Swine Colibacillosis: Global Epidemiologic and Antimicrobial Scenario
Abstract
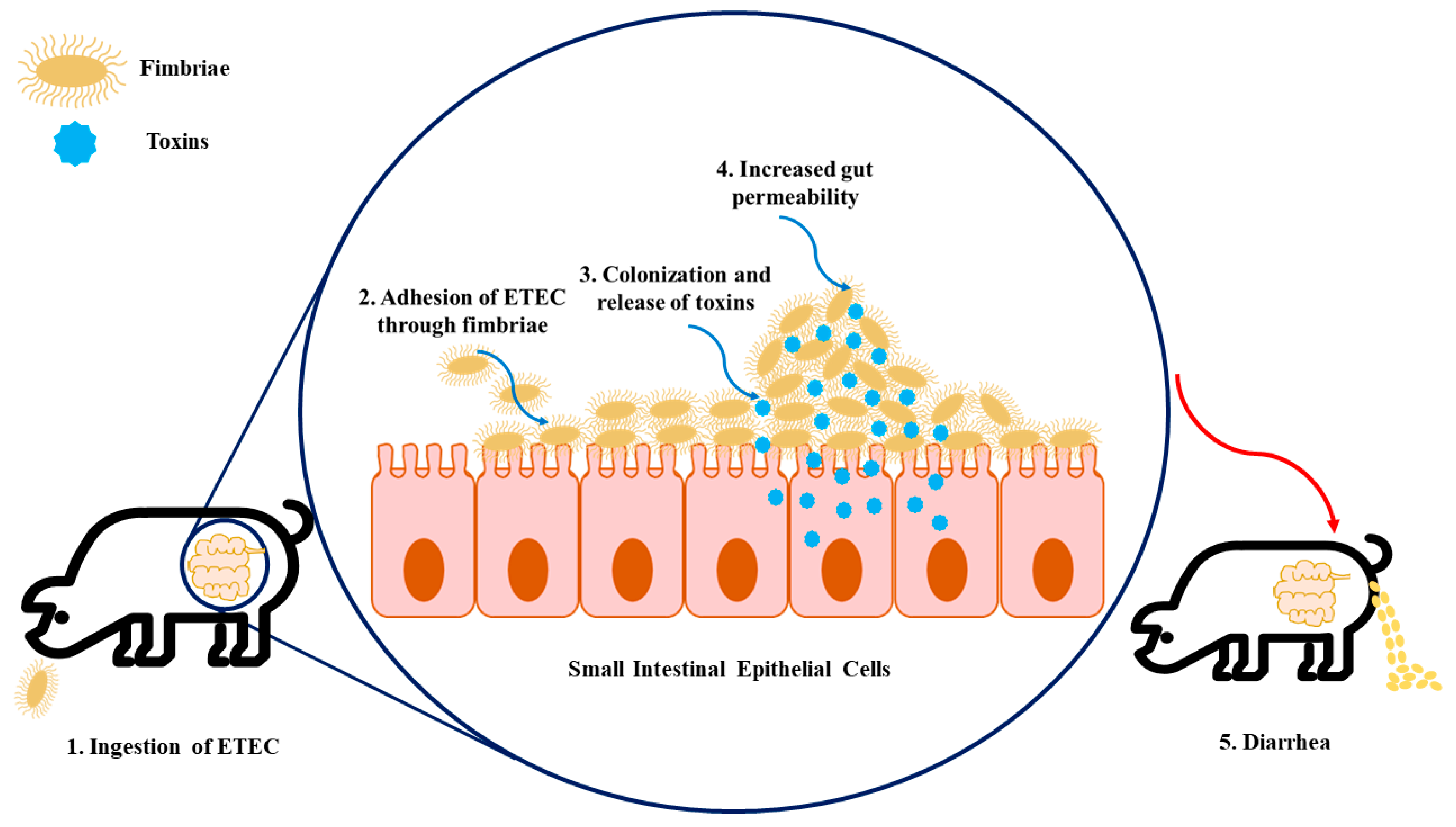
:1. Introduction
2. Etiology
2.1. ETEC Virulence Factors and Their Impact on Trigger Colibacillosis Infection
| Adhesins | Toxins | Serotypes | Disease |
|---|---|---|---|
| F5, F6, F41 | STa | O8, O9, O20, O64, O101 | Neonatal diarrhea |
| F4 | STa, STb, LT, EAST1, α-hemolysin b | O8, O138, O141, O145, O147, O149, O157 | Neonatal diarrhea Diarrhea in young pigs preweaning |
| F4, AIDA a, unknown | STa, STb, LT, EAST1, α-hemolysin b | O8, O138, O139, O141, O147, O149, O157 | PWD |
| F18, AIDA a | STa, STb, LT, Stx (or VT) c, EAST1, α-hemolysin b | O8, O138, O139, O141, O147, O149, O157 | PWD |
2.1.1. Neonatal ETEC
2.1.2. Postweaning ETEC
3. Global Epidemiology of Swine Enteric Colibacillosis: Prevalence, Diversity, and Outbreaks
| Country (n = Number of Isolates) | Percentage (Number) of Positive Isolates (%) | Reference | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fimbriae | Toxins | |||||||||
| F4 | F5 | F6 | F18 | F41 | LT | STa | STb | Stx2e | ||
| Australia (n = 104) | 38.5–96.3 | - | - | 0–15.4 | - | 62.1–92.3 | 64.8–92.3 | 83.7–100 | - | [26] |
| Belgium and The Netherlands (n = 100) | 51.0 | 1.0 | 1.0 | 42.0 | - | 14.0 | 22.0 | 30.0 | 5.0 | [18] |
| Denmark (n = 219) | 44.7 | - | 0.9 | 39.3 | - | 61.6 | 26.5 | 77.6 | - | [28] |
| France (n = 91) | 47.3 | - | - | 35.2 | - | 45.1 | 40.7 | 76.9 | 19.8 | [18] |
| Germany (n = 64) | 14.1 | - | - | 14.1 | - | 9.4 | 26.6 | 57.8 | 3.1 | [18] |
| Italy (n = 84) | 59.3 | 1.2 | 1.2 | 38.1 | 1.2 | 56.0 | 63.1 | 71.4 | 9.5 | [18] |
| Poland (n = 40) | 22.5 | - | - | 61.9 | - | 22.5 | 72.5 | 77.5 | 17.5 | [30] |
| Spain (n = 181) | 38.2 | 4.8 | 1.1 | 43.5 | 2.7 | 66.1 | 50.5 | 74.7 | 13.5 | [22] |
| Spain a (n = 277) | 27.7–40.5 | 16.7 | 11.9 | 51.5 | 16.7 | - | - | - | 10 | [25] |
| Slovakia (n = 101) | 19 | 0.9 | 5 | 35 | 0.9 | 20 | 26 | 46 | 5 | [27] |
| Uganda b (n = 83) | 8.4 | - | - | - | - | - | 1.2 | 26.5 | 2.4 | [34] |
| United States (n = 175) | 41.7 | - | - | 53.1 | - | 52.6 | 38.2 | 96 | - | [29] |
| Zimbabwe (n = 1984) | 28.4 | 22.3 | 1.5 | 25.4 | 22.3 | 50 | 73 | 16 | 27 | [35] |
4. Antimicrobial Prevalence in Enteric Colibacillosis Treatment
| Antimicrobial Class/Other Designations | Antimicrobial Agents | % Resistant Rates (n = Swine Isolates) | Country/City | Year/Time Range of the Study | Reference |
|---|---|---|---|---|---|
| Penicillins | Ampicillin | 85.9 (n = 608) | China/Shanghai | 2009–2021 | [48] |
| 75.4 (n = 481) | Spain/Lugo | 2006–2016 | [23] | ||
| 71.9 (n = 694) | Austria/Vienna | 2016–2018 | [49] | ||
| 84.8 (n = 455) | China/Beijing | 2014–2016 | [42] | ||
| 27.9 (n = 129) | China/Tibet | 2012 | [43] | ||
| 60.7 (n = 89) | Denmark/Frederiksberg C | 2014 | [46] | ||
| 81.4 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| 60.86 (n = 23) | Bangladesh/Tangail | 2018 | [50] | ||
| 86.4 (n = 118) | Korea | 2016–2017 | [40] | ||
| 48.3 (n = 90) | Denmark | 2018–2019 | [51] | ||
| 89.1 (n = 55) | United States | 2013–2014 | [44] | ||
| 34.5 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Ampicillin-sulbactam | 64.6 (n = 481) | Spain/Lugo | 2006–2016 | [23] | |
| Ticarcillin | 73.8 (n = 481) | Spain/Lugo | 2006–2016 | [23] | |
| 81.4 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| β-lactam combination agents | Amoxicillin/clavulanic acid | 42.3 (n = 608) | China/Shanghai | 2009–2021 | [48] |
| 84.63 (n = 455) | China/Beijing | 2014–2016 | [42] | ||
| 11.76 (n = 135) | Santa Catarina/ Brazil | 2016–2017 | [52] | ||
| 33.5 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| 82.6 (n = 23) | Bangladesh/Tangail | 2018 | [50] | ||
| 5.1 (n = 118) | Korea | 2016–2017 | [40] | ||
| 1.1 (n = 90) | Denmark | 2018–2019 | [51] | ||
| 9.5 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Ampicillin/ sulbactam | 70.8 (n = 161) | Spain/Lugo | 2005–2017 | [22] | |
| 5 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Penicillins + β-lactamase inhibitors | Piperacillin/ tazobactam | 0.6 (n = 161) | Spain/Lugo | 2005–2017 | [22] |
| Cephalosporins | Ceftiofur | 22.5 (n = 608) | China/Shanghai | 2009–2021 | [48] |
| 52.63 (n = 455) | China/Beijing | 2014–2016 | [42] | ||
| 25 (n = 135) | Santa Catarina/ Brazil | 2016–2017 | [52] | ||
| 10.9 (n = 129) | China/Tibet | 2012 | [43] | ||
| 25.5 (n = 55) | United States | 2013–2014 | [44] | ||
| Cefepime | 9.2 (n = 481) | Spain/Lugo | 2006–2016 | [23] | |
| 7.5 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| 2.5 (n = 118) | Korea | 2016–2017 | [40] | ||
| 4.2 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Cefazolin | 60.82 (n = 455) | China/Beijing | 2014–2016 | [42] | |
| 10.6 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| 10.2 (n = 118) | Korea | 2016–2017 | [40] | ||
| Cefuroxime | 8.7 (n = 161) | Spain/Lugo | 2005–2017 | [22] | |
| Cefotaxime | 10.6 (n = 161) | Spain/Lugo | 2005–2017 | [22] | |
| 9.1 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Ceftazidime | 5 (n = 161) | Spain/Lugo | 2005–2017 | [22] | |
| 3 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Cephalothin | 64.4 (n = 118) | Korea | 2016–2017 | [40] | |
| 41.7 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Cefoxitin | 3.4 (n = 118) | Korea | 2016–2017 | [40] | |
| 1.8 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Ceftriaxone | 6 (n = 168) | China/Shenzhen | 2009–2014 | [45] | |
| Carbapenems | Ceftazidime | 1.9 (n = 608) | China/Shanghai | 2009–2021 | [48] |
| 1.5 (n = 481) | Spain/Lugo | 2006–2016 | [23] | ||
| 5.9 (n = 694) | Austria/Vienna | 2016–2018 | [49] | ||
| Meropenem | 0.3 (n = 608) | China/Shanghai | 2009–2021 | [48] | |
| Aminoglycosides | Kanamycin | 63.74 (n = 455) | China/Beijing | 2014–2016 | [42] |
| 3.6 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Spectinomycin | 65.7 (n = 608) | China/Shanghai | 2009–2021 | [48] | |
| 2.3 (n = 129) | China/Tibet | 2012 | [43] | ||
| 18 (n = 89) | Denmark/Frederiksberg C | 2014 | [46] | ||
| 43.6 (n = 55) | United States | 2013–2014 | [44] | ||
| 55.6 (n = 90) | Denmark | 2018–2019 | [51] | ||
| Gentamicin | 37.2 (n = 608) | China/Shanghai | 2009–2021 | [48] | |
| 47.7 (n = 481) | Spain/Lugo | 2006–2016 | [23] | ||
| 7.7 (n = 694) | Austria/Vienna | 2016–2018 | [49] | ||
| 57.31 (n = 455) | China/Beijing | 2014–2016 | [42] | ||
| 32.35 (n = 135) | Santa Catarina/ Brazil | 2016–2017 | [52] | ||
| 6.9 (n = 129) | China/Tibet | 2012 | [43] | ||
| 14.6 (n = 89) | Denmark/Frederiksberg C | 2014 | [46] | ||
| 58.4 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| 36.4 (n = 118) | Korea | 2016–2017 | [40] | ||
| 32.7 (n = 55) | United States | 2013–2014 | [44] | ||
| 6.7 (n = 90) | Denmark | 2018–2019 | [51] | ||
| 5.4 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Tobramycin | 47.7 (n = 481) | Spain/Lugo | 2006–2016 | [23] | |
| 6.2 (n = 694) | Austria/Vienna | 2016–2018 | [49] | ||
| 54.7 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| Streptomycin | 40.35 (n = 455) | China/Beijing | 2014–2016 | [42] | |
| 16.2 (n = 129) | China/Tibet | 2012 | [43] | ||
| 29.2 (n = 89) | Denmark/Frederiksberg C | 2014 | [46] | ||
| 86.4 (n = 118) | Korea | 2016–2017 | [40] | ||
| 68.9 (n = 90) | Denmark | 2018–2019 | [51] | ||
| 18.5 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Amikacin | 15.2 (n = 455) | China/Beijing | 2014–2016 | [42] | |
| 1.2 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Apramycin | 14.6 (n = 89) | Denmark/Frederiksberg C | 2014 | [46] | |
| 8.9 (n = 90) | Denmark | 2018–2019 | [51] | ||
| Neomycin | 50 (n = 118) | Korea | 2016–2017 | [40] | |
| 49.1 (n = 55) | United States | 2013–2014 | [44] | ||
| 25.6 (n = 90) | Denmark | 2018–2019 | [51] | ||
| Tetracyclines | Doxycycline | 85.9 (n = 608) | China/Shanghai | 2009–2021 | [48] |
| 62.7 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| Tetracycline | 91.6 (n = 608) | China/Shanghai | 2009–2021 | [48] | |
| 67.7 (n = 694) | Austria/Vienna | 2016–2018 | [49] | ||
| 83.63 (n = 455) | China/Beijing | 2014–2016 | [42] | ||
| 40.4 (n = 129) | China/Tibet | 2012 | [43] | ||
| 47.2 (n = 89) | Denmark/Frederiksberg C | 2014 | [46] | ||
| 65.21 (n = 23) | Bangladesh/Tangail | 2018 | [50] | ||
| 86.4 (n = 118) | Korea | 2016–2017 | [40] | ||
| 56.7 (n = 90) | Denmark | 2018–2019 | [51] | ||
| 21.4 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Minocycline | 41.5 (n = 481) | Spain/Lugo | 2006–2016 | [23] | |
| 52.2 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| Chlortetracycline | 80 (n = 55) | United States | 2013–2014 | [44] | |
| Oxytetracycline | 94.5 (n = 55) | United States | 2013–2014 | [44] | |
| Sulfonamides | Sulfisoxazole | 85.4 (n = 608) | China/Shanghai | 2009–2021 | [48] |
| Sulphaamethoxazole | 75.2 (n = 608) | China/Shanghai | 2009–2021 | [48] | |
| 69.7 (n = 89) | Denmark/Frederiksberg C | 2014 | [46] | ||
| 67.8 (n = 90) | Denmark | 2018–2019 | [51] | ||
| Sulfadimethoxine | 61.8 (n = 55) | United States | 2013–2014 | [44] | |
| Fluoroquinolones | Enrofloxacin | 41.3 (n = 608) | China/Shanghai | 2009–2021 | [48] |
| 72.51135(n = 455) | China/Beijing | 2014–2016 | [42] | ||
| 54.41 (n = 135) | Santa Catarina/Brazil | 2016–2017 | [52] | ||
| 58.2 (n = 55) | United States | 2013–2014 | [44] | ||
| Ofloxacin | 39 (n = 608) | China/Shanghai | 2009–2021 | [48] | |
| Ciprofloxacin | 61.5 (n = 161) | Spain/Lugo | 2005–2017 | [22] | |
| 26.3 (n = 118) | Korea | 2016–2017 | [40] | ||
| 9.8 (n = 41, farm 1); 8.8% (n = 34, farm 2); 21.7% (n = 23, farm 3); 39.6% (n = 48, farm 4); 3.4% (n = 58, farm 5); 50% (n = 24, farm 6); 70% (n = 10, farm 7) | Germany Mecklenburg–Western Pomerania | 2018 | [53] | ||
| 12.3 (n = 481) | Spain/Lugo | 2006–2016 | [23] | ||
| 16.4 (n = 694) | Austria/Vienna | 2016–2018 | [49] | ||
| 60.82 (n = 455) | China/Beijing | 2014–2016 | [42] | ||
| 7.8 (n = 129) | China/Tibet | 2012 | [43] | ||
| 47.82 (n = 23) | Bangladesh/Tangail | 2018 | [50] | ||
| 3.6 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Levofloxacin | 55.3 (n = 161) | Spain/Lugo | 2005–2017 | [22] | |
| 3.6 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Polymyxins | Colistin | 21.9 (n = 608) | China/Shanghai | 2009–2021 | [48] |
| 76.4 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| 5.9 (n = 118) | Korea | 2016–2017 | [40] | ||
| Phosphonic | Fosfomycin | 4.6 (n = 481) | Spain/Lugo | 2006–2016 | [23] |
| 2.0 (n = 694) | Austria/Vienna | 2016–2018 | [49] | ||
| 1.9 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| Phenicols | Florfenicol | 77.78 (n = 455) | China/Beijing | 2014–2016 | [42] |
| 27.9 (n = 129) | China/Tibet | 2012 | [43] | ||
| 40 (n = 55) | United States | 2013–2014 | [44] | ||
| 92.6 (n = 608) | China/Shanghai | 2009–2021 | [48] | ||
| Chloramphenicol | 58.5 (n = 481) | Spain/Lugo | 2006–2016 | [23] | |
| 18.5 (n = 694) | Austria/Vienna | 2016–2018 | [49] | ||
| 76.61 (n = 455) | China/Beijing | 2014–2016 | [42] | ||
| 57.8 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| 88.1 (n = 118) | Korea | 2016–2017 | [40] | ||
| 16.7 (n = 90) | Denmark | 2018–2019 | [51] | ||
| 1.2 (n = 168) | China/Shenzhen | 2009–2014 | [46] | ||
| Trimethoprim | 69.7 (n = 89) | Denmark/Frederiksberg C | 2014 | [47] | |
| 53.3 (n = 90) | Denmark | 2018–2019 | [51] | ||
| 13.1 (n = 168) | China/Shenzhen | 2009–2014 | [46] | ||
| Folate pathway inhibitors | Trimethoprim-sulfamethoxazole | 72.3 (n = 481) | Spain/Lugo | 2006–2016 | [42] |
| 49.5 (n = 694) | Austria/Vienna | 2016–2018 | [49] | ||
| 85.55 (n = 455) | China/Beijing | 2014–2016 | [43] | ||
| 75 (n = 135) | Santa Catarina/Brazil | 2016–2017 | [52] | ||
| 19.4 (n = 129) | China/Tibet | 2012 | [44] | ||
| 59.6 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| 56.8 (n = 118) | Korea | 2016–2017 | [40] | ||
| 30.9 (n = 55) | United States | 2013–2014 | [44] | ||
| 13.1 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Quinolone | Nalidixic acid | 60 (n = 481) | Spain/Lugo | 2006–2016 | [23] |
| 90.05 (n = 455) | China/Beijing | 2014–2016 | [42] | ||
| 19.4 (n = 129) | China/Tibet | 2012 | [43] | ||
| 87.6 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| 73.91 (n = 23) | Bangladesh/Tangail | 2018 | [50] | ||
| 61.9 (n = 118) | Korea | 2016–2017 | [40] | ||
| 8.9 (n = 90) | Denmark | 2018–2019 | [51] | ||
| 77.4 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Levofloxacin | 10.8 (n = 481) | Spain/Lugo | 2006–2016 | [23] | |
| Norfloxacin | 24.6 (n = 118) | Korea | 2016–2017 | [40] | |
| Monobactam | Aztreonam | 2.2 (n = 694) | Austria/Vienna | 2016–2018 | [49] |
| 8.1 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| Glycylcyclines | Tigecycline | 1.9 (n = 161) | Spain/Lugo | 2005–2017 | [22] |
| Quindoxin | Olaquindox | 39.77 (n = 455) | China/Beijing | 2014–2016 | [42] |
| Polymyxin | 20.47 (n = 455) | China/Beijing | 2014–2016 | [42] | |
| Nitrofurans | Nitrofurantoin | 2.34 (n = 455) | China/Beijing | 2014–2016 | [42] |
| 9.3 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| ESBL-producing isolates | 10.6 (n = 161) | Spain/Lugo | 2005–2017 | [22] | |
| MDR (≥3 categories) | 91.3 (n = 161) | Spain/Lugo | 2005–2017 | [22] | |
| MDR (≥6 categories) | 59 (n = 161) | Spain/Lugo | 2005–2017 | [22] | |
| Macrolide | Azithromycin | 78.26 (n = 23) | Bangladesh/Tangail | 2018 | [50] |
| Erythromycin | 47.82 (n = 23) | Bangladesh/Tangail | 2018 | [50] | |
| Tilmicosin | 100 (n = 55) | United States | 2013–2014 | [44] | |
| Lincomycin | Clindamycin | 100 (n = 55) | United States | 2013–2014 | [44] |
| 3-MDR | Isolates resistant to penicillin, and cephalosporins, and at least one other class of antibiotics | 36.6 (n = 41, farm 1); 32.4 (n = 34, farm 2); 87 (n = 23, farm 3); 95.8 (n = 48, farm 4); 22.4 (n = 58, farm 5); 95.8 (n = 24, farm 6); 90 (n = 10, farm 7) | Germany Mecklenburg–Western Pomerania | 2018 | [53] |
| 5-MDR | Isolates resistant to penicillin and cephalosporins and at least three other classes of antibiotics | 4.9 (n = 41, farm 1); 5.9 (n = 34, farm 2); 17.4 (n = 23, farm 3); 14.6 (n = 48, farm 4); 1.7 (n = 58, farm 5); 8.3 (n = 24, farm 6) 0 (n = 10, farm 7) | Germany Mecklenburg–Western Pomerania | 2018 | [53] |
5. Prevalence of AMR-Associated Resistance Genes in ETEC
| Antibiotic Group | Gene | % Prevalence (n = Swine Isolates) | Country/City | Year/Time Range of the Study | Reference |
|---|---|---|---|---|---|
| Aminoglycosides | aph (phosphotransferases) | 64.4% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s | [51] |
| aphA1 | 27.1% (N = 70) | Australia | 1999–2005 | [56] | |
| aadA (nucleotidyltransferases) | 63.3% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s | [51] | |
| 58.6% (n = 70) | Australia | 1999–2005 | [56] | ||
| aac (acetyltransferases) | 10% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s | [51] | |
| aac(3)-II | 18.3% (N = 71) a | Korea | 2004–2007 | [62] | |
| aac(3)-III | 31% (N = 71) a | ||||
| aac(3)-IV | 47.1% (N = 70) | Australia | 1999–2005 | [56] | |
| ant(2″)-I | 7% (N = 71) a | Korea | 2004–2007 | [62] | |
| armA | 2.8% (N = 71) a | ||||
| ant(3)-I | 93.3% (N = 104) | Australia | 1999–2005 | [26] | |
| aac(3)-IV | 47.1% (N = 104) | ||||
| aphA-I | 27.9% (N = 104) | ||||
| strA | 8% (N = 119) b | Switzerland | 2014–2015 | [24] | |
| 50% (N = 70) | Australia | 1999–2005 | [56] | ||
| strB | 16% (N = 119) b | Switzerland | 2014–2015 | [24] | |
| 55.7% (N = 70) | Australia | 1999–2005 | [56] | ||
| Beta-lactams | blaTEM-1-A | 4.4% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s | [51] |
| blaTEM-1-B | 46.7% (N = 90) | ||||
| blaTEM-30 | 1.1% (N = 90) | ||||
| blaTEM-1 | 87% (N = 119) b | Switzerland | 2014–2015 | [24] | |
| blaTEM | 43.3% (N = 104) | Australia | 1999–2005 | [26] | |
| 38% (N = 199) | United States | 2007–2008 | [66] | ||
| 40% (N = 70) | Australia | 1999–2005 | [56] | ||
| blaCMY-2 | 34% (N = 199) | United States | 2007–2008 | [66] | |
| Lincosamides | Inu(F) | 5.6 % (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s | [51] |
| Inu(G) | 5.6 % (N = 90) | ||||
| Macrolides | mdf(A) | 100% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s | [51] |
| mph(A) | 8.9% (N = 90) | ||||
| mph(B) | 7.8% (N = 90) | ||||
| erm(B) | 10% (N = 90) | ||||
| ereA | 7.1% (N = 70) | Australia | 1999–2005 | [56] | |
| Phenicols | catA1 | 3.3% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s | [51] |
| 67% (N = 119) b | Switzerland | 2014–2015 | [24] | ||
| cmlA1 | 8.9% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s | [51] | |
| floR | 5.6% (N = 90) | ||||
| catAIII | 50% (N = 119) b | Switzerland | 2014–2015 | [24] | |
| catI | 9.6% (N = 104) | Australia | 1999–2005 | [26] | |
| catII | 1% (N = 104) | ||||
| cmlA | 31.7% (N = 104) | ||||
| 12.9% (N = 70) | Australia | 1999–2005 | [56] | ||
| Polymyxins | mcr-1 | 26.4% (N = 186) | Spain | 2006–2017 | [69] |
| mcr-4 | 72.8% (N = 186) | ||||
| mcr-5 | 3.6% (N = 186) | ||||
| Sulfonamides | sul1 | 33.3 % (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s | [51] |
| 57% (N = 119) b | Switzerland | 2014–2015 | [24] | ||
| 65.4% (N = 104) | Australia | 1999–2005 | [26] | ||
| 57.1% (N = 70) | Australia | 1999–2005 | [62] | ||
| sul2 | 46.7% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s | [51] | |
| 64% (N = 119) b | Switzerland | 2014–2015 | [24] | ||
| 20.2% (N = 104) | Australia | 1999–2005 | [26] | ||
| 21.4% (N = 70) | Australia | 1999–2005 | [62] | ||
| sul3 | 10% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s | [51] | |
| 31% (N = 119) b | Switzerland | 2014–2015 | [24] | ||
| Tetracycline | tet(A) | 44.4% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s | [51] |
| 65% (N = 119) b | Switzerland | 2014–2015 | [24] | ||
| 44.2% (N = 104) | Australia | 1999–2005 | [26] | ||
| 35.7% (N = 70) | Australia | 1999–2005 | [56] | ||
| tet(B) | 14.4% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s | [51] | |
| 23% (N = 119) b | Switzerland | 2014–2015 | [24] | ||
| 28.8% (N = 104) | Australia | 1999–2005 | [26] | ||
| 7.1% (N = 70) | Australia | 1999–2005 | [56] | ||
| tet(C) | 35% (N = 119) b | Switzerland | 2014–2015 | [24] | |
| 16.3% (N = 104) | Australia | 1999–2005 | [26] | ||
| 5.7% (N = 70) | Australia | 1999–2005 | [56] | ||
| tet(D) | 3% (N = 119) b | Switzerland | 2014–2015 | [24] | |
| tet(E) | 2% (N = 119) b | Switzerland | 2014–2015 | [24] | |
| tet(X) | 1.1% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s | [51] | |
| Trimethoprim | dfrA1 | 37.8% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s | [51] |
| 59% (N = 119) b | Switzerland | 2014–2015 | [24] | ||
| dfrA5 | 2.2% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s | [51] | |
| 7% (N = 119) b | Switzerland | 2014–2015 | [24] | ||
| dfrA7 | 7% (N = 119) b | Switzerland | 2014–2015 | [24] | |
| dfrA12 | 8.9% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s | [51] | |
| 10% (N = 119) b | Switzerland | 2014–2015 | [24] | ||
| dfrA13 | 7% (N = 119) b | Switzerland | 2014–2015 | [24] | |
| dfrA14 | 5.6% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s | [51] | |
| 5% (N = 119) b | Switzerland | 2014–2015 | [24] | ||
| dfrA17 | 2.2% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s | [51] | |
| 5% (N = 119) b | Switzerland | 2014–2015 | [24] | ||
| dfrA19 | 7% (N = 119) b | Switzerland | 2014–2015 | [24] | |
| dhfrI | 1.9% (N = 104) | Australia | 1999–2005 | [26] | |
| dhfrV | 31.7% (N = 104) | Australia | 1999–2005 | [26] | |
| 25.7% (N = 70) | Australia | 1999–2005 | [56] | ||
| dhfrXIII | 30.8% (N = 104) | Australia | 1999–2005 | [26] |
Horizontal Gene Transfer
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fairbrother, J.M.; Nadeau, É. Colibacillosis. In Diseases of Swine; Zimmerman, J.J., Karriker, L.A., Ramirez, A., Schwartz, K.J., Stevenson, G.W., Zhang, J., Eds.; John Wiley & Son: Hoboken, NJ, USA, 2019; pp. 807–834. [Google Scholar]
- Luppi, A. Swine Enteric Colibacillosis: Diagnosis, Therapy and Antimicrobial Resistance. Porcine Health Manag. 2017, 3, 16. [Google Scholar] [CrossRef]
- Fairbrother, J.M.; Nadeau, É.; Gyles, C.L. Escherichia coli in Postweaning Diarrhea in Pigs: An Update on Bacterial Types, Pathogenesis, and Prevention Strategies. Anim. Health Res. Rev. 2005, 6, 17–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro, J.; Barros, M.M.; Araújo, D.; Campos, A.M.; Oliveira, R.; Silva, S.; Almeida, C. Swine Enteric Colibacillosis: Current Treatment Avenues and Future Directions. Front. Vet. Sci. 2022, 9, 981207. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Madec, J.-Y.; Lupo, A.; Schink, A.-K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial Resistance in Escherichia coli. Microbiol. Spectr. 2018, 6, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Javadi, M.; Bouzari, S.; Oloomi, M. Horizontal Gene Transfer and the Diversity of Escherichia coli. In Escherichia coli—Recent Advances on Physiology, Pathogenesis and Biotechnological Applications; InTech: London, UK, 2017; pp. 318–331. [Google Scholar]
- Nielsen, S.S.; Bicout, D.J.; Calistri, P.; Canali, E.; Drewe, J.A.; Garin-Bastuji, B.; Gonzales Rojas, J.L.; Gortázar, C.; Herskin, M.; Michel, V.; et al. Assessment of Listing and Categorisation of Animal Diseases within the Framework of the Animal Health Law (Regulation (EU) No 2016/429): Antimicrobial-Resistant Escherichia coli in Dogs and Cats, Horses, Swine, Poultry, Cattle, Sheep and Goats. EFSA J. 2022, 20, e07311. [Google Scholar]
- Gyles, C.L.; Fairbrother, J.M. Escherichia coli. In Pathogenesis of Bacterial Infections in Animals; Gyles, C.L., Prescott, J.F., Songer, J.G., Thoen, C.O., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2010; pp. 267–279. ISBN 9780813812373. [Google Scholar]
- Loayza, F.; Graham, J.P.; Trueba, G. Factors Obscuring the Role of E. coli from Domestic Animals in the Global Antimicrobial Resistance Crisis: An Evidence-Based Review. Int. J. Environ. Res. Public Health 2020, 17, 3061. [Google Scholar] [CrossRef]
- Valat, C.; Drapeau, A.; Beurlet, S.; Bachy, V.; Boulouis, H.-J.; Pin, R.; Cazeau, G.; Madec, J.-Y.; Haenni, M. Pathogenic Escherichia coli in Dogs Reveals the Predominance of ST372 and the Human-Associated ST73 Extra-Intestinal Lineages. Front. Microbiol. 2020, 11, 580. [Google Scholar] [CrossRef]
- Kim, K.; Song, M.; Liu, Y.; Ji, P. Enterotoxigenic Escherichia coli Infection of Weaned Pigs: Intestinal Challenges and Nutritional Intervention to Enhance Disease Resistance. Front. Immunol. 2022, 13, 885253. [Google Scholar] [CrossRef] [PubMed]
- Burgos, Y.; Beutin, L. Common Origin of Plasmid Encoded Alpha-Hemolysin Genes in Escherichia coli. BMC Microbiol. 2010, 10, 193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubreuil, J.D.; Isaacson, R.E.; Schifferli, D.M. Animal Enterotoxigenic Escherichia coli. EcoSal Plus 2016, 7, 1–47. [Google Scholar] [CrossRef] [Green Version]
- Dubreuil, J.D. Enterotoxigenic Escherichia coli and Probiotics in Swine: What the Bleep Do We Know? Biosci. Microbiota Food Health 2017, 36, 75–90. [Google Scholar] [CrossRef] [Green Version]
- Hampson, D.J.; Fu, Z.F.; Robertson, I.D. Investigation of the Source of Haemolytic Escherichia coli Infecting Weaned Pigs. Epidemiol. Infect. 1987, 99, 149–153. [Google Scholar] [CrossRef] [Green Version]
- Moredo, F.A.; Piñeyro, P.E.; Márquez, G.C.; Sanz, M.; Colello, R.; Etcheverría, A.; Padola, N.L.; Quiroga, M.A.; Perfumo, C.J.; Galli, L.; et al. Enterotoxigenic Escherichia coli Subclinical Infection in Pigs: Bacteriological and Genotypic Characterization and Antimicrobial Resistance Profiles. Foodborne Pathog. Dis. 2015, 12, 704–711. [Google Scholar] [CrossRef]
- van Breda, L.K.; Dhungyel, O.P.; Ward, M.P. Antibiotic Resistant Escherichia coli in Southeastern Australian Pig Herds and Implications for Surveillance. Zoonoses Public Health 2018, 65, e1–e7. [Google Scholar] [CrossRef] [PubMed]
- Luppi, A.; Gibellini, M.; Gin, T.; Vangroenweghe, F.; Vandenbroucke, V.; Bauerfeind, R.; Bonilauri, P.; Labarque, G.; Hidalgo, Á. Prevalence of Virulence Factors in Enterotoxigenic Escherichia coli Isolated from Pigs with Post-Weaning Diarrhoea in Europe. Porc. Health Manag. 2016, 2, 20. [Google Scholar] [CrossRef] [Green Version]
- Dors, A.; Czyzewska-Dors, E.; Wasyl, D.; Pomorska-Mól, M. Prevalence and Factors Associated with the Occurrence of Bacterial Enteropathogens in Suckling Piglets in Farrow-to-Finish Herds. Vet. Rec. 2016, 179, 598. [Google Scholar] [CrossRef] [PubMed]
- Ogundare, S.T.; Fasanmi, O.G.; Fasina, F.O. Risk Factors for Prevalence of Enterotoxigenic Escherichia coli (ETEC) in Diarrheic and Non-Diarrheic Neonatal and Weaner Pigs, South Africa. Biomed. Environ. Sci. 2018, 31, 149–154. [Google Scholar] [PubMed]
- Mohlatlole, R.P.; Madoroba, E.; Muchadeyi, F.C.; Chimonyo, M.; Kanengoni, A.T.; Dzomba, E.F. Virulence Profiles of Enterotoxigenic, Shiga Toxin and Enteroaggregative Escherichia coli in South African Pigs. Trop. Anim. Health Prod. 2013, 45, 1399–1405. [Google Scholar] [CrossRef]
- García-Meniño, I.; García, V.; Alonso, M.P.; Blanco, J.E.; Blanco, J.; Mora, A. Clones of Enterotoxigenic and Shiga Toxin-Producing Escherichia coli Implicated in Swine Enteric Colibacillosis in Spain and Rates of Antibiotic Resistance. Vet. Microbiol. 2021, 252, 108924. [Google Scholar] [CrossRef]
- García-Meniño, I.; García, V.; Mora, A.; Díaz-Jiménez, D.; Flament-Simon, S.C.; Alonso, M.P.; Blanco, J.E.; Blanco, M.; Blanco, J. Swine Enteric Colibacillosis in Spain: Pathogenic Potential of Mcr-1 ST10 and ST131 E. coli Isolates. Front. Microbiol. 2018, 9, 2659. [Google Scholar] [CrossRef]
- Brand, P.; Brawand, S.G.; Perreten, V. Pathotyping and Antibiotic Resistance of Porcine Enterovirulent Escherichia coli Strains from Switzerland (2014–2015). Schweiz. Arch. Tierheilkd. 2017, 159, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Marchant, M.; Moreno, M.A. Dynamics and Diversity of Escherichia coli in Animals and System Management of the Manure on a Commercial Farrow-to-Finish Pig Farm. Appl. Environ. Microbiol. 2013, 79, 853–859. [Google Scholar] [CrossRef] [Green Version]
- Smith, M.G.; Jordan, D.; Chapman, T.A.; Chin, J.J.C.; Barton, M.D.; Do, T.N.; Fahy, V.A.; Fairbrother, J.M.; Trott, D.J. Antimicrobial Resistance and Virulence Gene Profiles in Multi-Drug Resistant Enterotoxigenic Escherichia coli Isolated from Pigs with Post-Weaning Diarrhoea. Vet. Microbiol. 2010, 145, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Vu Khac, H.V.; Holoda, E.; Pilipcinec, E.; Blanco, M.; Blanco, J.E.; Mora, A.; Dahbi, G.; López, C.; González, E.A.; Blanco, J. Serotypes, Virulence Genes, and PFGE Profiles of Escherichia coli Isolated from Pigs with Postweaning Diarrhoea in Slovakia. BMC Vet. Res. 2006, 2, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frydendahl, K. Prevalence of Serogroups and Virulence Genes in Escherichia coli Associated with Postweaning Diarrhoea and Edema Disease in Pigs and a Comparison of Diagnostic Approaches. Vet. Microbiol. 2002, 85, 169–182. [Google Scholar] [CrossRef]
- Post, K.W.; Bosworth, B.T.; Knoth Post DVM, K.W.; Bosworth, B.T.; Knoth, J.L. Frequency of Virulence Factors in Escherichia Coli Isolated from Pigs with Postweaning Diarrhea and Edema Disease in North Caroline. Swine Health and Produc. 2000, 8, 1–2. [Google Scholar]
- Osek, J.; Gallien, P.; Truszczynä, M.; Protz, D. The Use of Polymerase Chain Reaction for Determination of Virulence Factors of Escherichia coli Strains Isolated from Pigs in Poland. Comp. Immunol. Microbiol. Infect. Dis. 1999, 22, 163–174. [Google Scholar] [CrossRef]
- Kusumoto, M.; Hikoda, Y.; Fujii, Y.; Murata, M.; Miyoshi, H.; Ogura, Y.; Gotoh, Y.; Iwata, T.; Hayashi, T.; Akiba, M. Emergence of a Multidrug-Resistant Shiga Toxin-Producing Enterotoxigenic Escherichia coli Lineage in Diseased Swine in Japan. J. Clin. Microbiol. 2016, 54, 1074–1081. [Google Scholar] [CrossRef] [Green Version]
- Francis, D.H. Enterotoxigenic Escherichia coli Infection in Pigs and Its Diagnosis. J. Swine Health Prod. 2002, 10, 171–175. [Google Scholar]
- Baldo, V.; Salogni, C.; Giovannini, S.; D’Incau, M.; Boniotti, M.B.; Birbes, L.; Pitozzi, A.; Formenti, N.; Grassi, A.; Pasquali, P.; et al. Pathogenicity of Shiga Toxin Type 2e Escherichia Coli in Pig Colibacillosis. Front. Vet. Sci. 2020, 7, 545818. [Google Scholar] [CrossRef]
- Ikwap, K.; Larsson, J.; Jacobson, M.; Owiny, D.O.; Nasinyama, G.W.; Nabukenya, I.; Mattsson, S.; Aspan, A.; Erume, J. Prevalence of Adhesin and Toxin Genes in E. coli Strains Isolated from Diarrheic and Non-Diarrheic Pigs from Smallholder Herds in Northern and Eastern Uganda. BMC Microbiol. 2016, 16, 178. [Google Scholar] [CrossRef] [Green Version]
- Madoroba, E.; van Driessche, E.; de Greve, H.; Mast, J.; Ncube, I.; Read, J.; Beeckmans, S. Prevalence of Enterotoxigenic Escherichia coli Virulence Genes from Scouring Piglets in Zimbabwe. Trop. Anim. Health Prod. 2009, 41, 1539–1547. [Google Scholar] [CrossRef]
- Pissetti, C.; Kich, J.D.; Allen, H.K.; Navarrete, C.; de Freitas Costa, E.; Morés, N.; Cardoso, M. Antimicrobial Resistance in Commensal Escherichia coli and Enterococcus spp. Isolated from Pigs Subjected to Different Antimicrobial Administration Protocols. Res. Vet. Sci. 2021, 137, 174–185. [Google Scholar] [CrossRef]
- Public Health Producer Guide. Antibiotics on the Farm: What You Need to Know about New Regulations Overview FDA’s New Regulations; National Pork Board: Des Moines, IA, USA, 2015; 800-456-7675.
- Holman, D.B.; Chénier, M.R. Antimicrobial Use in Swine Production and Its Effect on the Swine Gut Microbiota and Antimicrobial Resistance. Can. J. Microbiol. 2015, 61, 785–798. [Google Scholar] [CrossRef] [PubMed]
- Diário Oficial da União Instrução Normativa N47, de 22 de Novembro de 2016. Available online: http://www.in.gov.br/autenticidade.html (accessed on 11 January 2023).
- Do, K.H.; Byun, J.W.; Lee, W.K. Virulence Genes and Antimicrobial Resistance of Pathogenic Escherichia coli Isolated from Diarrheic Weaned Piglets in Korea. J. Anim. Sci. Technol. 2020, 62, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.Y.; Guo, L.; Su, J.H.; Zhu, Y.H.; Jiao, L.G.; Wang, J.F. Frequency of Diarrheagenic Virulence Genes and Characteristics in Escherichia coli Isolates from Pigs with Diarrhea in China. Microorganisms 2019, 7, 308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Wu, D.; Liu, K.; Suolang, S.; He, T.; Liu, X.; Wu, C.; Wang, Y.; Lin, D. Investigation of Antimicrobial Resistance in Escherichia coli and Enterococci Isolated from Tibetan Pigs. PLoS ONE 2014, 9, e95623. [Google Scholar] [CrossRef] [Green Version]
- Jiang, F.; Wu, Z.; Zheng, Y.; Frana, T.S.; Sahin, O.; Zhang, Q.; Li, G. Genotypes and Antimicrobial Susceptibility Profiles of Hemolytic Escherichia coli from Diarrheic Piglets. Foodborne Pathog. Dis. 2019, 16, 94–103. [Google Scholar] [CrossRef]
- Li, Y.; Luo, Q.; Shi, X.; Lin, Y.; Qiu, Y.; Lv, D.; Jiang, Y.; Chen, Q.; Jiang, M.; Ma, H.; et al. Phenotypic and Genotypic Characterization of Clinical Enterotoxigenic Escherichia coli Isolates from Shenzhen, China. Foodborne Pathog. Dis. 2017, 14, 333–340. [Google Scholar] [CrossRef]
- Rosager, W.N.; Peter, N.J.; Erik Lind, J.S.; Svend, H.; Matthew, D.; Steen, P.K. Comparison of Antimicrobial Resistance in E. coli Isolated from Rectal and Floor Samples in Pens with Diarrhoeic Nursery Pigs in Denmark. Prev. Vet. Med. 2017, 147, 42–49. [Google Scholar] [CrossRef] [Green Version]
- Stannarius, C.; Bürgi, E.; Regula, G.; Zychowska, M.A.; Zweifel, C.; Stephan, R. Antimicrobial Resistance in Escherichia coli Strains Isolated from Swiss Weaned Pigs and Sows. Schweiz. Arch. Tierheilkd. 2009, 151, 119–125. [Google Scholar] [CrossRef] [Green Version]
- Lv, C.; Shang, J.; Zhang, W.; Sun, B.; Li, M.; Guo, C.; Zhou, N.; Guo, X.; Huang, S.; Zhu, Y. Dynamic Antimicrobial Resistant Patterns of Escherichia coli from Healthy Poultry and Swine over 10 Years in Chongming Island, Shanghai. Infect. Dis. Poverty 2022, 11, 98. [Google Scholar] [CrossRef]
- Renzhammer, R.; Loncaric, I.; Roch, F.F.; Pinior, B.; Käsbohrer, A.; Spergser, J.; Ladinig, A.; Unterweger, C. Prevalence of Virulence Genes and Antimicrobial Resistances in E. coli Associated with Neonatal Diarrhea, Postweaning Diarrhea, and Edema Disease in Pigs from Austria. Antibiotics 2020, 9, 208. [Google Scholar] [CrossRef]
- Rahman, M.M.; Ahmed, P.; Kar, A.; Sakib, N.; Shibly, A.Z.; Zohora, F.T.; Hasan, M.N. Prevalence, Antimicrobial Resistance, and Pathogenic Potential of Enterotoxigenic and Enteropathogenic Escherichia coli Associated with Acute Diarrheal Patients in Tangail, Bangladesh. Foodborne Pathog. Dis. 2020, 17, 434–439. [Google Scholar] [CrossRef]
- García, V.; Gambino, M.; Pedersen, K.; Haugegaard, S.; Olsen, J.E.; Herrero-Fresno, A. F4- and F18-Positive Enterotoxigenic Escherichia coli Isolates from Diarrhea of Postweaning Pigs: Genomic Characterization. Appl. Environ. Microbiol. 2020, 86, e01913-20. [Google Scholar] [CrossRef] [PubMed]
- Brisola, M.C.; Crecencio, R.B.; Bitner, D.S.; Frigo, A.; Rampazzo, L.; Stefani, L.M.; Faria, G.A. Escherichia coli Used as a Biomarker of Antimicrobial Resistance in Pig Farms of Southern Brazil. Sci. Total Environ. 2019, 647, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Meissner, K.; Sauter-Louis, C.; Heiden, S.E.; Schaufler, K.; Tomaso, H.; Conraths, F.J.; Homeier-Bachmann, T. Extended-Spectrum ß-Lactamase-Producing Escherichia coli in Conventional and Organic Pig Fattening Farms. Microorganisms 2022, 10, 603. [Google Scholar] [CrossRef] [PubMed]
- Boerlin, P.; Travis, R.; Gyles, C.L.; Reid-Smith, R.; Janecko, N.; Lim, H.; Nicholson, V.; McEwen, S.A.; Friendship, R.; Archambault, M. Antimicrobial Resistance and Virulence Genes of Escherichia coli Isolates from Swine in Ontario. Appl. Environ. Microbiol. 2005, 71, 6753–6761. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.M.; Jiang, H.X.; Liao, X.P.; Liu, J.H.; Zhang, W.J.; Zhang, H.; Jiang, Z.G.; Lü, D.H.; Xiang, R.; Liu, Y.H. Antimicrobial Resistance, Virulence Genes, and Phylogenetic Background in Escherichia coli Isolates from Diseased Pigs. FEMS Microbiol. Lett. 2010, 306, 15–21. [Google Scholar] [CrossRef] [Green Version]
- Shepard, S.M.; Danzeisen, J.L.; Isaacson, R.E.; Seemann, T.; Achtman, M.; Johnson, T.J. Genome Sequences and Phylogenetic Analysis of K88- and F18-Positive Porcine Enterotoxigenic Escherichia coli. J. Bacteriol. 2012, 194, 395–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, T.J.; Nolan, L.K. Pathogenomics of the Virulence Plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 2009, 73, 750–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes, T.A.T.; Elias, W.P.; Scaletsky, I.C.A.; Guth, B.E.C.; Rodrigues, J.F.; Piazza, R.M.F.; Ferreira, L.C.S.; Martinez, M.B. Diarrheagenic Escherichia coli. Braz. J. Microbiol. 2016, 47, 3–30. [Google Scholar] [CrossRef] [Green Version]
- Olasz, F.; Fekete, P.Z.; Blum-Oehler, G.; Boldogkoi, Z.; Nagy, B. Characterization of an F18+ Enterotoxigenic Escherichia coli Strain from Post Weaning Diarrhoea of Swine, and of Its Conjugative Virulence Plasmid PTC. FEMS Microbiol. Lett. 2005, 244, 281–289. [Google Scholar] [CrossRef]
- Goswami, P.S.; Gyles, C.L.; Friendship, R.M.; Poppe, C.; Kozak, G.K.; Boerlin, P. Effect of Plasmid PTENT2 on Severity of Porcine Post-Weaning Diarrhoea Induced by an O149 Enterotoxigenic Escherichia coli. Vet. Microbiol. 2008, 131, 400–405. [Google Scholar] [CrossRef]
- Martínez, J.L.; Baquero, F. Interactions among Strategies Associated with Bacterial Infection: Pathogenicity, Epidemicity, and Antibiotic Resistance. Clin. Microbiol. Rev. 2002, 15, 647–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, M.-J.; Lim, S.-K.; Nam, H.-M.; Kim, A.-R.; Jung, S.-C.; Kim, M.-N. Apramycin and Gentamicin Resistances in Indicator and Clinical Escherichia coli Isolates from Farm Animals in Korea. Foodborne Pathog. Dis. 2011, 8, 119–123. [Google Scholar] [CrossRef]
- Bortolaia, V.; Guardabassi, L.; Trevisani, M.; Bisgaard, M.; Venturi, L.; Bojesen, A.M. High Diversity of Extended-Spectrum β-Lactamases in Escherichia coli Isolates from Italian Broiler Flocks. Antimicrob. Agents Chemother. 2010, 54, 1623–1626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diestra, K.; Juan, C.; Curiao, T.; Moyá, B.; Miró, E.; Oteo, J.; Coque, T.M.; Pérez-Vázquez, M.; Campos, J.; Cantón, R.; et al. Characterization of Plasmids Encoding BlaESBL and Surrounding Genes in Spanish Clinical Isolates of Escherichia coli and Klebsiella pneumoniae. J. Antimicrob. Chemother. 2009, 63, 60–66. [Google Scholar] [CrossRef] [Green Version]
- Cloeckaert, A.; Praud, K.; Doublet, B.; Bertini, A.; Carattoli, A.; Butaye, P.; Imberechts, H.; Bertrand, S.; Collard, J.M.; Arlet, G.; et al. Dissemination of an Extended-Spectrum-β-Lactamase BlaTEM-52 Gene-Carrying IncI1 Plasmid in Various Salmonella enterica Serovars Isolated from Poultry and Humans in Belgium and France between 2001 and 2005. Antimicrob. Agents Chemother. 2007, 51, 1872–1875. [Google Scholar] [CrossRef] [Green Version]
- Johnson, T.J.; Shepard, S.M.; Rivet, B.; Danzeisen, J.L.; Carattoli, A. Comparative Genomics and Phylogeny of the IncI1 Plasmids: A Common Plasmid Type among Porcine Enterotoxigenic Escherichia coli. Plasmid 2011, 66, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.C.; Turnidge, J.; Collignon, P.; Looke, D.; Barton, M.; Gottlieb, T. Control of Fluoroquinolone Resistance through Successful Regulation, Australia. Emerg. Infect. Dis. 2012, 18, 1453–1460. [Google Scholar] [CrossRef] [PubMed]
- Jordan, D.; Chin, J.J.C.; Fahy, V.A.; Barton, M.D.; Smith, M.G.; Trott, D.J. Antimicrobial Use in the Australian Pig Industry: Results of a National Survey. Aust. Vet. J. 2009, 87, 222–229. [Google Scholar] [CrossRef] [PubMed]
- García, V.; García-Meniño, I.; Mora, A.; Flament-Simon, S.C.; Díaz-Jiménez, D.; Blanco, J.E.; Alonso, M.P.; Blanco, J. Co-Occurrence of Mcr-1, Mcr-4 and Mcr-5 Genes in Multidrug-Resistant ST10 Enterotoxigenic and Shiga Toxin-Producing Escherichia coli in Spain (2006–2017). Int. J. Antimicrob. Agents 2018, 52, 104–108. [Google Scholar] [CrossRef]
- Sahl, J.W.; Steinsland, H.; Redman, J.C.; Angiuoli, S.v.; Nataro, J.P.; Sommerfelt, H.; Rasko, D.A. A Comparative Genomic Analysis of Diverse Clonal Types of Enterotoxigenic Escherichia coli Reveals Pathovar-Specific Conservation. Infect. Immun. 2011, 79, 950–960. [Google Scholar] [CrossRef] [Green Version]
- Rasko, D.A.; Rosovitz, M.J.; Myers, G.S.A.; Mongodin, E.F.; Fricke, W.F.; Gajer, P.; Crabtree, J.; Sebaihia, M.; Thomson, N.R.; Chaudhuri, R.; et al. The Pangenome Structure of Escherichia coli: Comparative Genomic Analysis of E. coli Commensal and Pathogenic Isolates. J. Bacteriol. 2008, 190, 6881–6893. [Google Scholar] [CrossRef] [Green Version]
- Cooper, K.K.; Mandrell, R.E.; Louie, J.W.; Korlach, J.; Clark, T.A.; Parker, C.T.; Huynh, S.; Chain, P.S.; Ahmed, S.; Carter, M.Q. Comparative Genomics of Enterohemorrhagic Escherichia coli O145:H28 Demonstrates a Common Evolutionary Lineage with Escherichia coli O157:H7. BMC Genom. 2014, 15, 17. [Google Scholar] [CrossRef] [Green Version]
- Ogura, Y.; Ooka, T.; Iguchi, A.; Toh, H.; Asadulghani, M.; Oshima, K.; Kodama, T.; Abe, H.; Nakayama, K.; Kurokawa, K.; et al. Comparative Genomics Reveal the Mechanism of the Parallel Evolution of O157 and Non-O157 Enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. USA 2009, 106, 17939–17944. [Google Scholar] [CrossRef] [Green Version]
- Nyholm, O.; Halkilahti, J.; Wiklund, G.; Okeke, U.; Paulin, L.; Auvinen, P.; Haukka, K.; Siitonen, A. Comparative Genomics and Characterization of Hybrid Shigatoxigenic and Enterotoxigenic Escherichia coli (STEC/ETEC) Strains. PLoS ONE 2015, 10, e0135936. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Savarino, S.J.; Venkatesan, M.M. Subtractive Hybridization and Optical Mapping of the Enterotoxigenic Escherichia coli H10407 Chromosome: Isolation of Unique Sequences and Demonstration of Significant Similarity to the Chromosome of E. coli K-12. Microbiology 2006, 152, 1041–1054. [Google Scholar] [CrossRef] [Green Version]
- von Mentzer, A.; Connor, T.R.; Wieler, L.H.; Semmler, T.; Iguchi, A.; Thomson, N.R.; Rasko, D.A.; Joffre, E.; Corander, J.; Pickard, D.; et al. Identification of Enterotoxigenic Escherichia coli (ETEC) Clades with Long-Term Global Distribution. Nat. Genet. 2014, 46, 1321–1326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, S.S.; Bicout, D.J.; Calistri, P.; Canali, E.; Drewe, J.A.; Garin-Bastuji, B.; Gonzales Rojas, J.L.; Gortazar Schmidt, C.; Herskin, M.; Michel, V.; et al. Assessment of Animal Diseases Caused by Bacteria Resistant to Antimicrobials: Swine. EFSA J. 2021, 19, e06955. [Google Scholar] [PubMed]
| Country | Prevalence of ETEC (%) (n = Number of Isolates) | Sampling Information/Origin | Period | Reference |
|---|---|---|---|---|
| Argentina | 15.2 (n = 990) | 11 farms with no history or clinical signs of colibacillosis | 2015 | [16] |
| Australia | 58.8 (n = 325) | 22 pig herds | 2013–2014 | [17] |
| Belgium and the Netherlands | 36.4 (n = 160) | 88 farms | 2012–2014 | [18] |
| France | 64.8 (n = 455) | 91 farms | 2012–2014 | [18] |
| Germany | 47.1 (n = 99) | 17 farms | 2012–2014 | [18] |
| Italy | 81.0 (n = 159) | 84 farms | 2012–2014 | [18] |
| Poland | 30 (n = 386) a | 70 pig herds | 2011–2013 | [19] |
| South Africa | 72.0 (n = 228) | 8 piggeries of different sizes (16–650 sow units) and production systems: large-scale commercial (>250 sow units), medium-scale commercial (51–250 sow units), and emerging small-scale pig farms (<50 sow units) | 2015–2016 | [20] |
| South Africa | 18.6 (n = 263) | 263 neonatal and post-weaned pigs | 2013 | [21] |
| Spain | 86.5 (n = 186) | 50 different Spanish farms | 2005–2017 | [22] |
| Spain | 67.0 (n = 499) | 179 outbreaks | 2008–2018 | [23] |
| Switzerland | 50.4 (n = 131) | 115 pigs suffering from diarrhea | 2014–2015 | [24] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barros, M.M.; Castro, J.; Araújo, D.; Campos, A.M.; Oliveira, R.; Silva, S.; Outor-Monteiro, D.; Almeida, C. Swine Colibacillosis: Global Epidemiologic and Antimicrobial Scenario. Antibiotics 2023, 12, 682. https://doi.org/10.3390/antibiotics12040682
Barros MM, Castro J, Araújo D, Campos AM, Oliveira R, Silva S, Outor-Monteiro D, Almeida C. Swine Colibacillosis: Global Epidemiologic and Antimicrobial Scenario. Antibiotics. 2023; 12(4):682. https://doi.org/10.3390/antibiotics12040682
Chicago/Turabian StyleBarros, Maria Margarida, Joana Castro, Daniela Araújo, Ana Maria Campos, Ricardo Oliveira, Sónia Silva, Divanildo Outor-Monteiro, and Carina Almeida. 2023. "Swine Colibacillosis: Global Epidemiologic and Antimicrobial Scenario" Antibiotics 12, no. 4: 682. https://doi.org/10.3390/antibiotics12040682
APA StyleBarros, M. M., Castro, J., Araújo, D., Campos, A. M., Oliveira, R., Silva, S., Outor-Monteiro, D., & Almeida, C. (2023). Swine Colibacillosis: Global Epidemiologic and Antimicrobial Scenario. Antibiotics, 12(4), 682. https://doi.org/10.3390/antibiotics12040682












