Insight into the Mechanisms of Carbapenem Resistance in Klebsiella pneumoniae: A Study on IS26 Integrons, Beta-Lactamases, Porin Modifications, and Plasmidome Analysis
Abstract
:1. Introduction
2. Results
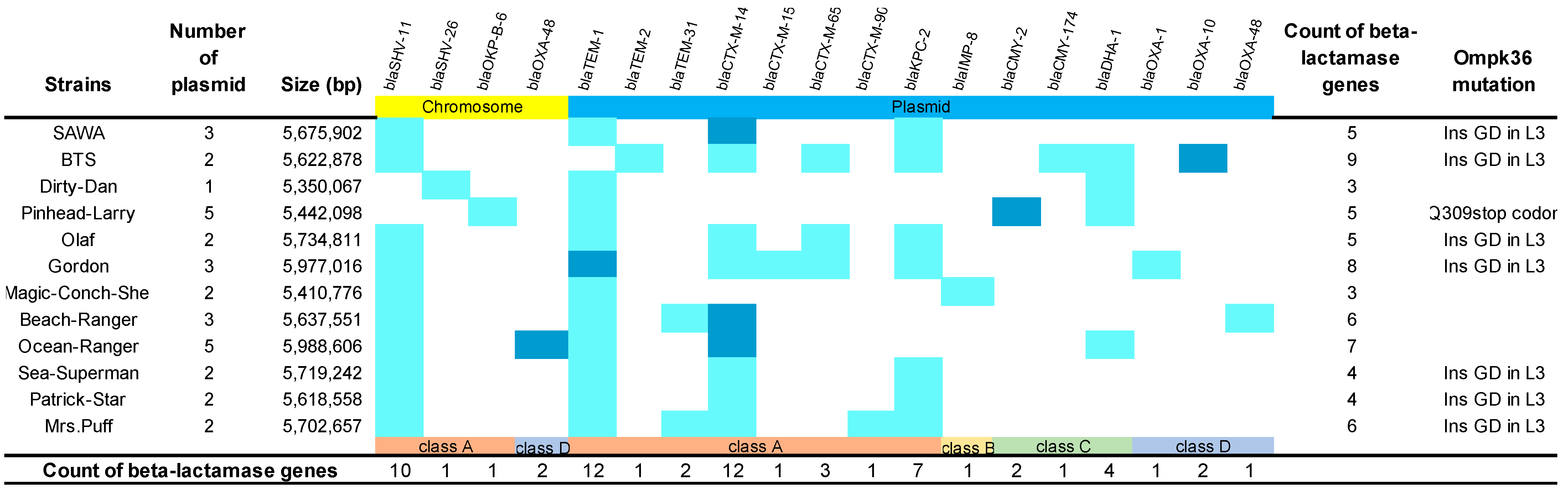
2.1. Distribution of Carbapenem-Resistant Determinants in K. pneumoniae
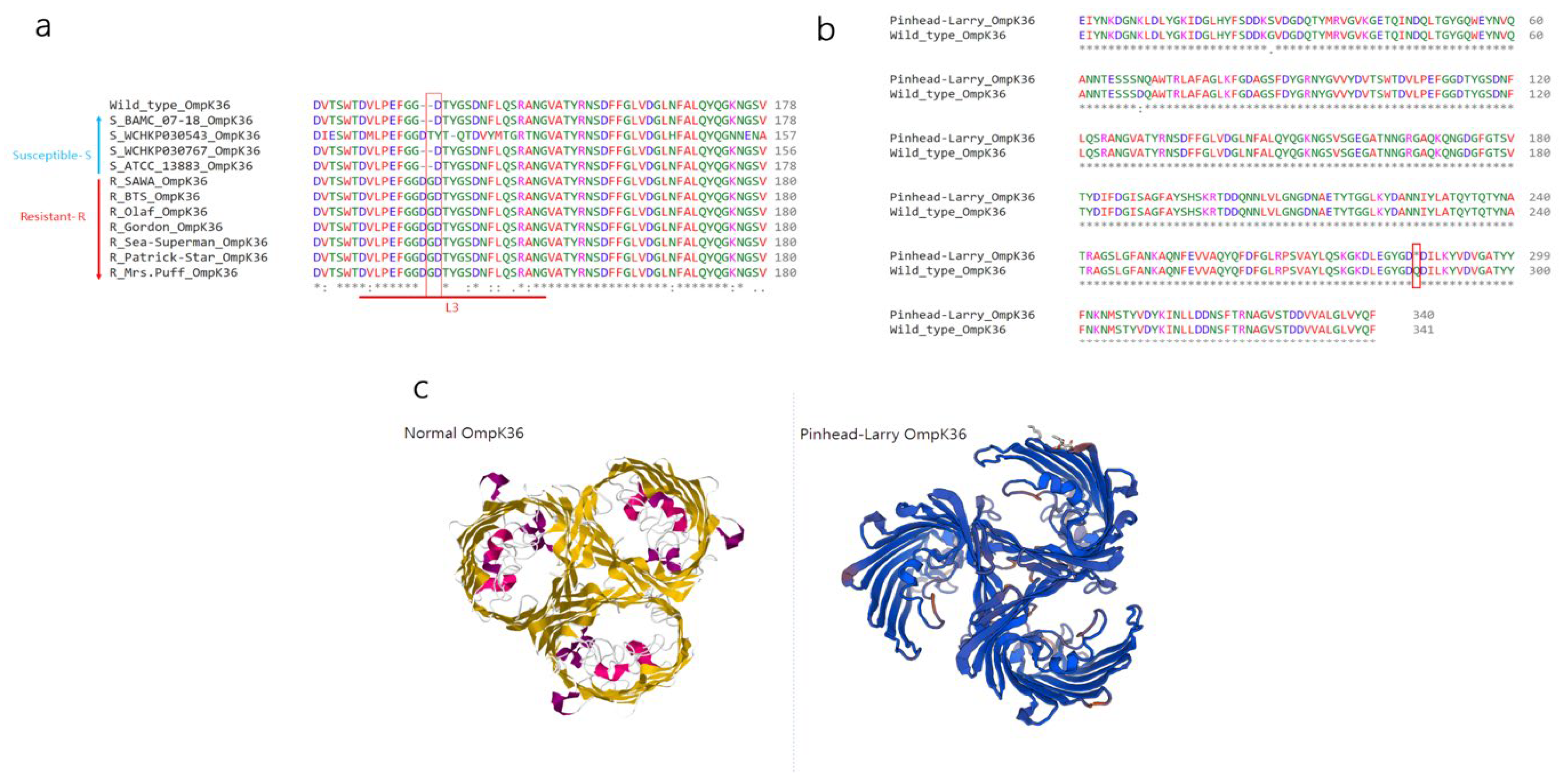
2.2. Investigation of Novel Stop Codon Associated with Carbapenemase-Negative CRKP
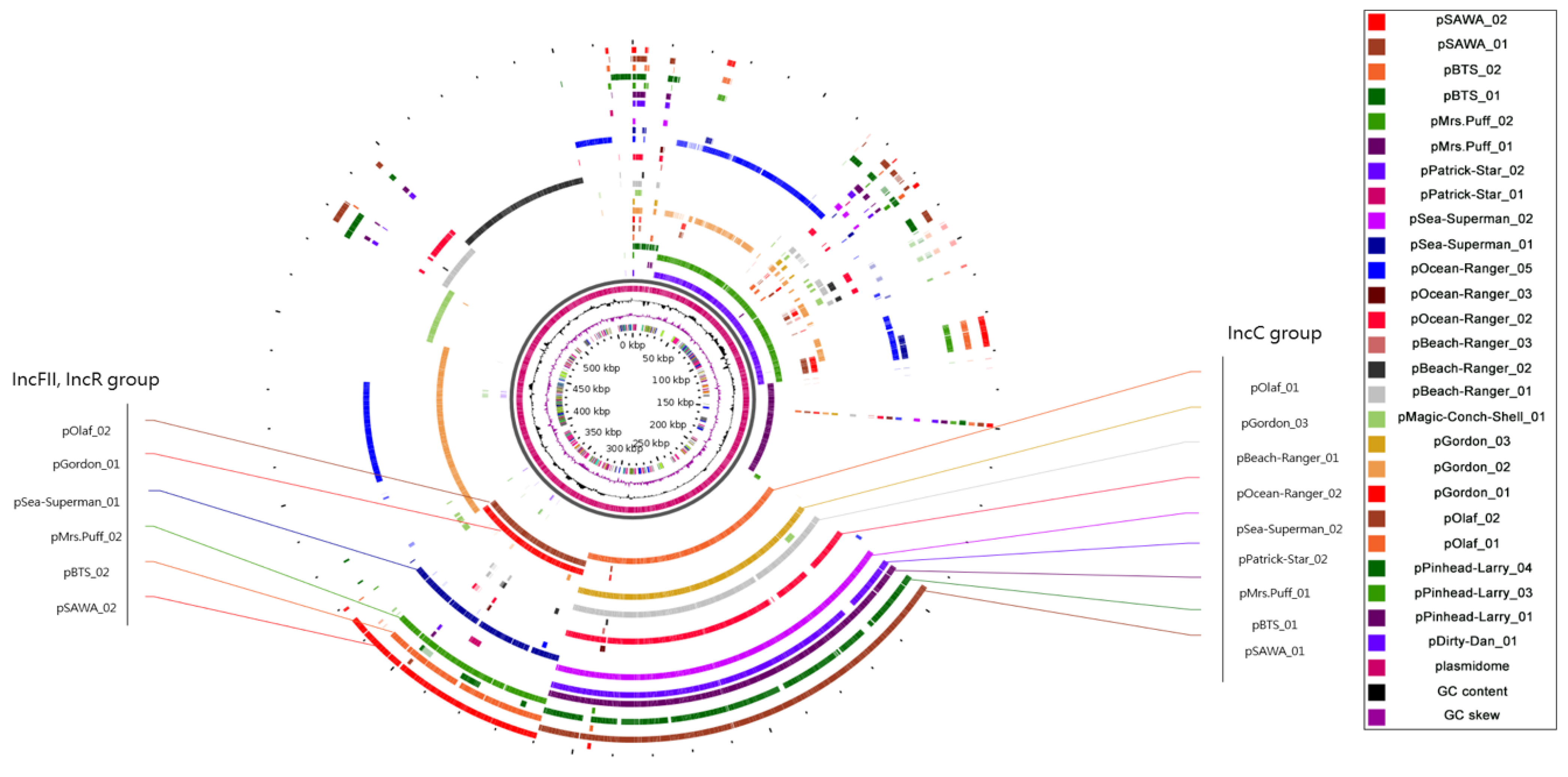
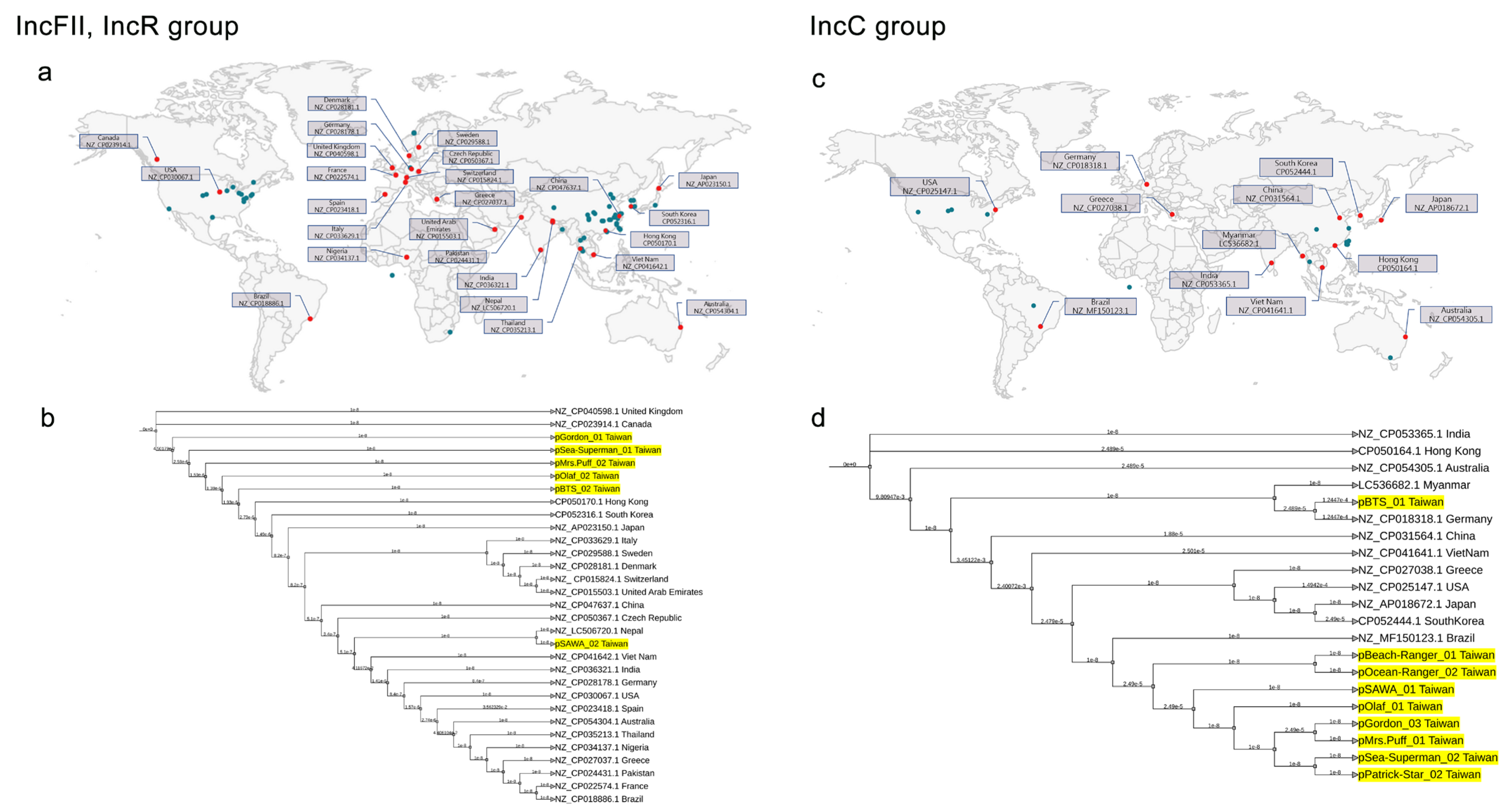
2.3. Plasmidome Analysis Reveals the Importance of IncFII/IncR and IncC in CRKP
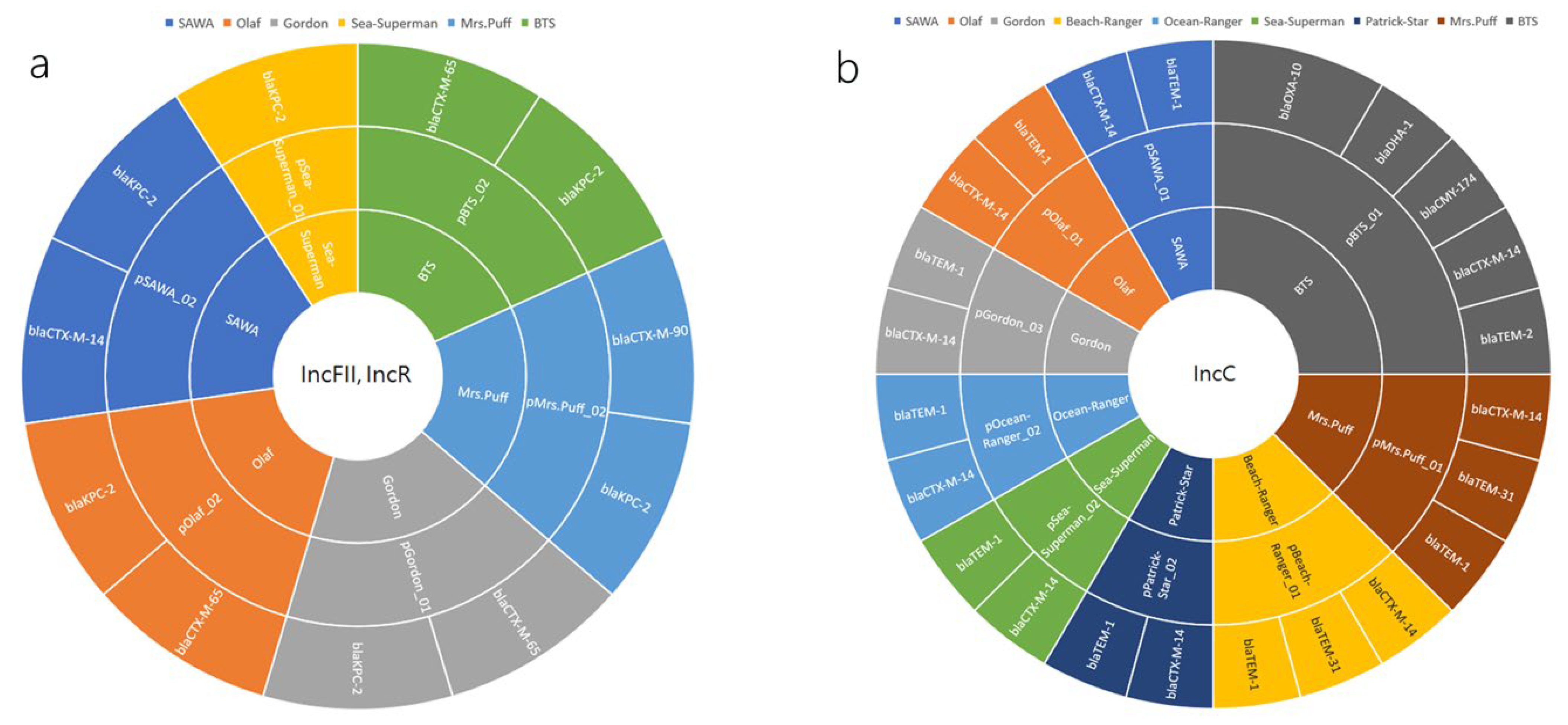
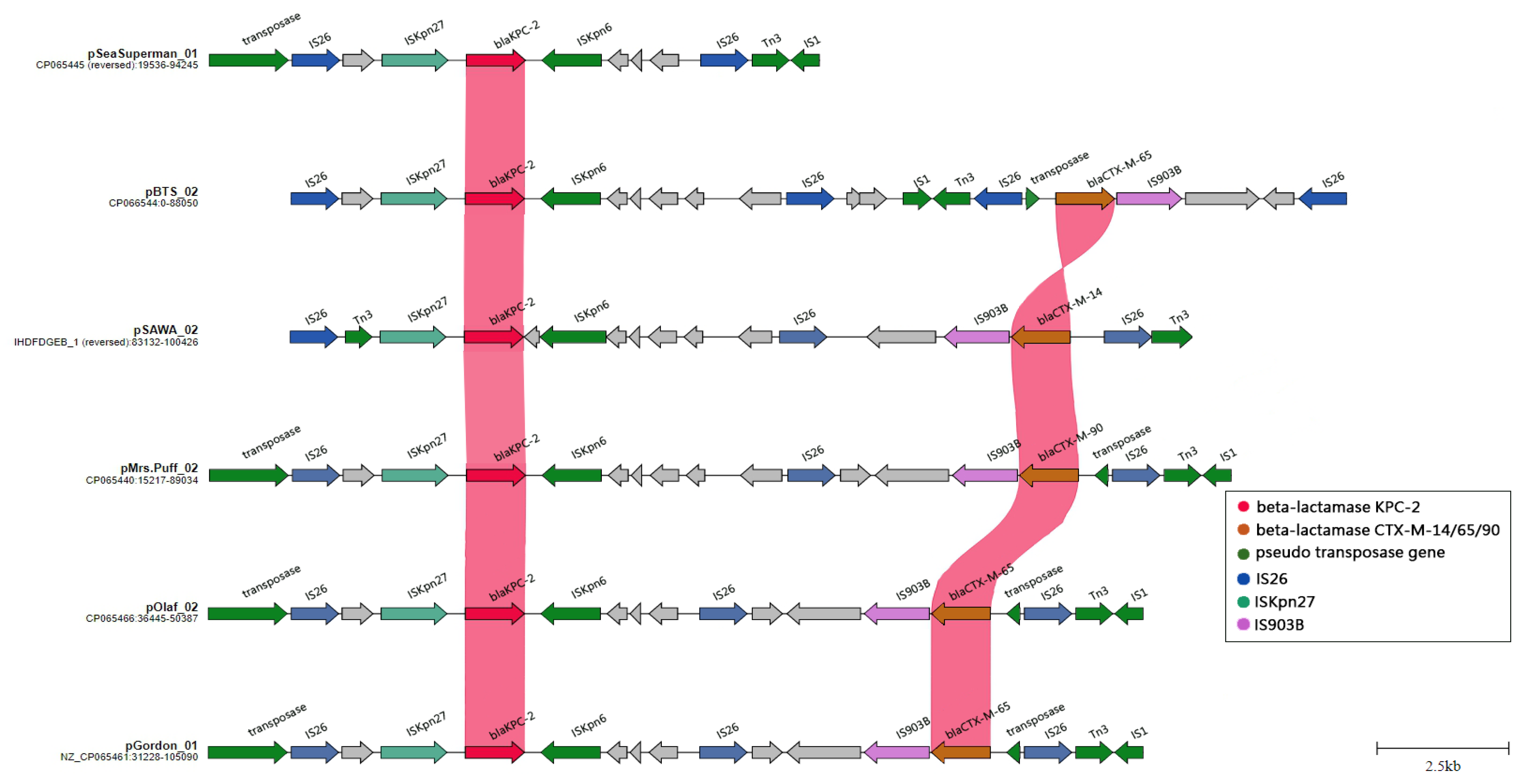
2.4. Plasmid-Mediated Dissemination of β-Lactamase Resistance Determinants in CRKP: A Closer Look at IncFII/IncR and IncC Plasmid Groups
2.5. Global Phylogenetic Analysis of Plasmids in CRKP: Evidence of Endemic IncC in Taiwan
2.6. The Evolution and Expansion of IS26 Integrons: A Driving Force in Carbapenem Resistance
3. Discussion
4. Materials and Methods
4.1. Bacterial Isolates and Antibiotic Susceptibility Testing
4.2. DNA Extraction, Whole-Genome Sequencing, Assembly, and Prediction of Antimicrobial Resistance Determinants
4.3. Analysis of Plasmidome
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Van Duin, D.; Paterson, D.L. Multidrug-Resistant Bacteria in the Community: An Update. Infect. Dis. Clin. 2020, 34, 709–722. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.L.C.; Romano, M.; Kerry, L.E.; Kwong, H.S.; Low, W.W.; Brett, S.J.; Clements, A.; Beis, K.; Frankel, G. OmpK36-mediated Carbapenem resistance attenuates ST258 Klebsiella pneumoniae in vivo. Nat. Commun. 2019, 10, 3957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-David, D.; Kordevani, R.; Keller, N.; Tal, I.; Marzel, A.; Gal-Mor, O.; Maor, Y.; Rahav, G. Outcome of carbapenem resistant Klebsiella pneumoniae bloodstream infections. Clin. Microbiol. Infect. 2012, 18, 54–60. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Li, Y.; Song, N.; Chen, Y. Risk factors for carbapenem-resistant Klebsiella pneumoniae infection: A meta-analysis. J. Glob. Antimicrob. Resist. 2020, 21, 306–313. [Google Scholar] [CrossRef] [PubMed]
- David, S.; Reuter, S.; Harris, S.R.; Glasner, C.; Feltwell, T.; Argimon, S.; Abudahab, K.; Goater, R.; Giani, T.; Errico, G.; et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat. Microbiol. 2019, 4, 1919–1929. [Google Scholar] [CrossRef]
- Ramadan, R.A.; Bedawy, A.M.; Negm, E.M.; Hassan, T.H.; Ibrahim, D.A.; ElSheikh, S.M.; Amer, R.M. Carbapenem-Resistant Klebsiella pneumoniae Among Patients with Ventilator-Associated Pneumonia: Evaluation of Antibiotic Combinations and Susceptibility to New Antibiotics. Infect. Drug Resist. 2022, 15, 3537–3548. [Google Scholar] [CrossRef]
- Bulman, Z.P.; Krapp, F.; Pincus, N.B.; Wenzler, E.; Murphy, K.R.; Qi, C.; Ozer, E.A.; Hauser, A.R. Genomic Features Associated with the Degree of Phenotypic Resistance to Carbapenems in Carbapenem-Resistant Klebsiella pneumoniae. mSystems 2021, 6, e0019421. [Google Scholar] [CrossRef]
- Ghafourian, S.; Sadeghifard, N.; Soheili, S.; Sekawi, Z. Extended Spectrum Beta-lactamases: Definition, Classification and Epidemiology. Curr. Issues Mol. Biol. 2015, 17, 11–21. [Google Scholar]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-spectrum beta-lactamases: An update on their characteristics, epidemiology and detection. JAC Antimicrob. Resist. 2021, 3, dlab092. [Google Scholar] [CrossRef]
- Codjoe, F.S.; Donkor, E.S. Carbapenem Resistance: A Review. Med. Sci. 2017, 6, 1. [Google Scholar] [CrossRef] [Green Version]
- Temkin, E.; Adler, A.; Lerner, A.; Carmeli, Y. Carbapenem-resistant Enterobacteriaceae: Biology, epidemiology, and management. Ann. N. Y. Acad. Sci. 2014, 1323, 22–42. [Google Scholar] [CrossRef]
- Bush, K.; Jacoby, G.A. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- David, S.; Wong, J.L.C.; Sanchez-Garrido, J.; Kwong, H.S.; Low, W.W.; Morecchiato, F.; Giani, T.; Rossolini, G.M.; Brett, S.J.; Clements, A.; et al. Widespread emergence of OmpK36 loop 3 insertions among multidrug-resistant clones of Klebsiella pneumoniae. PLoS Pathog. 2022, 18, e1010334. [Google Scholar] [CrossRef]
- Zeng, L.; Zhang, J.; Hu, K.; Li, J.; Wang, J.; Yang, C.; Huang, W.; Yin, L.; Zhang, X. Microbial Characteristics and Genomic Analysis of an ST11 Carbapenem-Resistant Klebsiella pneumoniae Strain Carrying bla (KPC-2) Conjugative Drug-Resistant Plasmid. Front. Public Health 2021, 9, 809753. [Google Scholar] [CrossRef]
- Qu, D.; Shen, Y.; Hu, L.; Jiang, X.; Yin, Z.; Gao, B.; Zhao, Y.; Yang, W.; Yang, H.; Han, J.; et al. Comparative analysis of KPC-2-encoding chimera plasmids with multi-replicon IncR:Inc(pA1763-KPC):IncN1 or IncFII(pHN7A8):Inc(pA1763-KPC):IncN1. Infect. Drug Resist. 2019, 12, 285–296. [Google Scholar] [CrossRef] [Green Version]
- Hirabayashi, A.; Dao, T.D.; Takemura, T.; Hasebe, F.; Trang, L.T.; Thanh, N.H.; Tran, H.H.; Shibayama, K.; Kasuga, I.; Suzuki, M. A Transferable IncC-IncX3 Hybrid Plasmid Cocarrying bla(NDM-4), tet(X), and tmexCD3-toprJ3 Confers Resistance to Carbapenem and Tigecycline. mSphere 2021, 6, e0059221. [Google Scholar] [CrossRef]
- Roch, M.; Sierra, R.; Sands, K.; Martins, W.; Schrenzel, J.; Walsh, T.R.; Gales, A.C.; Andrey, D.O. Vertical and horizontal dissemination of an IncC plasmid harbouring rmtB 16S rRNA methylase gene, conferring resistance to plazomicin, among invasive ST258 and ST16 KPC-producing Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 2021, 24, 183–189. [Google Scholar] [CrossRef]
- Farzana, R.; Jones, L.S.; Rahman, M.A.; Sands, K.; van Tonder, A.J.; Portal, E.; Criollo, J.M.; Parkhill, J.; Guest, M.F.; Watkins, W.J.; et al. Genomic Insights into the Mechanism of Carbapenem Resistance Dissemination in Enterobacterales From a Tertiary Public Heath Setting in South Asia. Clin. Infect. Dis. 2023, 76, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Shropshire, W.C.; Aitken, S.L.; Pifer, R.; Kim, J.; Bhatti, M.M.; Li, X.; Kalia, A.; Galloway-Peña, J.; Sahasrabhojane, P.; Arias, C.A.; et al. IS26-mediated amplification of blaOXA-1 and blaCTX-M-15 with concurrent outer membrane porin disruption associated with de novo carbapenem resistance in a recurrent bacteraemia cohort. J. Antimicrob. Chemother. 2021, 76, 385–395. [Google Scholar] [CrossRef]
- Adler, M.; Anjum, M.; Andersson, D.I.; Sandegren, L. Influence of acquired β-lactamases on the evolution of spontaneous carbapenem resistance in Escherichia coli. J. Antimicrob. Chemother. 2013, 68, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Goessens, W.H.F.; van der Bij, A.K.; van Boxtel, R.; Pitout, J.D.D.; van Ulsen, P.; Melles, D.C.; Tommassen, J. Antibiotic trapping by plasmid-encoded CMY-2 β-lactamase combined with reduced outer membrane permeability as a mechanism of carbapenem resistance in Escherichia coli. Antimicrob. Agents Chemother. 2013, 57, 3941–3949. [Google Scholar] [CrossRef] [Green Version]
- Van Boxtel, R.; Wattel, A.A.; Arenas, J.; Goessens, W.H.F.; Tommassen, J. Acquisition of Carbapenem Resistance by Plasmid-Encoded-AmpC-Expressing Escherichia coli. Antimicrob. Agents Chemother. 2017, 61, e01413-16. [Google Scholar] [CrossRef] [Green Version]
- Shropshire, W.C.; Konovalova, A.; McDaneld, P.; Gohel, M.; Strope, B.; Sahasrabhojane, P.; Tran, C.N.; Greenberg, D.; Kim, J.; Zhan, X.; et al. Systematic Analysis of Mobile Genetic Elements Mediating β-lactamase Gene Amplification in Non-Carbapenemase-Producing Carbapenem Resistant Enterobacterales Bloodstream Infections. bioRxiv 2022, 7, e00476-22. [Google Scholar] [CrossRef]
- Wong, J.L.C.; David, S.; Sanchez-Garrido, J.; Woo, J.Z.; Low, W.W.; Morecchiato, F.; Giani, T.; Rossolini, G.M.; Beis, K.; Brett, S.J.; et al. Recurrent emergence of Klebsiella pneumoniae carbapenem resistance mediated by an inhibitory ompK36 mRNA secondary structure. Proc. Natl. Acad. Sci. USA 2022, 119, e2203593119. [Google Scholar] [CrossRef]
- Mike, L.A.; Stark, A.J.; Forsyth, V.S.; Vornhagen, J.; Smith, S.N.; Bachman, M.A.; Mobley, H.L.T. A systematic analysis of hypermucoviscosity and capsule reveals distinct and overlapping genes that impact Klebsiella pneumoniae fitness. PLoS Pathog. 2021, 17, e1009376. [Google Scholar] [CrossRef] [PubMed]
- Choby, J.E.; Howard-Anderson, J.; Weiss, D.S. Hypervirulent Klebsiella pneumoniae—Clinical and molecular perspectives. J. Intern. Med. 2020, 287, 283–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.M.; Ko, W.C.; Ho, M.W.; Lee, Y.L.; Hsueh, P.R. In vitro activity of imipenem/relebactam, meropenem/vaborbactam and comparators against Pseudomonas aeruginosa in Taiwan: Results from the Study for Monitoring Antimicrobial Resistance Trends (SMART) in 2020. J. Infect. 2023, 86, 66–117. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.H.; Ma, L.; Huang, L.Y.; Yeh, K.M.; Lin, J.C.; Siu, L.K.; Chang, F.Y. Molecular epidemiology and resistance patterns of bla(OXA-48)Klebsiella pneumoniae and Escherichia coli: A nationwide multicenter study in Taiwan. J. Microbiol. Immunol. Infect. 2021, 54, 665–672. [Google Scholar] [CrossRef]
- Durso, L.M.; Cook, K.L. Impacts of antibiotic use in agriculture: What are the benefits and risks? Curr. Opin. Microbiol. 2014, 19, 37–44. [Google Scholar] [CrossRef]
- Hernando-Amado, S.; Coque, T.M.; Baquero, F.; Martinez, J.L. Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nat. Microbiol. 2019, 4, 1432–1442. [Google Scholar] [CrossRef]
- Kotsakis, S.D.; Flach, C.-F.; Razavi, M.; Larsson, D.G.J. Characterization of the First OXA-10 Natural Variant with Increased Carbapenemase Activity. Antimicrob. Agents Chemother. 2019, 63, e01817-18. [Google Scholar] [CrossRef] [Green Version]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Vaser, R.; Sović, I.; Nagarajan, N.; Šikić, M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017, 27, 737–746. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.-T.; Liu, P.-Y.; Shih, P.-W. Homopolish: A method for the removal of systematic errors in nanopore sequencing by homologous polishing. Genome Biol. 2021, 22, 95. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef] [PubMed]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [Green Version]
- Bell, G.; MacLean, C. The Search for ‘Evolution-Proof’ Antibiotics. Trends Microbiol. 2018, 26, 471–483. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tseng, C.-H.; Huang, Y.-T.; Mao, Y.-C.; Lai, C.-H.; Yeh, T.-K.; Ho, C.-M.; Liu, P.-Y. Insight into the Mechanisms of Carbapenem Resistance in Klebsiella pneumoniae: A Study on IS26 Integrons, Beta-Lactamases, Porin Modifications, and Plasmidome Analysis. Antibiotics 2023, 12, 749. https://doi.org/10.3390/antibiotics12040749
Tseng C-H, Huang Y-T, Mao Y-C, Lai C-H, Yeh T-K, Ho C-M, Liu P-Y. Insight into the Mechanisms of Carbapenem Resistance in Klebsiella pneumoniae: A Study on IS26 Integrons, Beta-Lactamases, Porin Modifications, and Plasmidome Analysis. Antibiotics. 2023; 12(4):749. https://doi.org/10.3390/antibiotics12040749
Chicago/Turabian StyleTseng, Chien-Hao, Yao-Ting Huang, Yan-Chiao Mao, Chung-Hsu Lai, Ting-Kuang Yeh, Chung-Mei Ho, and Po-Yu Liu. 2023. "Insight into the Mechanisms of Carbapenem Resistance in Klebsiella pneumoniae: A Study on IS26 Integrons, Beta-Lactamases, Porin Modifications, and Plasmidome Analysis" Antibiotics 12, no. 4: 749. https://doi.org/10.3390/antibiotics12040749
APA StyleTseng, C.-H., Huang, Y.-T., Mao, Y.-C., Lai, C.-H., Yeh, T.-K., Ho, C.-M., & Liu, P.-Y. (2023). Insight into the Mechanisms of Carbapenem Resistance in Klebsiella pneumoniae: A Study on IS26 Integrons, Beta-Lactamases, Porin Modifications, and Plasmidome Analysis. Antibiotics, 12(4), 749. https://doi.org/10.3390/antibiotics12040749





