Antimicrobial Resistance Linked to Septic System Contamination in the Indiana Lake Michigan Watershed
Abstract
:1. Introduction
2. Results
2.1. E. coli Concentrations in Water Samples
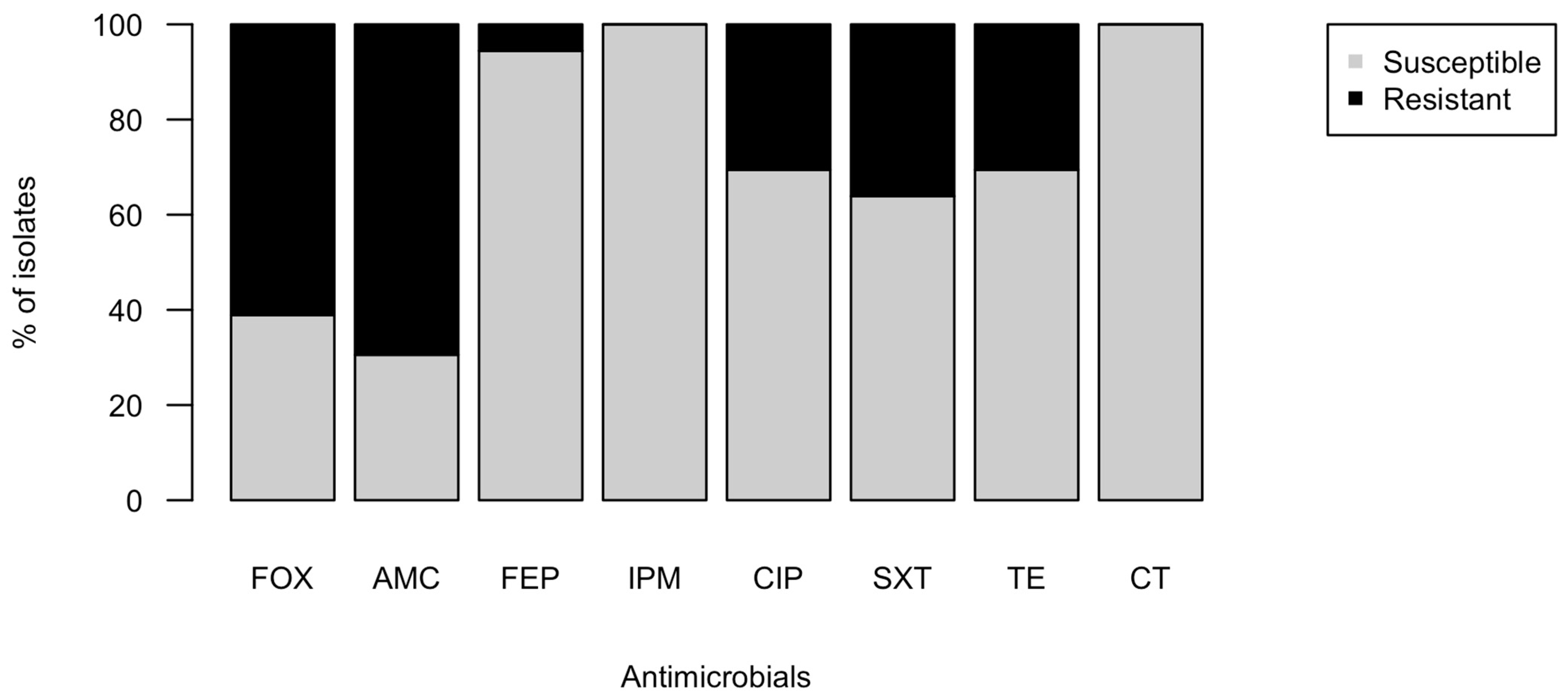
2.2. Phenotypic Antimicrobial Resistance of Isolates
2.3. Genetic Determinants of Antimicrobial Resistance in Isolates and the Environment
2.4. Environmental Factors Contributing to Antimicrobial Resistance
3. Discussion
3.1. Septic Systems Threaten Environmental Water Quality in the Lake Michigan Watershed
3.2. Antibiotic Resistance Genes (ARG) and Antibiotic Resistant E. coli in Septic Effluent
3.3. Mobile Resistance Genes in Environmental Isolates Increase Potential Health Risks
4. Materials and Methods
4.1. Study Sites, Sample Collection
4.2. Water Quality Assessment
4.3. Sample Processing and Purification of Cefotaxime-Resistant Escherichia coli Isolates
4.4. Minimum Inhibitory Concentration and Multi-Drug Resistance Tests
4.5. Environmental DNA and Plasmid Extractions
4.6. qPCR Assays for Antibiotic Resistance Genes and Human Fecal Marker Genes
4.7. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thakuria, B. The Beta Lactam Antibiotics as an Empirical Therapy in a Developing Country: An Update on Their Current Status and Recommendations to Counter the Resistance against Them. J. Clin. Diagn. Res. 2013, 7, 1207–1214. [Google Scholar] [CrossRef] [PubMed]
- Tipper, D.J.; Strominger, J.L. Mechanism of action of penicillins: A proposal based on their structural similarity to acyl-D-alanyl-D-alanine. Proc. Natl. Acad. Sci. USA 1965, 54, 1133–1141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanders, C.C.; Sanders, W.E. Emergence of Resistance to Cefamandole: Possible Role of Cefoxitin-Inducible Beta-Lactamases. Antimicrob. Agents Chemother. 1979, 15, 792–797. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Ma, X.; Liu, Y.; Yi, X.; Du, G.; Li, J. Fate of antibiotics, antibiotic-resistant bacteria, and cell-free antibiotic-resistant genes in full-scale membrane bioreactor wastewater treatment plants. Bioresour. Technol. 2020, 302, 122825. [Google Scholar] [CrossRef] [PubMed]
- Wilson, H.; Török, M.E. Extended-spectrum β-lactamase-producing and carbapenemase-producing Enterobacteriaceae. Microb. Genom. 2018, 4, e000197. [Google Scholar] [CrossRef] [PubMed]
- Dunne, E.F. Emergence of Domestically Acquired Ceftriaxone-Resistant Salmonella Infections Associated With AmpC β-Lactamase. JAMA 2000, 284, 3151. [Google Scholar] [CrossRef] [Green Version]
- Gekenidis, M.-T.; Kläui, A.; Smalla, K.; Drissner, D. Transferable Extended-Spectrum β-Lactamase (ESBL) Plasmids in Enterobacteriaceae from Irrigation Water. Microorganisms 2020, 8, 978. [Google Scholar] [CrossRef]
- Ndlovu, T.; Le Roux, M.; Khan, W.; Khan, S. Co-Detection of Virulent Escherichia coli Genes in Surface Water Sources. PLoS ONE 2015, 10, e0116808. [Google Scholar] [CrossRef]
- Stange, C.; Sidhu, J.P.S.; Tiehm, A.; Toze, S. Antibiotic resistance and virulence genes in coliform water isolates. Int. J. Hyg. Environ. Health 2016, 219, 823–831. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.K.; Das, S.; Kumar, S.; Gajamer, V.R.; Najar, I.N.; Lepcha, Y.D.; Tiwari, H.K.; Singh, S. Distribution of Antibiotic-Resistant Enterobacteriaceae Pathogens in Potable Spring Water of Eastern Indian Himalayas: Emphasis on Virulence Gene and Antibiotic Resistance Genes in Escherichia coli. Front. Microbiol. 2020, 11, 581072. [Google Scholar] [CrossRef]
- Ma, Y.; Chen, J.; Fong, K.; Nadya, S.; Allen, K.; Laing, C.; Ziebell, K.; Topp, E.; Carroll, L.M.; Wiedmann, M.; et al. Antibiotic Resistance in Shiga Toxigenic Escherichia coli Isolates from Surface Waters and Sediments in a Mixed Use Urban Agricultural Landscape. Antibiotics 2021, 10, 237. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.L. The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc. R. Soc. B Biol. Sci. 2009, 276, 2521–2530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dougherty, J.A.; Swarzenski, P.W.; Dinicola, R.S.; Reinhard, M. Occurrence of Herbicides and Pharmaceutical and Personal Care Products in Surface Water and Groundwater around Liberty Bay, Puget Sound, Washington. J. Environ. Qual. 2010, 39, 1173–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattioli, M.C.; Benedict, K.M.; Murphy, J.; Kahler, A.; Kline, K.E.; Longenberger, A.; Mitchell, P.K.; Watkins, S.; Berger, P.; Shanks, O.C.; et al. Identifying septic pollution exposure routes during a waterborne norovirus outbreak—A new application for human-associated microbial source tracking qPCR. J. Microbiol. Methods 2021, 180, 106091. [Google Scholar] [CrossRef]
- Geary, P.; Lucas, S. Contamination of estuaries from failing septic tank systems: Difficulties in scaling up from monitored individual systems to cumulative impact. Environ. Sci. Pollut. Res. 2019, 26, 2132–2144. [Google Scholar] [CrossRef]
- Rowles III, L.S.; Hossain, A.I.; Ramirez, I.; Durst, N.J.; Ward, P.M.; Kirisits, M.J.; Araiza, I.; Lawler, D.F.; Saleh, N.B. Seasonal contamination of well-water in flood-prone colonias and other unincorporated U.S. communities. Sci. Total Environ. 2020, 740, 140111. [Google Scholar] [CrossRef]
- Fong, T.-T.; Mansfield, L.S.; Wilson, D.L.; Schwab, D.J.; Molloy, S.L.; Rose, J.B. Massive Microbiological Groundwater Contamination Associated with a Waterborne Outbreak in Lake Erie, South Bass Island, Ohio. Environ. Health Perspect. 2007, 115, 856–864. [Google Scholar] [CrossRef] [Green Version]
- Amin, N.; Liu, P.; Foster, T.; Rahman, M.; Miah, M.R.; Ahmed, G.B.; Kabir, M.; Raj, S.; Moe, C.L.; Willetts, J. Pathogen flows from on-site sanitation systems in low-income urban neighborhoods, Dhaka: A quantitative environmental assessment. Int. J. Hyg. Environ. Health 2020, 230, 113619. [Google Scholar] [CrossRef]
- Stec, J.; Kosikowska, U.; Mendrycka, M.; Stępień-Pyśniak, D.; Niedźwiedzka-Rystwej, P.; Bębnowska, D.; Hrynkiewicz, R.; Ziętara-Wysocka, J.; Grywalska, E. Opportunistic Pathogens of Recreational Waters with Emphasis on Antimicrobial Resistance-A Possible Subject of Human Health Concern. Int. J. Environ. Res. Public Health 2022, 19, 7308. [Google Scholar] [CrossRef]
- Solaiman, S.; Handy, E.; Brinks, T.; Goon, K.; Bollinger, C.; Sapkota, A.R.; Sharma, M.; Micallef, S.A. Extended Spectrum β-Lactamase Activity and Cephalosporin Resistance in Escherichia coli from U.S. Mid-Atlantic Surface and Reclaimed Water. Appl. Environ. Microbiol. 2022, 88, e00837-22. [Google Scholar] [CrossRef]
- Withers, P.J.; Jordan, P.; May, L.; Jarvie, H.P.; Deal, N.E. Do septic tank systems pose a hidden threat to water quality? Front. Ecol. Environ. 2014, 12, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Johnson, N. Homeowners’ Knowledge & Awareness of Septic Systems and Barriers to Septic System Maintenance in Northwest Indiana: Information to Enhance Agency Outreach and Education Efforts. Master’s Thesis, Natural Resources and Environmental Sciences, University of Illinois at Urbana-Champaign, Urbana, IL, USA, 2016. [Google Scholar]
- Great Lakes Scientific Advisory Board. Groundwater in the Great Lakes Basin; International Joint Commisssion: Windsor, ON, Canada, 2016. [Google Scholar]
- Tan, B.; Ng, C.; Nshimyimana, J.P.; Loh, L.L.; Gin, K.Y.-H.; Thompson, J.R. Next-generation sequencing (NGS) for assessment of microbial water quality: Current progress, challenges, and future opportunities. Front. Microbiol. 2015, 6, 1027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verhougstraete, M.P.; Martin, S.L.; Kendall, A.D.; Hyndman, D.W.; Rose, J.B. Linking fecal bacteria in rivers to landscape, geochemical, and hydrologic factors and sources at the basin scale. Proc. Natl. Acad. Sci. USA 2015, 112, 10419–10424. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, W.; Neller, R.; Katouli, M. Evidence of septic system failure determined by a bacterial biochemical fingerprinting method. J. Appl. Microbiol. 2005, 98, 910–920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nevers, M.B.; Byappanahalli, M.N.; Shively, D.; Buszka, P.M.; Jackson, P.R.; Phanikumar, M.S. Identifying and Eliminating Sources of Recreational Water Quality Degradation along an Urban Coast. J. Environ. Qual. 2018, 47, 1042–1050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dila, D.K.; Corsi, S.R.; Lenaker, P.L.; Baldwin, A.K.; Bootsma, M.J.; McLellan, S.L. Patterns of Host-Associated Fecal Indicators Driven by Hydrology, Precipitation, and Land Use Attributes in Great Lakes Watersheds. Environ. Sci. Technol. 2018, 52, 11500–11509. [Google Scholar] [CrossRef] [PubMed]
- Uluseker, C.; Kaster, K.M.; Thorsen, K.; Basiry, D.; Shobana, S.; Jain, M.; Kumar, G.; Kommedal, R.; Pala-Ozkok, I. A Review on Occurrence and Spread of Antibiotic Resistance in Wastewaters and in Wastewater Treatment Plants: Mechanisms and Perspectives. Front. Microbiol. 2021, 12, 717809. [Google Scholar] [CrossRef]
- Blaak, H.; Lynch, G.; Italiaander, R.; Hamidjaja, R.A.; Schets, F.M.; de Husman, A.M.R. Multidrug-Resistant and Extended Spectrum Beta-Lactamase-Producing Escherichia coli in Dutch Surface Water and Wastewater. PLoS ONE 2015, 10, e0127752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thornton, C.N.; Tanner, W.D.; VanDerslice, J.A.; Brazelton, W.J. Localized effect of treated wastewater effluent on the resistome of an urban watershed. GigaScience 2020, 9, giaa125. [Google Scholar] [CrossRef]
- Damashek, J.; Westrich, J.R.; McDonald, J.M.B.; Teachey, M.E.; Jackson, C.R.; Frye, J.G.; Lipp, E.K.; Capps, K.A.; Ottesen, E.A. Non-point source fecal contamination from aging wastewater infrastructure is a primary driver of antibiotic resistance in surface waters. Water Res. 2022, 222, 118853. [Google Scholar] [CrossRef]
- Li, L.-G.; Huang, Q.; Yin, X.; Zhang, T. Source tracking of antibiotic resistance genes in the environment—Challenges, progress, and prospects. Water Res. 2020, 185, 116127. [Google Scholar] [CrossRef]
- Burch, T.R.; Stokdyk, J.P.; Firnstahl, A.D.; Kieke, B.A.; Cook, R.M.; Opelt, S.A.; Spencer, S.K.; Durso, L.M.; Borchardt, M.A. Microbial source tracking and land use associations for antibiotic resistance genes in private wells influenced by human and livestock fecal sources. J. Environ. Qual. 2022. [Google Scholar] [CrossRef] [PubMed]
- Taggar, G.; Rehman, M.A.; Yin, X.; Lepp, D.; Ziebell, K.; Handyside, P.; Boerlin, P.; Diarra, M.S. Antimicrobial-Resistant E. coli from Surface Waters in Southwest Ontario Dairy Farms. J. Environ. Qual. 2018, 47, 1068–1078. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Bai, X.; Jing, L.; Chen, R.; Teng, Y. Characterization of antibiotic resistance genes in the sediments of an urban river revealed by comparative metagenomics analysis. Sci. Total Environ. 2019, 653, 1513–1521. [Google Scholar] [CrossRef] [PubMed]
- CDC. Antibiotic Resistance Threats in the United States; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019.
- Kurittu, P.; Khakipoor, B.; Aarnio, M.; Nykäsenoja, S.; Brouwer, M.; Myllyniemi, A.-L.; Vatunen, E.; Heikinheimo, A. Plasmid-Borne and Chromosomal ESBL/AmpC Genes in Escherichia coli and Klebsiella pneumoniae in Global Food Products. Front. Microbiol. 2021, 12, 592291. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Kämpfer, P.; Nordmann, P. Chromosome-encoded Ambler class A beta-lactamase of Kluyvera georgiana, a probable progenitor of a subgroup of CTX-M extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 2002, 46, 4038–4040. [Google Scholar] [CrossRef] [Green Version]
- Talukdar, P.K.; Rahman, M.; Rahman, M.; Nabi, A.; Islam, Z.; Hoque, M.M.; Endtz, H.P.; Islam, M.A. Antimicrobial resistance, virulence factors and genetic diversity of Escherichia coli isolates from household water supply in Dhaka, Bangladesh. PLoS ONE 2013, 8, e61090. [Google Scholar] [CrossRef] [Green Version]
- Alawi, M.; Torrijos, T.V.; Walsh, F. Plasmid-mediated antimicrobial resistance in drinking water. Environ. Adv. 2022, 8, 100191. [Google Scholar] [CrossRef]
- Bortolaia, V.; Hansen, K.H.; Nielsen, C.A.; Fritsche, T.R.; Guardabassi, L. High diversity of plasmids harbouring blaCMY-2 among clinical Escherichia coli isolates from humans and companion animals in the upper Midwestern USA. J. Antimicrob. Chemother. 2014, 69, 1492–1496. [Google Scholar] [CrossRef] [Green Version]
- Anjum, M.; Madsen, J.S.; Nesme, J.; Jana, B.; Wiese, M.; Jasinskytė, D.; Nielsen, D.S.; Sørensen, S.J.; Dalsgaard, A.; Moodley, A.; et al. Fate of CMY-2-Encoding Plasmids Introduced into the Human Fecal Microbiota by Exogenous Escherichia coli. Antimicrob. Agents Chemother. 2019, 63, e02528-18. [Google Scholar] [CrossRef] [Green Version]
- CLSI. CLSI Supplement M100, 30th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Kanwar, N.; Scott, H.M.; Norby, B.; Loneragan, G.H.; Vinasco, J.; Cottell, J.L.; Chalmers, G.; Chengappa, M.M.; Bai, J.; Boerlin, P. Impact of treatment strategies on cephalosporin and tetracycline resistance gene quantities in the bovine fecal metagenome. Sci. Rep. 2014, 4, 5100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subirats, J.; Royo, E.; Balcázar, J.L.; Borrego, C.M. Real-time PCR assays for the detection and quantification of carbapenemase genes (bla KPC, bla NDM, and bla OXA-48) in environmental samples. Environ. Sci. Pollut. Res. 2017, 24, 6710–6714. [Google Scholar] [CrossRef] [PubMed]
- Nõlvak, H.; Truu, M.; Tiirik, K.; Oopkaup, K.; Sildvee, T.; Kaasik, A.; Mander, Ü.; Truu, J. Dynamics of Antibiotic Resistance Genes and Their Relationships with System Treatment Efficiency in a Horizontal Subsurface Flow Constructed Wetland. Sci. Total Environ. 2013, 461–462, 636–644. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, Z.; Feng, Y.; Hu, H.; Yu, Y.; Qiu, L.; Liu, H.; Guo, Z.; Huang, J.; Du, C.; et al. Molecular Detection of the mcr Genes by Multiplex PCR. Infect. Drug Resist. 2020, 13, 3463–3468. [Google Scholar] [CrossRef] [PubMed]
- Njage, P.M.K.; Buys, E. A High Resolution DNA Melting Curve Analysis for the Rapid and Efficient Molecular Diagnostics of Extended Spectrum β-Lactamase Determinants from Foodborne Escherichia coli. Microorganisms 2020, 8, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, H.C.; Haugland, R.A.; Varma, M.; Millen, H.T.; Borchardt, M.A.; Field, K.G.; Walters, W.A.; Knight, R.; Sivaganesan, M.; Kelty, C.A.; et al. Improved HF183 Quantitative Real-Time PCR Assay for Characterization of Human Fecal Pollution in Ambient Surface Water Samples. Appl. Environ. Microbiol. 2014, 80, 3086–3094. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2022. [Google Scholar]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef] [Green Version]



| Site Name | Sample ID | E. coli | % CTXR | HB |
|---|---|---|---|---|
| (MPN/100 mL) | (of N Tested a) | (CN/100 mL) | ||
| Burns Ditch | E00307 | 139.6 | 40.0 (10) | 102 |
| E00308 | 113.7 | 30.0 (10) | 219 | |
| Coffee Creek | E00324 | 1020 b | 8.3 (12) | 40 |
| E00340 | 980.4 | 54.5 (11) | 0 | |
| E00341 | 980.4 | 100.0 (12) | 58 | |
| Damon Run | E00337 | 770.1 | 50.0 (4) | 387 |
| Deer Creek | E00215 | >2419.6 c | 11.1 (9) | 6.14 × 105 |
| E00303 | 613.1 | 12.5 (8) | 1416 | |
| E00306 | 270 | 25.0 (4) | 78 | |
| E00346 | 344.8 | 100.0 (12) | 2528 | |
| Dunes Creek | E00210 | 193.6 | 8.3 (12) | 857 |
| E00211 | 307.6 | 16.7 (12) | 159 | |
| Lee Creek | E00330 | >2419.6 | 100.0 (2) | 0 |
| Little Calumet | E00349 | 103.9 | 11.1 (9) | 221 |
| Long Beach | E00195 | 450 | 8.3 (12) | 0 |
| E00198 | 399 | 9.1 (11) | 289 | |
| E00310 | 2 | 66.7 (3) d | 0 | |
| E00313 | 20 | 33.3 (3) | 0 | |
| E00315 | 145 | 25.0 (8) | 29 | |
| E00316 | 1 | 100.0 (2) d | 102 | |
| Salt Creek | E00335 | 816.4 | 63.6 (11) | 3980 |
| E00336 | 920.8 | 9.1 (11) | 8467 | |
| Sand Creek | E00343 | 387.3 | 75.0 (12) | 0 |
| E00344 | 2419.6 | 66.7 (12) | DNQ e | |
| Smith Ditch | E00332 | >2419.6 | 27.3 (3) | 193 |
| Trail Creek | E00205 | 285.6 | 100 (1) | 1571 |
| E00209 | 182.8 | 9.1 (11) | 839 | |
| E00221 | 488.4 | 8.3 (12) | 554 | |
| E00276 | 272.3 | 8.3 (12) | DNQ | |
| Septic reference | P00041 | 1.46 × 106 | 72.7 (11) | 7.06 × 106 |
| Isolate ID | Resistance Phenotype | CTX MIC | FEP MIC |
|---|---|---|---|
| I00646 | AMC-CTX | 48 < x < 64 | 0.5 < x < 0.75 |
| I00729 | AMC-CTX | 8 < x < 16 | 0.5 < x < 1 |
| I00735 | AMC-CTX | 8 < x < 16 | 0.25 < x < 0.5 |
| I00600 | AMC-CTX-FOX-CIP-SXT-TE | 12 < x < 16 | 1.5 < x < 2 |
| I00643 | AMC-CTX-FOX-CIP-SXT-TE | 16 < x < 24 | 1.5 < x < 2 |
| I00644 | AMC-FOX-CTX | 16 < x < 24 | 0.125 < x < 0.19 |
| I00645 | AMC-FOX-CTX | 16 < x < 32 | 0.25 < x < 0.5 |
| I00647 | AMC-FOX-CTX | 12 < x < 16 | 0.38 < x < 0.5 |
| I00648 | AMC-FOX-CTX | 32 < x < 48 | 0.75 < x < 1 |
| I00649 | AMC-FOX-CTX | 16 < x < 24 | 0.5 < x < 0.75 |
| I00650 | AMC-FOX-CTX | 12 < x < 16 | 0.75 < x < 1 |
| I00651 | AMC-FOX-CTX | 16 < x < 32 | 0.25 < x < 0.5 |
| I00730 | AMC-FOX-CTX | 6 < x < 8 | 0.25 < x < 0.38 |
| I00731 | AMC-FOX-CTX | 8 < x < 16 | 0.5 < x < 1 |
| I00732 | AMC-FOX-CTX | 16 < x < 32 | 1 < x < 2 |
| I00733 | AMC-FOX-CTX | 8 < x < 16 | 0.25 < x < 0.5 |
| I00734 | AMC-FOX-CTX | 8 < x < 16 | 0.5 < x < 1 |
| I00736 | AMC-FOX-CTX | 8 < x < 16 | 0.125 < x < 0.25 |
| I00737 | AMC-FOX-CTX | 32 < x < 48 | 1 < x < 1.5 |
| I00738 | AMC-FOX-CTX | 6 < x < 8 | 0.38 < x < 0.5 |
| I00739 | AMC-FOX-CTX | 6 < x < 8 | 0.125 < x < 0.19 |
| I00740 | AMC-FOX-CTX | 6 < x < 8 | 0.38 < x < 0.5 |
| I00652 | CTX-FEP-CIP-SXT-TE | 24 < x < 32 | 1.5 < x < 2 |
| I00657 | CTX-FEP-CIP-SXT-TE | 12 < x < 16 | 4 < x < 6 |
| I00653 | CTX-CIP-SXT-TE | 16 < x < 24 | 2 < x < 3 |
| I00654 | CTX-CIP-SXT-TE | 32 < x < 48 | 4 < x < 6 |
| I00655 | CTX-CIP-SXT-TE | 16 < x < 24 | 2 < x < 3 |
| I00656 | CTX-CIP-SXT-TE | 24 < x < 32 | 2 < x < 3 |
| I00659 | CTX-CIP-SXT-TE | 24 < x < 32 | 2 < x < 3 |
| I00613 | CTX-SXT | 8 < x < 12 | 0.75 < x < 1 |
| I00614 | CTX-SXT | 8 < x < 12 | 1 < x < 1.5 |
| Sample ID | Site | Isolates | CTX 1–15 | CTX 9–14 | SHV-2 | CMY-2 | NDM | KPC | qnrS | mcr-1 |
|---|---|---|---|---|---|---|---|---|---|---|
| E00205 | Trail Creek | |||||||||
| I00600 | ||||||||||
| E00209 | Trail Creek | |||||||||
| I00613 | ||||||||||
| E00276 | Trail Creek | |||||||||
| I00643 | ||||||||||
| E00215 | Deer Creek | |||||||||
| I00614 | ||||||||||
| E00303 | Deer Creek | |||||||||
| I00652 | ||||||||||
| E00306 | Deer Creek | |||||||||
| I00653 | ||||||||||
| E00346 | Deer Creek | |||||||||
| I00729 | ||||||||||
| I00730 | ||||||||||
| I00731 | ||||||||||
| I00732 | ||||||||||
| I00733 | ||||||||||
| I00734 | ||||||||||
| I00735 | ||||||||||
| I00736 | ||||||||||
| I00737 | ||||||||||
| I00738 | ||||||||||
| I00739 | ||||||||||
| I00740 | ||||||||||
| E00307 | Burns Ditch | |||||||||
| I00654 | ||||||||||
| I00655 | ||||||||||
| I00656 | ||||||||||
| I00657 | ||||||||||
| E00308 | Burns Ditch | |||||||||
| I00659 | ||||||||||
| P00041 | Septic | |||||||||
| I00644 | ||||||||||
| I00645 | ||||||||||
| I00646 | ||||||||||
| I00647 | ||||||||||
| I00648 | ||||||||||
| I00649 | ||||||||||
| I00650 | ||||||||||
| I00651 |
| Assay | Primer/ Probe | Sequence | Cycles | Ref. |
|---|---|---|---|---|
| CTX 1–15 | fwd | CGCAAATACTTTATCGTGCTGAT | 95 °C for 3 min, 40 cycles of 95 °C for 5 s, 57 °C for 30 s, and 95 °C for 60 s. Final elongation at 72 °C for 7 min | [48] |
| rev | GATTCGGTTCGCTTTCACTTT | |||
| CTX 9–14 | fwd | GCTCATCGATACCGCAGATAAT | 95 °C for 3 min, 40 cycles of 95 °C for 5 s, 57 °C for 30 s, and 95 °C for 60 s. Final elongation at 72 °C for 7 min | [48] |
| rev | CCGCCATAACTTTACTGGTACT | |||
| SHV-2 | fwd | CTGGAGCGAAAGATCCACTATC | 95 °C for 3 min, 40 cycles of 95 °C for 5 s, 57 °C for 30 s, and 95 °C for 60 s. Final elongation at 72 °C for 7 min | [49] |
| rev | CGCTGTTATCGCTCATGGTAA | |||
| CMY-2 | fwd | AGGGAAGCCCGTACACGTT | 95 °C for 10 min, 40 cycles of 95 °C for 10 s, 52 °C for 30 s, and 79 °C for 17 s | [45] |
| rev | GCTGGATTTCACGCCATAGG | |||
| NDM | fwd | GATTGCGACTTATGCCAATG | 95 °C for 3 min, 40 cycles of 95 °C for 30 s and 60 °C for 60 s | [46] |
| rev | TCGATCCCAACGGTGATATT | |||
| KPC | fwd | CAGCTCATTCAAGGGCTTTC | 95 °C for 3 min, 40 cycles of 95 °C for 30 s and 60 °C for 45 s | [46] |
| rev | GGCGGCGTTATCACTGTATT | |||
| qnrS | fwd | GTGAGTAATCGTATGTACTTTTG | 95 °C for 3 min, 40 cycles of 95 °C for 45 s, 52 °C for 45 s, and 72 °C for 60 s. Final elongation at 72 °C for 10 min | [47] |
| rev | AAACACCTCGACTTAAGTCT | |||
| mcr-1 | fwd | TCCAAAATGCCCTACAGACC | 94 °C for 4 min, 40 cycles of 94 °C for 5 s, 59 °C for 15. Final elongation at 72 °C for 5 min | [48] |
| rev | GCCACCACAGGCAGTAAAAT | |||
| HF183 | fwd | ATCATGAGTTCACATGTCCG | 95 °C for 10 min, 40 cycles of 95 °C for 15 s, 60 °C for 1 min | [50] |
| HB287R | rev | CTTCCTCTCAGAACCCCTATCC | ||
| BacP234 | probe | FAM-CTAATGGAACGCATCCC-MGB |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sidhu, A.S.; Mikolajczyk, F.N.; Fisher, J.C. Antimicrobial Resistance Linked to Septic System Contamination in the Indiana Lake Michigan Watershed. Antibiotics 2023, 12, 569. https://doi.org/10.3390/antibiotics12030569
Sidhu AS, Mikolajczyk FN, Fisher JC. Antimicrobial Resistance Linked to Septic System Contamination in the Indiana Lake Michigan Watershed. Antibiotics. 2023; 12(3):569. https://doi.org/10.3390/antibiotics12030569
Chicago/Turabian StyleSidhu, Angad S., Faith N. Mikolajczyk, and Jenny C. Fisher. 2023. "Antimicrobial Resistance Linked to Septic System Contamination in the Indiana Lake Michigan Watershed" Antibiotics 12, no. 3: 569. https://doi.org/10.3390/antibiotics12030569






