Antibacterial Properties of Ethacridine Lactate and Sulfmethoxazole Loaded Functionalized Graphene Oxide Nanocomposites
Abstract
1. Introduction
2. Results and Discussion
2.1. FTIR Analysis
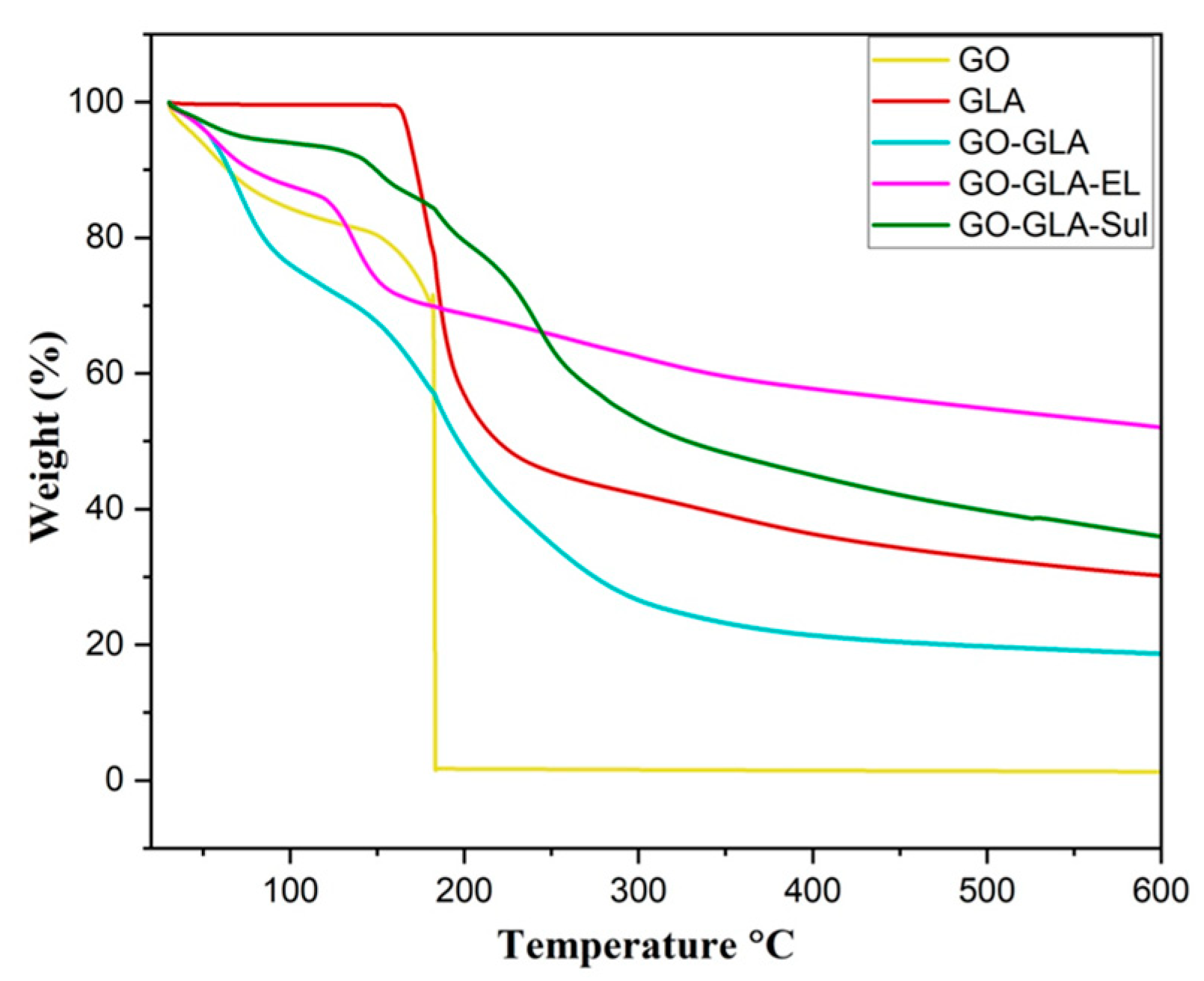
2.2. Thermogravimetric Analysis (TGA)
2.3. Zeta Potential
2.4. Morphological Analysis
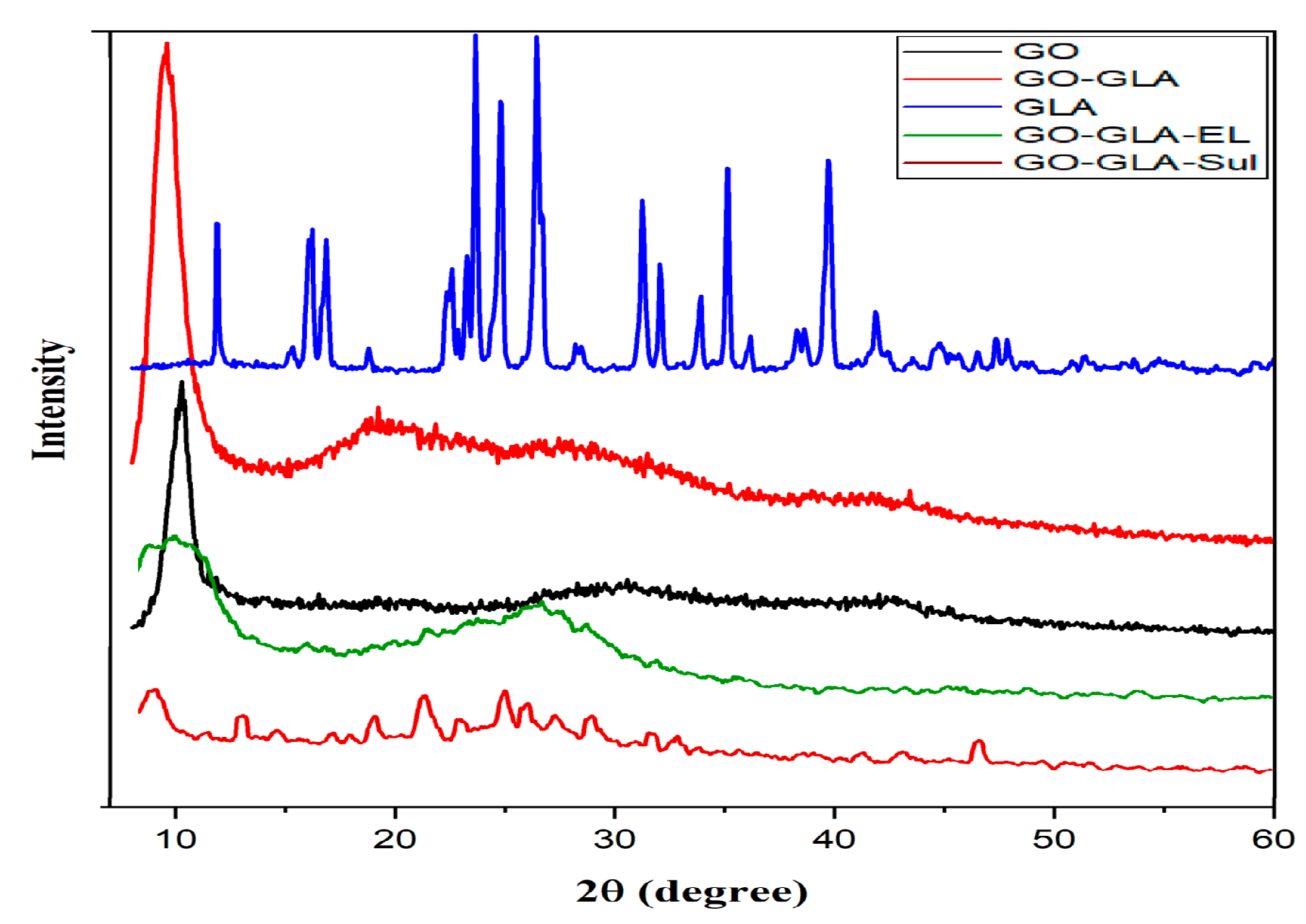
2.5. X-ray Diffraction Analysis
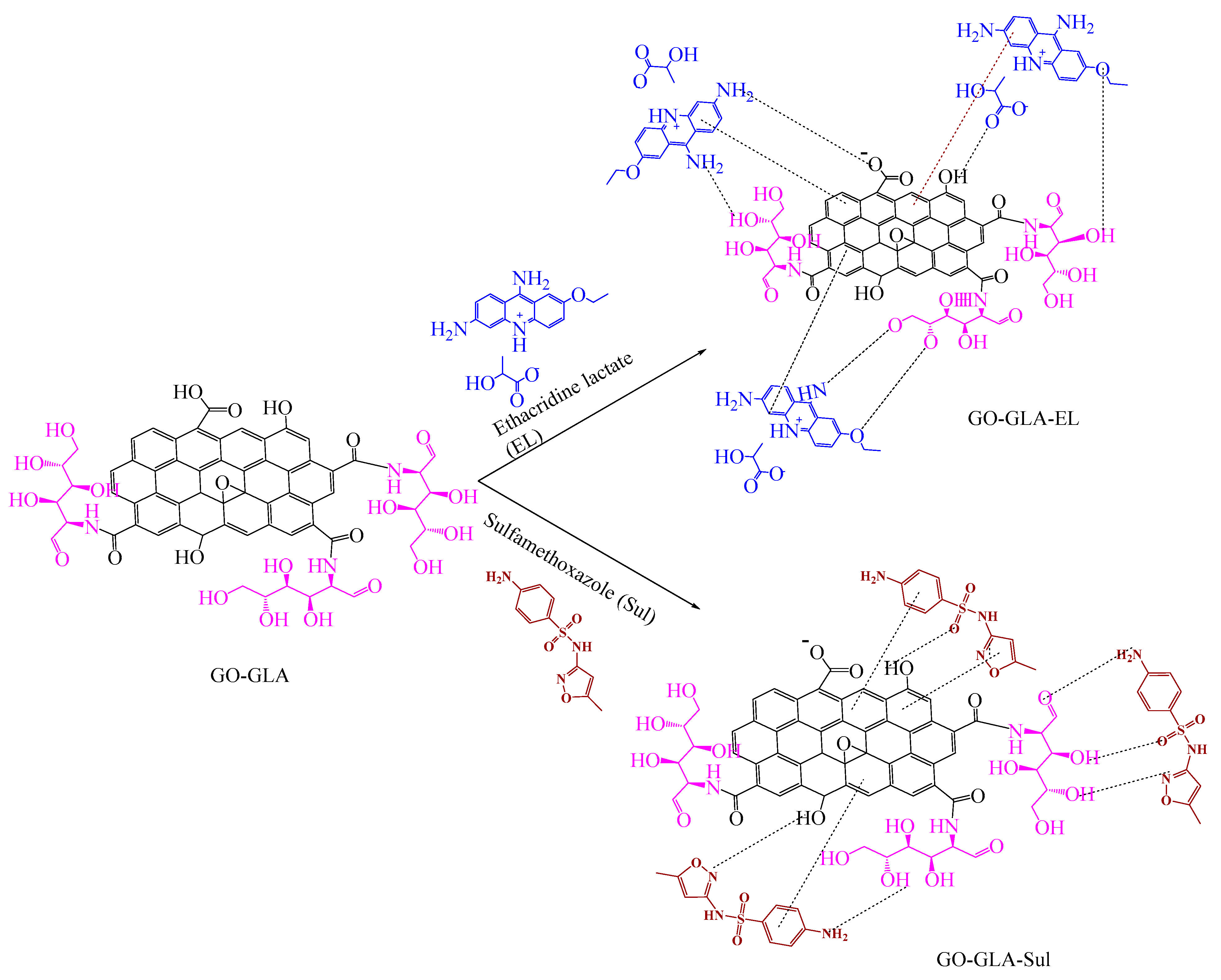
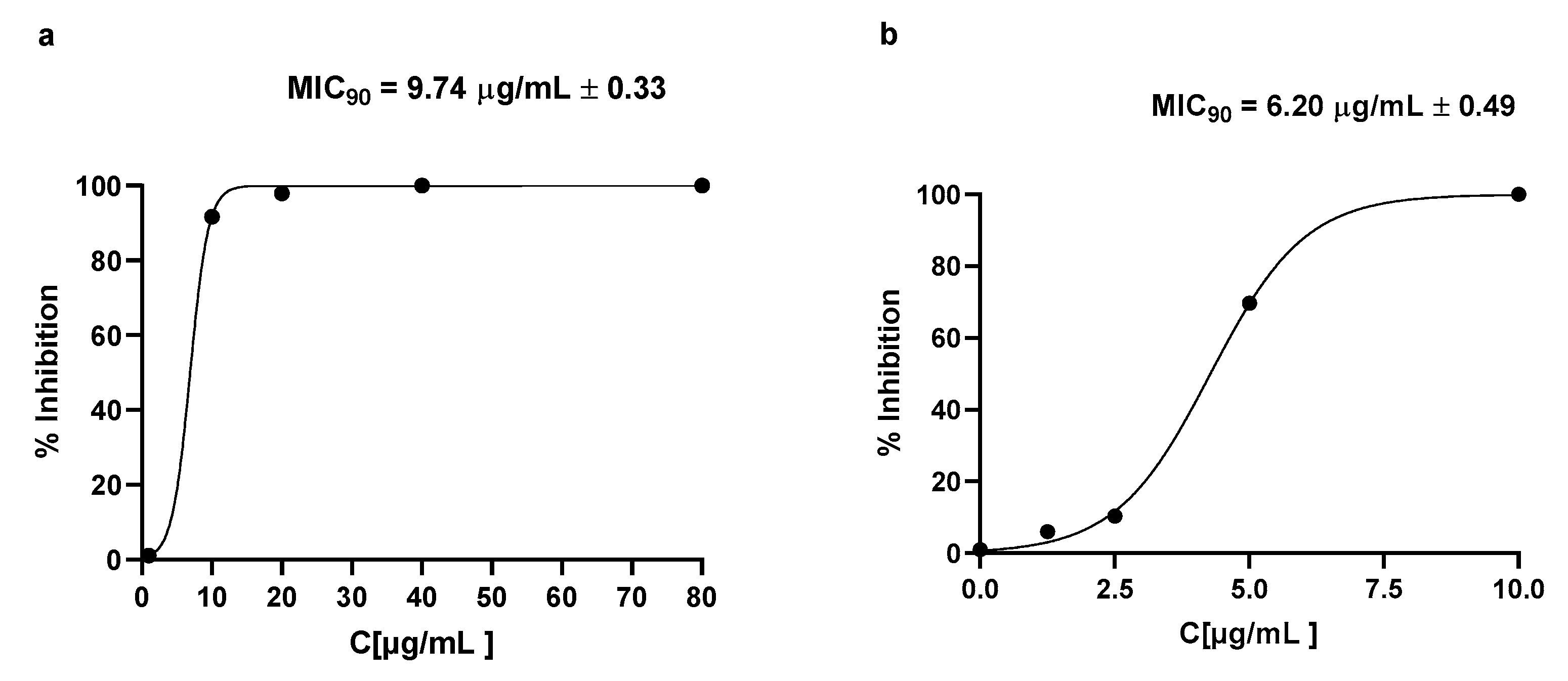
2.6. Drug Loading
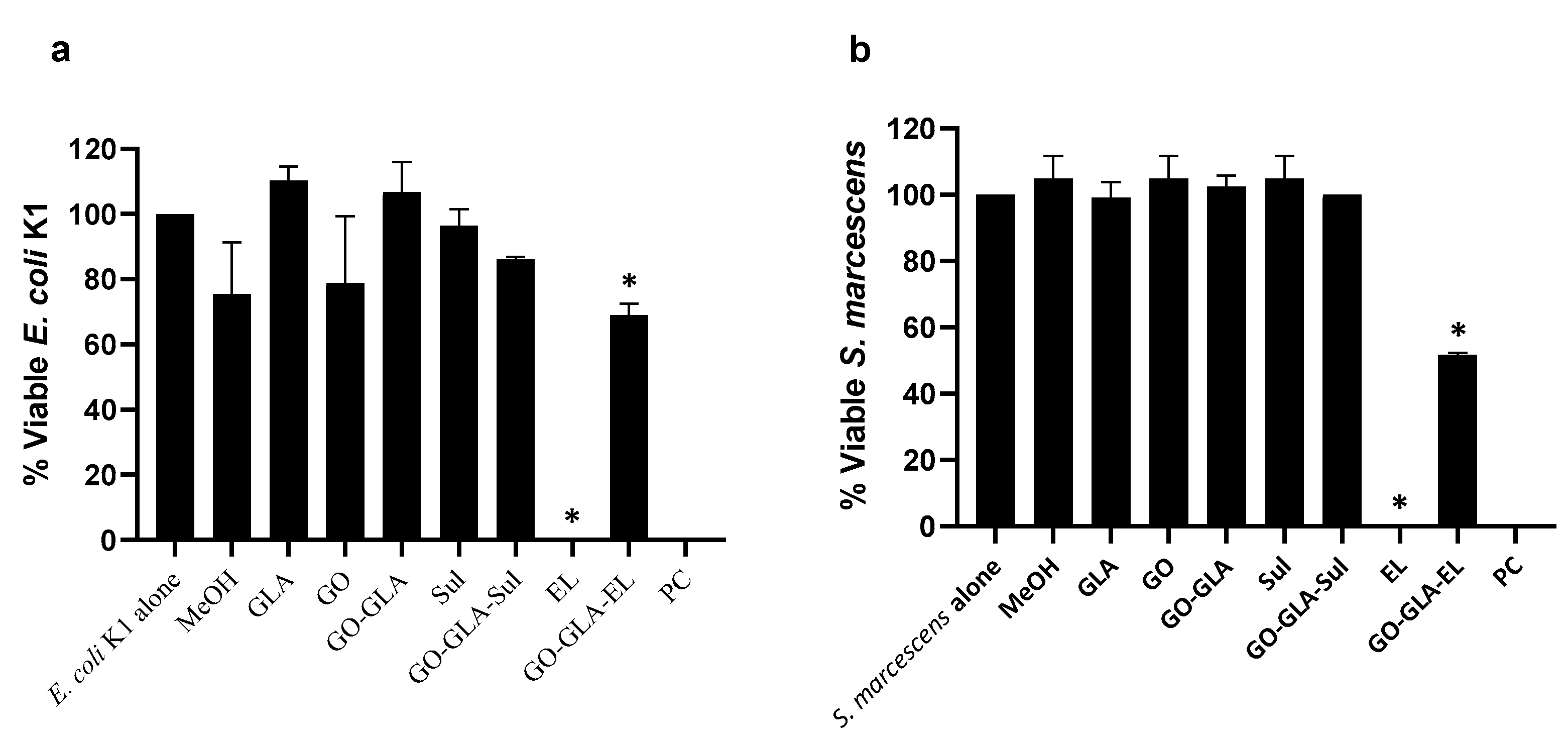
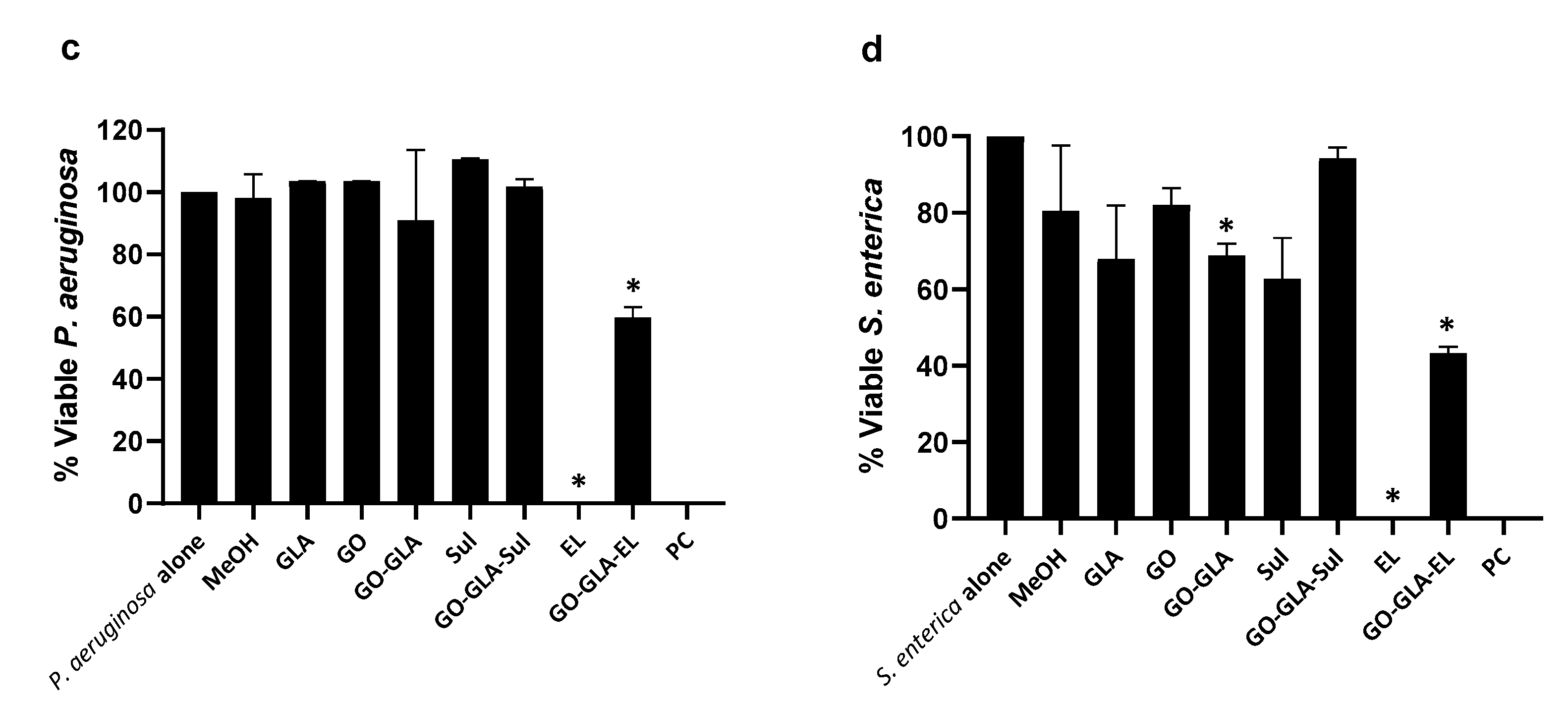
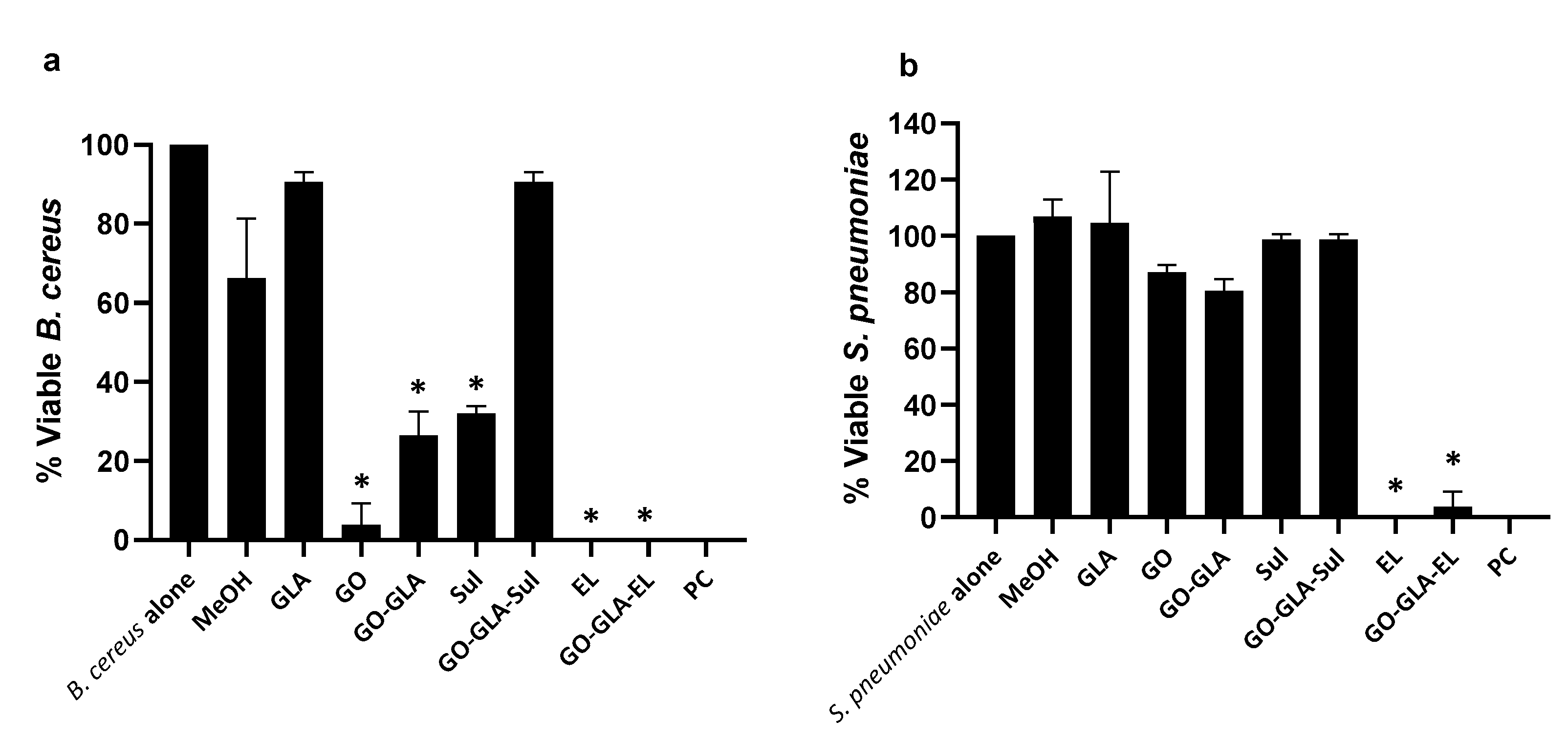
2.7. Graphene Oxide–GlucosamineDrugs Combination Demonstrated Remarkable Antibacterial Activities
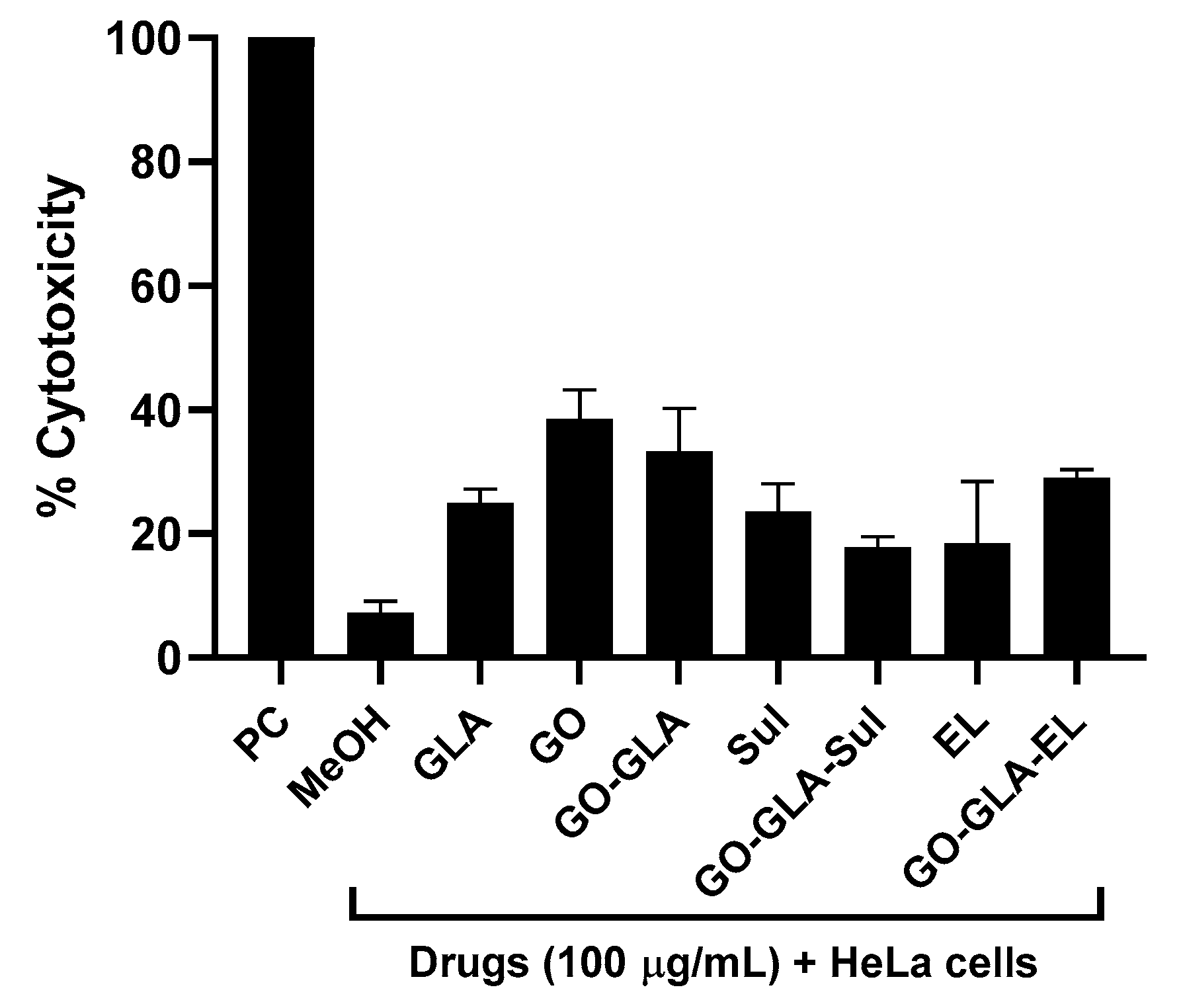
2.8. Drugs, Nanoparticles, and Drug–NPS Combination Revealed Limited Cytotoxic Effects against Human Cells
3. Materials and Methods
3.1. Synthesis of Graphene Oxide (GO)
3.2. Synthesis of Glucosamine Functionalized Graphene Oxide Nanoparticle (GO–GLA)
3.3. EL Loaded GO–GLA and Sul Loaded GO–GLA
3.4. Characterization Using FTIR
3.5. Morphological Analysis
3.6. Zeta Potential
3.7. Thermogravimetric Analysis (TGA)
3.8. PXRD Analysis
3.9. Bacteria Used in This Study
3.10. Cultures of Henrietta Lacks (HeLa) Cervical Cancer Cells
3.11. Bactericidal Assays
3.12. Cytotoxicity Assays
3.13. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lamberte, L.E.; van Schaik, W. Antibiotic resistance in the commensal human gut microbiota. Curr. Opin. Microbiol. 2022, 68, 102150. [Google Scholar] [CrossRef] [PubMed]
- Munir, M.U.; Ahmad, M.M. Nanomaterials aiming to tackle antibiotic-resistant bacteria. Pharmaceutics 2022, 14, 582. [Google Scholar] [CrossRef] [PubMed]
- Legese, M.H.; Asrat, D.; Swedberg, G.; Hasan, B.; Mekasha, A.; Getahun, T.; Worku, M.; Shimber, E.T.; Getahun, S.; Ayalew, T.; et al. Sepsis: Emerging pathogens and antimicrobial resistance in Ethiopian referral hospitals. Antimicrob. Resist. Infect. Control. 2022, 11, 83. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Li, B.; Jiang, X.T.; Wang, Y.L.; Xia, Y.; Li, A.D.; Zhang, T. Catalogue of antibiotic resistome and host-tracking in drinking water deciphered by a large scale survey. Microbiome 2017, 5, 154. [Google Scholar] [CrossRef]
- Navarro, A.; Sanseverino, I.; Cappelli, F.; Lahm, A.; Niegowska, M.; Fabbri, M.; Paracchini, V.; Petrillo, M.; Skejo, H.; Valsecchi, S.; et al. Study of antibiotic resistance in freshwater ecosystems with low anthropogenic impact. Sci. Total Environ. 2023, 857, 159378. [Google Scholar] [CrossRef]
- Xu, C.; Kong, L.; Gao, H.; Cheng, X.; Wang, X. A Review of Current Bacterial Resistance to Antibiotics in Food Animals. Front. Microbiol. 2022, 13, 1458. [Google Scholar] [CrossRef]
- Lewis, K. The science of antibiotic discovery. Cell 2020, 181, 29–45. [Google Scholar] [CrossRef]
- Azizi-lalabadi, M.; Hashemi, H.; Feng, J.; Jafari, S.M. Carbon nanomaterials against pathogens; the antimicrobial activity of carbon nanotubes, graphene/graphene oxide, fullerenes, and their nanocomposites. Adv. Colloid Interface Sci. 2020, 284, 102250. [Google Scholar] [CrossRef]
- Power, A.; Chandra, S.; Chapman, J. Graphene, electrospun membranes and granular activated carbon for eliminating heavy metals, pesticides and bacteria in water and wastewater treatment processes. Analyst 2018, 143, 5629–5645. [Google Scholar]
- Loh, K.P.; Bao, Q.; Ang, P.K.; Yang, J. The chemistry of graphene. J. Mater. Chem. 2010, 20, 2277–2289. [Google Scholar] [CrossRef]
- Rajapaksha, P.; Orrell-Trigg, R.; Shah, D.; Cheeseman, S.; Vu, K.; Ngo, S.; Murdoch, B.; Choudhury, N.; Yin, H.; Cozzolino, D. Broad spectrum antibacterial zinc oxide-reduced graphene oxide nanocomposite for water depollution. Mater. Today Chem. 2023, 27, 101242. [Google Scholar] [CrossRef]
- Pramanik, A.; Gates, K.; Gao, Y.; Zhang, Q.; Han, F.X.; Begum, S.; Rightsell, C.; Sardar, D.; Ray, P.C. Composites composed of polydopamine nanoparticles, graphene oxide, and ε-poly-L-lysine for removal of waterborne contaminants and eradication of superbugs. ACS Appl. Nano Mater. 2019, 2, 3339–3347. [Google Scholar] [CrossRef]
- Jones, S.; Pramanik, A.; Kanchanapally, R.; Viraka Nellore, B.P.; Begum, S.; Sweet, C.; Ray, P.C. Multifunctional three-dimensional chitosan/gold nanoparticle/graphene oxide architecture for separation, label-free SERS identification of pharmaceutical contaminants, and effective killing of superbugs. ACS Sustain. Chem. Eng. 2017, 5, 7175–7187. [Google Scholar] [CrossRef]
- Veerapandian, M.; Lévaray, N.; Lee, M.H.; Giasson, S.; Zhu, X.X. Glucosamine-anchored graphene oxide nanosheets: Fabrication, ultraviolet irradiation, and electrochemical properties. ACS Appl. Mater. Interfaces 2015, 7, 14552–14556. [Google Scholar] [CrossRef] [PubMed]
- Shekhar, S.; Sharma, R.; Sharma, S.; Sharma, B.; Sarkar, A.; Jain, P. An exploration of electrocatalytic analysis and antibacterial efficacy of electrically conductive poly (D-glucosamine)/graphene oxide bionanohybrid. Carbohydr. Polym. 2020, 240, 116242. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, C.S.; Geske, T.; Schmolz, M. A topical wound disinfectant (ethacridine lactate) differentially affects the production of immunoregulatory cytokines in human whole-blood cultures. Wounds 2005, 17, 213. [Google Scholar]
- Alhbou, Y.; Karsli, B.; Sümer, T. Comparision of the efficiency of ethacridine lactate and hypochlorous acid during the early period of wound healing in rats. Kocatepe Vet. J. 2021, 14, 366–375. [Google Scholar] [CrossRef]
- Wainwright, M. Acridine—A neglected antibacterial chromophore. J. Antimicrob. Chemother. 2001, 47, 1–13. [Google Scholar] [CrossRef]
- Sahyon, H.A.; Ramadan, E.N.; Mashaly, M.M. Synergistic effect of quercetin in combination with sulfamethoxazole as new antibacterial agent: In vitro and in vivo study. Pharm. Chem. J. 2019, 53, 803–813. [Google Scholar] [CrossRef]
- Akbar, N.; Gul, J.; Siddiqui, R.; Shah, M.R.; Khan, N.A. Moxifloxacin and sulfamethoxazole-based nanocarriers exhibit potent antibacterial activities. Antibiotics 2021, 10, 964. [Google Scholar] [CrossRef]
- Masters, P.A.; O’bryan, T.A.; Zurlo, J.; Miller, D.Q.; Joshi, N. Trimethoprim-sulfamethoxazole revisited. Arch. Intern. Med. 2003, 163, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Hummers, W.S., Jr.; Offeman, R.E. Preparation of graphitic oxide. J. Am. Chem. Soc. 1958, 80, 1339. [Google Scholar] [CrossRef]
- Li, D.; He, X.; Zhao, L.; Li, H.; Zhang, X.; Chen, J.; Xu, J. Ultrafast charge transfer dynamics of Rhodamine B with graphene oxide. J. Chem. Phys. 2022, 157, 214701. [Google Scholar] [CrossRef] [PubMed]
- Avisar, D.; Primor, O.; Gozlan, I.; Mamane, H. Sorption of sulfonamides and tetracyclines to montmorillonite clay. Water Air Soil Pollut. 2010, 209, 439–450. [Google Scholar] [CrossRef]
- Kesavan, S.; Meena, K.S.; Sharmili, S.A.; Govindarajan, M.; Alharbi, N.S.; Kadaikunnan, S.; Khaled, J.M.; Alobaidi, A.S.; Alanzi, K.F.; Vaseeharan, B. Ulvan loaded graphene oxide nanoparticle fabricated with chitosan and d-mannose for targeted anticancer drug delivery. J. Drug Deliv. Sci. Technol. 2021, 65, 102760. [Google Scholar] [CrossRef]
- Veerapandian, M.; Neethirajan, S. Graphene oxide chemically decorated with Ag–Ru/chitosan nanoparticles: Fabrication, electrode processing and immunosensing properties. RSC Adv. 2015, 5, 75015–75024. [Google Scholar] [CrossRef]
- Marjanović, B.; Juranić, I.; Ćirić-Marjanović, G.; Mojović, M.; Pašti, I.; Janošević, A.; Trchová, M.; Holler, P.; Horský, J. Chemical oxidative polymerization of ethacridine. React. Funct. Polym. 2012, 72, 25–35. [Google Scholar] [CrossRef]
- Dzierzkowska, E.; ŚcisŁowska-Czarnecka, A.N.N.A.; Matwally, S.; Romaniszyn, D.; ChadziŃska, M.A.G.D.A.L.E.N.A.; Stodolak-Zych, E. Porous poly (lactic acid) based fibres as drug carriers in active dressings. Acta Bioeng. Biomech. 2020, 22, 185–197. [Google Scholar] [CrossRef]
- Mendes, C.; Valentini, G.; Chamorro Rengifo, A.F.; Pinto, J.M.; Silva, M.A.; Parize, A.L. Supersaturating drug delivery system of fixed drug combination: Sulfamethoxazole and trimethoprim. Expert Rev. Anti-Infect. Ther. 2019, 17, 841–850. [Google Scholar] [CrossRef]
- Mokhtar, M.; El Enein, S.A.; Hassaan, M.; Morsy, M.; Khalil, M. Thermally reduced graphene oxide: Synthesis, structural and electrical properties. Int. J. Nanoparticles Nanotechnol. 2017, 3, 8. [Google Scholar]
- Cheng, M.M.; Huang, L.J.; Wang, Y.X.; Zhao, Y.C.; Tang, J.G.; Wang, Y.; Zhang, Y.; Hedayati, M.; Kipper, M.J.; Wickramasinghe, S.R. Synthesis of graphene oxide/polyacrylamide composite membranes for organic dyes/water separation in water purification. J. Mater. Sci. 2019, 54, 252–264. [Google Scholar] [CrossRef]
- Dong, Y.; Niu, X.; Song, W.; Wang, D.; Chen, L.; Yuan, F.; Zhu, Y. Facile synthesis of vanadium oxide/reduced graphene oxide composite catalysts for enhanced hydroxylation of benzene to phenol. Catalysts 2016, 6, 74. [Google Scholar] [CrossRef]
- Petrikaite, V.; Tarasevišius, E.; Pavilonis, A. New ethacridine derivatives as the potential antifungal and antibacterial preparations. Medicina 2007, 43, 657. [Google Scholar] [CrossRef]
- Akbar, N.; Siddiqui, R.; Iqbal, M.; Sagathevan, K.; Kim, K.S.; Habib, F.; Khan, N.A. Gut bacteria of Rattus rattus (Rat) produce broad-spectrum antibacterial lipopeptides. ACS Omega 2021, 6, 12261–12273. [Google Scholar] [CrossRef]
- Akbar, N.; Kawish, M.; Jabri, T.; Khan, N.A.; Shah, M.R.; Siddiqui, R. Enhancing efficacy of existing antibacterials against selected multiple drug resistant bacteria using cinnamic acid-coated magnetic iron oxide and mesoporous silica nanoparticles. Pathog. Glob. Health 2022, 116, 438–454. [Google Scholar] [CrossRef]
- Akbar, N.; Khan, N.A.; Sagathevan, K.; Iqbal, M.; Tawab, A.; Siddiqui, R. Gut bacteria of Cuoraamboinensis (turtle) produce broad-spectrum antibacterial molecules. Sci. Rep. 2019, 9, 17012. [Google Scholar] [CrossRef]





| Bacteria | Strain |
|---|---|
| Escherichia coli K1 | MTCC 710859 (clinical isolate) |
| Serratia marcescens | MTTC 13880 (clinical isolate) |
| Pseudomonas aeruginosa | ATCC 10145 (clinical isolate) |
| Salmonella enterica | ATTC 14028 (clinical isolate) |
| Bacillus cereus | MTCC 131621 (clinical isolate) |
| Streptococcus pneumoniae | ATCC 33400 (clinical isolate) |
| Streptococcus pyogenes | ATCC 12344 (clinical isolate) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jabri, T.; Khan, N.A.; Makhlouf, Z.; Akbar, N.; Gul, J.; Shah, M.R.; Siddiqui, R. Antibacterial Properties of Ethacridine Lactate and Sulfmethoxazole Loaded Functionalized Graphene Oxide Nanocomposites. Antibiotics 2023, 12, 755. https://doi.org/10.3390/antibiotics12040755
Jabri T, Khan NA, Makhlouf Z, Akbar N, Gul J, Shah MR, Siddiqui R. Antibacterial Properties of Ethacridine Lactate and Sulfmethoxazole Loaded Functionalized Graphene Oxide Nanocomposites. Antibiotics. 2023; 12(4):755. https://doi.org/10.3390/antibiotics12040755
Chicago/Turabian StyleJabri, Tooba, Naveed Ahmed Khan, Zinb Makhlouf, Noor Akbar, Jasra Gul, Muhammad Raza Shah, and Ruqaiyyah Siddiqui. 2023. "Antibacterial Properties of Ethacridine Lactate and Sulfmethoxazole Loaded Functionalized Graphene Oxide Nanocomposites" Antibiotics 12, no. 4: 755. https://doi.org/10.3390/antibiotics12040755
APA StyleJabri, T., Khan, N. A., Makhlouf, Z., Akbar, N., Gul, J., Shah, M. R., & Siddiqui, R. (2023). Antibacterial Properties of Ethacridine Lactate and Sulfmethoxazole Loaded Functionalized Graphene Oxide Nanocomposites. Antibiotics, 12(4), 755. https://doi.org/10.3390/antibiotics12040755








