.3390/antibiotics12030522






