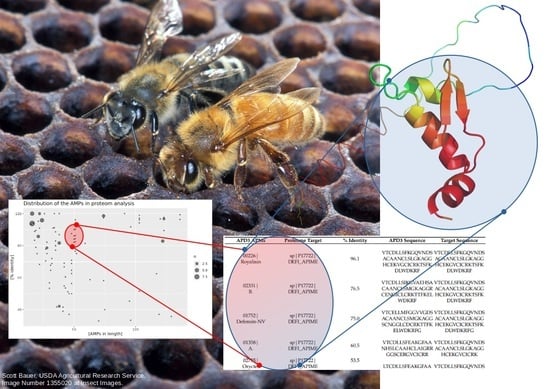
Assessing the Antimicrobial Properties of Honey Protein Components through In Silico Comparative Peptide Composition and Distribution Analysis
Abstract
:1. Introduction
2. Results
2.1. Evaluation of Reference AMPs Databases
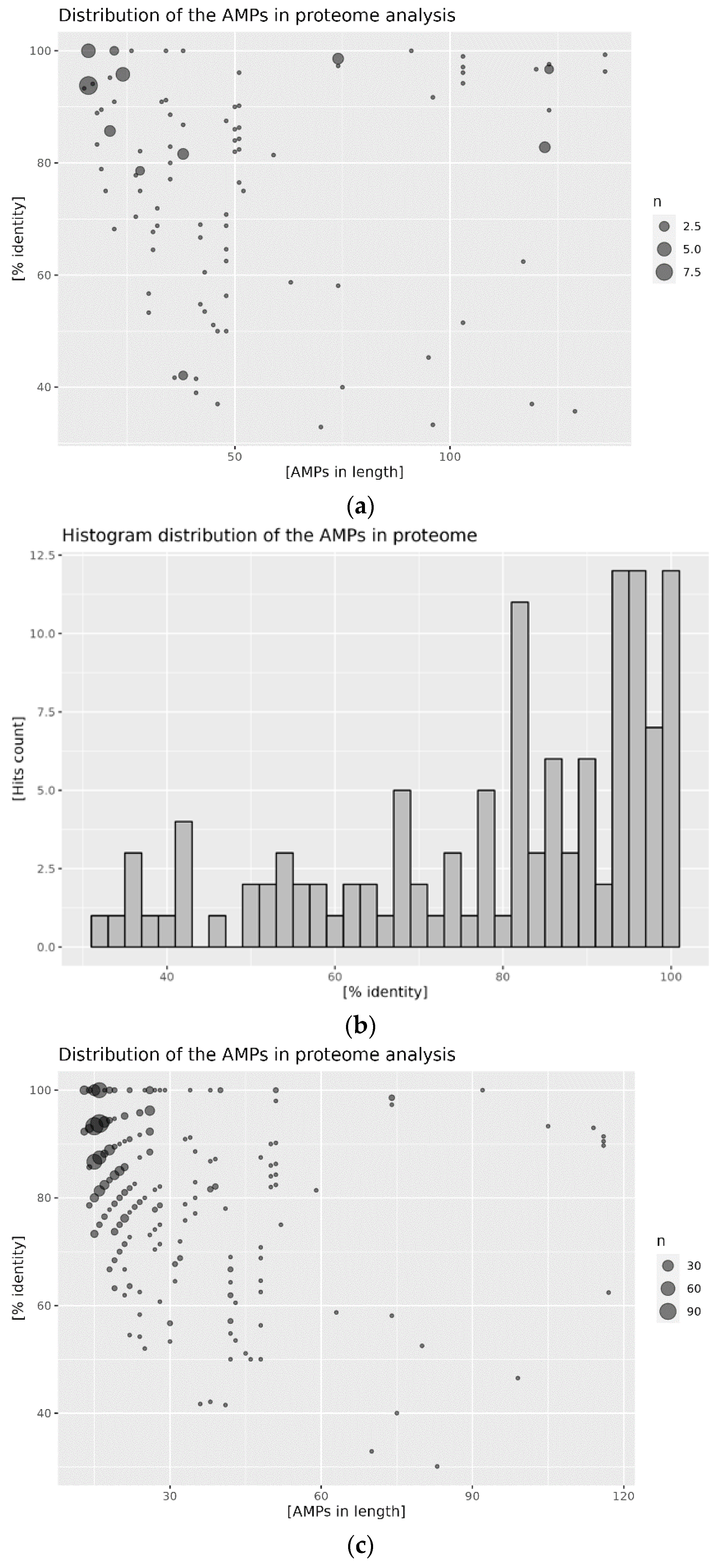
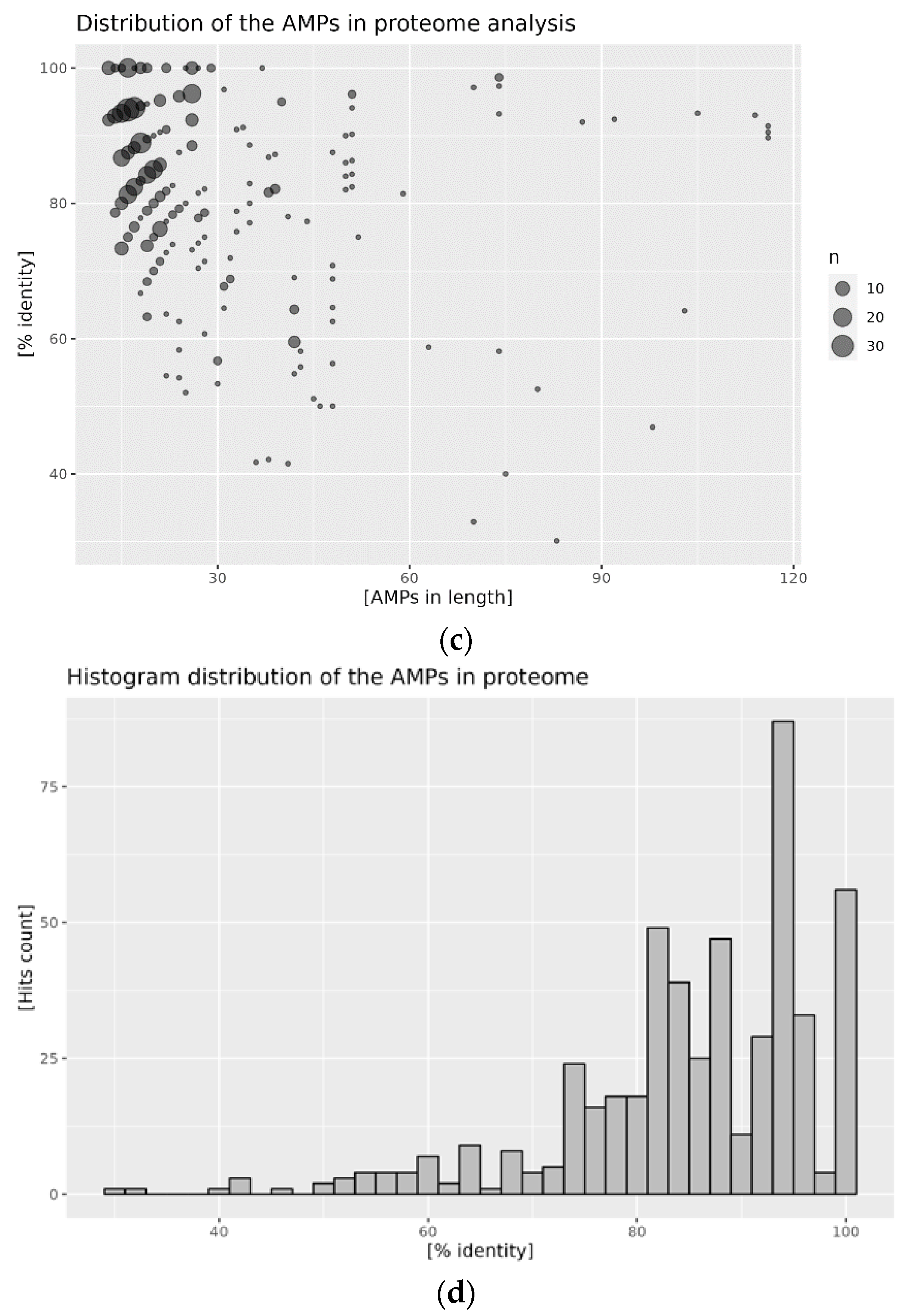
2.2. Distribution of AMPs in the Analyzed Proteomes
2.3. Mapping of AMPs on the Proteomic Target
3. Discussion
4. Materials and Methods
4.1. Data Sources
- Antimicrobial Peptide Database (APD3, https://aps.unmc.edu/ accessed on 23 January 2023) [37];
- Database of Antimicrobial Activity and Structure of Peptides (DBAASP, https://dbaasp.org/ accessed on 23 January 2023) [38];
- UniProt proteome sequences (https://uniprot.org/ accessed on 23 January 2023) reference proteomes section (https://www.uniprot.org/proteomes accessed on 23 January 2023) [39].
4.2. Alignment Programs
4.3. Data Visualization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yupanqui Mieles, J.; Vyas, C.; Aslan, E.; Humphreys, G.; Diver, C.; Bartolo, P. Honey: An Advanced Antimicrobial and Wound Healing Biomaterial for Tissue Engineering Applications. Pharmaceutics 2022, 14, 1663. [Google Scholar] [CrossRef] [PubMed]
- Kwakman, P.H.S.; Zaat, S.A.J. Antibacterial Components of Honey. IUBMB Life 2012, 64, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Almasaudi, S. The Antibacterial Activities of Honey. Saudi J. Biol. Sci. 2021, 28, 2188–2196. [Google Scholar] [CrossRef] [PubMed]
- Brudzynski, K. Effect of Hydrogen Peroxide on Antibacterial Activities of Canadian Honeys. Can. J. Microbiol. 2006, 52, 1228–1237. [Google Scholar] [CrossRef]
- Brudzynski, K.; Abubaker, K.; St-Martin, L.; Castle, A. Re-Examining the Role of Hydrogen Peroxide in Bacteriostatic and Bactericidal Activities of Honey. Front. Microbiol. 2011, 2, 213. [Google Scholar] [CrossRef]
- Bucekova, M.; Valachova, I.; Kohutova, L.; Prochazka, E.; Klaudiny, J.; Majtan, J. Honeybee Glucose Oxidase—Its Expression in Honeybee Workers and Comparative Analyses of Its Content and H2O2-Mediated Antibacterial Activity in Natural Honeys. Naturwissenschaften 2014, 101, 661–670. [Google Scholar] [CrossRef]
- Tonks, A.J.; Cooper, R.A.; Jones, K.P.; Blair, S.; Parton, J.; Tonks, A. Honey Stimulates Inflammatory Cytokine Production from Monocytes. Cytokine 2003, 21, 242–247. [Google Scholar] [CrossRef]
- Fontana, R.; Mendes, M.A.; De Souza, B.M.; Konno, K.; César, L.M.M.; Malaspina, O.; Palma, M.S. Jelleines: A Family of Antimicrobial Peptides from the Royal Jelly of Honeybees (Apis mellifera). Peptides 2004, 25, 919–928. [Google Scholar] [CrossRef]
- Brudzynski, K. A Current Perspective on Hydrogen Peroxide Production in Honey. A Review. Food Chem. 2020, 332, 127229. [Google Scholar] [CrossRef]
- Kwakman, P.H.S.; Velde, A.A.; de Boer, L.; Speijer, D.; Vandenbroucke-Grauls, M.J.C.; Zaat, S.A.J. How Honey Kills Bacteria. FASEB J. 2010, 24, 2576–2582. [Google Scholar] [CrossRef]
- Abdel-Azim, S.G.; Abdel-Azim, A.G.; Piasecki, B.P.; Abdel-Azim, G.A. Characterization of the Gain and Loss of Resistance to Antibiotics versus Tolerance to Honey as an Antimutagenic and Antimicrobial Medium in Extended-Time Serial Transfer Experiments. Pharmacogn. Res. 2019, 11, 147–154. [Google Scholar] [CrossRef]
- Proaño, A.; Coello, D.; Villacrés-Granda, I.; Ballesteros, I.; Debut, A.; Vizuete, K.; Brenciani, A.; Álvarez-Suarez, J.M. The osmotic action of sugar combined with hydrogen peroxide and bee-derived antibacterial peptide Defensin-1 is crucial for the antibiofilm activity of eucalyptus honey. LWT 2021, 136, 110379. [Google Scholar] [CrossRef]
- Estevinho, L.; Pereira, A.P.; Moreira, L.; Dias, L.G.; Pereira, E. Antioxidant and antimicrobial effects of phenolic compounds extracts of Northeast Portugal honey. Food Chem. Toxicol. 2008, 46, 3774–3779. [Google Scholar] [CrossRef] [PubMed]
- Bucekova, M.; Jardekova, L.; Juricova, V.; Bugarova, V.; Di Marco, G.; Gismondi, A.; Leonardi, D.; Farkasovska, J.; Godocikova, J.; Laho, M.; et al. Antibacterial Activity of Different Blossom Honeys: New Findings. Molecules 2019, 24, 1573. [Google Scholar] [CrossRef] [PubMed]
- Nolan, V.C.; Harrison, J.; Cox, J.A.G. Dissecting the antimicrobial composition of honey. Antibiotics 2019, 8, 251. [Google Scholar] [CrossRef]
- Rabie, E.; Serem, J.C.; Oberholzer, H.M.; Gaspar, A.R.M.; Bester, M.J. How methylglyoxal kills bacteria: An ultrastructural study. Ultrastruct. Pathol. 2016, 40, 107–111. [Google Scholar] [CrossRef]
- Deng, J.; Liu, R.; Lu, Q.; Hao, P.; Xu, A.; Zhang, J.; Tan, J. Biochemical properties, antibacterial and cellular antioxidant activities of buckwheat honey in comparison to manuka honey. Food Chem. 2018, 252, 243–249. [Google Scholar] [CrossRef]
- Girma, A.; Seo, W.; SheI, R.C. Antibacterial activity of varying UMF-graded Manuka honeys. PLoS ONE 2019, 14, e0224495. [Google Scholar] [CrossRef]
- Kwakman, P.H.S.; te Velde, A.A.; de Boer, L.; Vandenbroucke-Grauls, C.M.J.E.; Zaat, S.A.J. Two major medicinal honeys have different mechanisms of bactericidal activity. PLoS ONE 2011, 6, e17709. [Google Scholar] [CrossRef]
- Brudzynski, K.; Sjaarda, C. Honey glycoproteins containing antimicrobial peptides, jelleins of the Major Royal Jelly Protein 1, are responsible for the cell wall lytic and bactericidal activities of honey. PLoS ONE 2015, 10, e0120238. [Google Scholar] [CrossRef]
- Wang, X.; Chen, Y.; Hu, Y.; Zhou, J.; Chen, L.; Lu, X. Systematic Review of the Characteristic Markers in Honey of Various Botanical, Geographic, and Entomological Origins. ACS Food Sci. Technol. 2022, 2, 206–220. [Google Scholar] [CrossRef]
- Dzugan, M.; Tomczyk, M.; Sowa, P.; Grabek-Lejko, D. Antioxidant activity as biomarker of honey variety. Molecules 2018, 23, 2069. [Google Scholar] [CrossRef] [PubMed]
- Moniruzzaman, M.; Khalil, I.; Sulaiman, S.A.; Gan, S.H. Physicochemical and antioxidant properties of Malaysian honeys produced by Apis cerana, Apis dorsata and Apis mellifera. BMC Complement. Altern. Med. 2013, 13, 43. [Google Scholar] [CrossRef] [PubMed]
- Ewnetu, Y.; Lemma, W.; Birhane, N. Antibacterial effects of Apis mellifera and stingless bees honeys on susceptible and resistant strains of Escherichia coli, Staphylococcus aureus and Klebsiella pneumoniae in Gondar, Northwest Ethiopia. BMC Complement. Altern. Med. 2013, 13, 269. [Google Scholar] [CrossRef] [PubMed]
- Rozman, A.S.; Hashim, N.; Maringgal, B.; Abdan, K. A Comprehensive Review of Stingless Bee Products: Phytochemical Composition and Beneficial Properties of Honey, Propolis, and Pollen. Appl. Sci. 2022, 12, 6370. [Google Scholar] [CrossRef]
- Stan, D.; Enciu, A.M.; Mateescu, A.L.; Ion, A.C.; Brezeanu, A.C.; Stan, D.; Tanase, C. Natural Compounds with Antimicrobial and Antiviral Effect and Nanocarriers Used for Their Transportation. Front Pharm. 2021, 12, 2405. [Google Scholar] [CrossRef] [PubMed]
- Ullah, H.; Ali, S.; Ullah, H.; Ali, S. Classification of Anti-Bacterial Agents and Their Functions. Antibact. Agents 2017, 1–16. [Google Scholar] [CrossRef]
- Feknous, N.; Boumendjel, M. Natural bioactive compounds of honey and their antimicrobial activity. Czech J. Food Sci. 2022, 40, 163–178. [Google Scholar] [CrossRef]
- Danihlík, J.; Aronstein, K.; Petřivalský, M. Antimicrobial Peptides: A Key Component of Honey Bee Innate Immunity. J. Apic. Res. 2016, 54, 123–136. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 2559. [Google Scholar] [CrossRef]
- Ebenhan, T.; Gheysens, O.; Kruger, H.G.; Zeevaart, J.R.; Sathekge, M.M. Antimicrobial Peptides: Their Role as Infection-Selective Tracers for Molecular Imaging. Biomed. Res. Int. 2014, 2014, 867381. [Google Scholar] [CrossRef] [PubMed]
- Osorio, D.; Rondón-Villarreal, P.; Torres, R. Peptides: A Package for Data Mining of Antimicrobial Peptides. R J. 2015, 7, 4–14. [Google Scholar] [CrossRef]
- Waghu, F.H.; Idicula-Thomas, S. Collection of Antimicrobial Peptides Database and Its Derivatives: Applications and Beyond. Protein Sci. 2020, 29, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Fan, L.; Sun, J.; Lao, X.; Zheng, H. Computational Resources and Tools for Antimicrobial Peptides. J. Pept. Sci. 2017, 23, 4–12. [Google Scholar] [CrossRef]
- Ramazi, S.; Mohammadi, N.; Allahverdi, A.; Khalili, E.; Abdolmaleki, P. A Review on Antimicrobial Peptides Databases and the Computational Tools. Database 2022, 2022, 1–17. [Google Scholar] [CrossRef]
- Ma, L.; Zou, D.; Liu, L.; Shireen, H.; Abbasi, A.A.; Bateman, A.; Xiao, J.; Zhao, W.; Bao, Y.; Zhang, Z. Database Commons: A Catalog of Worldwide Biological Databases. Genom. Proteom. Bioinform. 2022. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD3: The Antimicrobial Peptide Database as a Tool for Research and Education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef]
- Pirtskhalava, M.; Amstrong, A.A.; Grigolava, M.; Chubinidze, M.; Alimbarashvili, E.; Vishnepolsky, B.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M. DBAASP v3: Database of Antimicrobial/Cytotoxic Activity and Structure of Peptides as a Resource for Development of New Therapeutics. Nucleic Acids Res. 2021, 49, D288–D297. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bye-A-Jee, H.; Cukura, A.; et al. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Stefanis, C.; Stavropoulou, E.; Giorgi, E.; Voidarou, C.; Constantinidis, T.C.; Vrioni, G.; Tsakris, A. Honey’s Antioxidant and Antimicrobial Properties: A Bibliometric Study. Antioxidants 2023, 12, 414. [Google Scholar] [CrossRef] [PubMed]
- Eghbaliferiz, S.; Iranshahi, M. Prooxidant Activity of Polyphenols, Flavonoids, Anthocyanins and Carotenoids: Updated Review of Mechanisms and Catalyzing Metals. Phytother. Res. 2016, 30, 1379–1391. [Google Scholar] [CrossRef]
- Albaridi, N.A. Antibacterial Potency of Honey. Int. J. Microbiol. 2019, 2019, 2464507. [Google Scholar] [CrossRef] [PubMed]
- Grecka, K.; Kús, P.M.; Worobo, R.W.; Szweda, P. Study of the Anti-Staphylococcal Potential of Honeys Produced in Northern Poland. Molecules 2018, 23, 260. [Google Scholar] [CrossRef] [PubMed]
- Breed, M.D.; Guzmán-Novoa, E.; Hunt, G.J. Defensive behavior of honey bees: Organization, Genetics, and Comparisons with Other Bees. Annu. Rev. Entomol. 2003, 49, 271–298. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Shi, M.; Chen, X.X. Antimicrobial Peptide Evolution in the Asiatic Honey Bee Apis cerana. PLoS ONE 2009, 4, e4239. [Google Scholar] [CrossRef]
- Muzammal, M.; Khan, M.A.; Mohaini, M.A.; Alsalman, A.J.; Al Hawaj, M.A.; Farid, A. In Silico Analysis of Honeybee Venom Protein Interaction with Wild Type and Mutant (A82V + P375S) Ebola Virus Spike Protein. Biologics 2022, 2, 45–55. [Google Scholar] [CrossRef]
- Buchfink, B.; Reuter, K.; Drost, H.G. Sensitive Protein Alignments at Tree-of-Life Scale Using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023; Available online: https://www.R-project.org/ (accessed on 29 March 2023).
- Posit Team. RStudio: Integrated Development Environment for R. Posit Software, PBC, Boston, MA, USA. 2023. Available online: http://www.posit.co/ (accessed on 29 March 2023).
- PyMOL. Pymol.Org. Available online: https://pymol.org/2/ (accessed on 29 March 2023).




| Honey Antimicrobial Component | Mechanism | Microorganisms | References |
|---|---|---|---|
| Sugars | Osmotic pressure | Staphylococcus aureus Pseudomonas aeruginosa | Abdel-Azim et al., 2019 [11] Proaño et al., 2021 [12] |
| Polyphenols | Antioxidant activity Immunomodulation H2O2 generation Inhibition of bacterial enzymes Membrane disruption Chelation of metal ions DNA/RNA/protein disorders | Escherichia coli Bacillus subtilis Staphylococcus aureus Staphylococcus lentus Pseudomonas aeruginosa Klebsiella pneumoniae | Estevinho et al., 2008 [13] Grecka et al., 2018 [14] Nolan et al., 2019 [15] |
| Methylgyoxal (MGO) | Alterations in bacterial structure, limiting bacterial motility and adherence | Staphylococcus aureus Pseudomonas aeruginosa Staphylococcus epidermidis Klebsiella pneumoniae Escherichia coli | Rabie et al., 2016 [16] Deng et al., 2018 [17] Girma et al., 2019 [18] |
| Glucose oxidase | H2O2 generation | Escherichia coli Bacillus subtilis Pseudomonas aeruginosa | Kwakman et al., 2011 [19] Bucekova et al., 2014 [6] Brudzynski and Sjaarda, 2015 [20] |
| Peptides (mainly defensin-1) | Immunomodulation, membrane disruption, and inhibition of bacterial cell wall synthesis | Staphylococcus aureus Pseudomonas aeruginosa Bacillus subtilis Escherichia coli | Kwakman et al., 2011 [19] Proaño et al., 2021 [12] |
| APD3 APMs | Proteome Target | % Identity | APD3 Sequence | Target Sequence |
|---|---|---|---|---|
| 00226| Royalisin | sp|P17722| DEFI_APIME | 96.1 | VTCDLLSFKGQVNDSACAANCLSLGKAGGHCEKVGCICRKTSFKDLWDKRF | VTCDLLSFKGQVNDSACAANCLSLGKAGGHCEKGVCICRKTSFKDLWDKRF |
| 02331| B. | sp|P17722| DEFI_APIME | 76.5 | VTCDLLSIKGVAEHSACAANCLSMGKAGGRCENGICLCRKTTFKELWDKRF | VTCDLLSFKGQVNDSACAANCLSLGKAGGHCEKGVCICRKTSFKDLWDKRF |
| 01752| Defensin-NV | sp|P17722| DEFI_APIME | 75.0 | VTCELLMFGGVVGDSACAANCLSMGKAGGSCNGGLCDCRKTTFKELWDKRFG | VTCDLLSFKGQVNDSACAANCLSLGKAGGHCEKGVCICRKTSFKDLWDKRFG |
| 01358| A. | sp|P17722| DEFI_APIME | 60.5 | VTCDLLSFEAKGFAANHSLCAAHCLAIGRRGGSCERGVCICRR | VTCDLLSFKGQVNDSACAANCLSLGKAGGHCEKGVCICRK |
| 02735| Oryctes | sp|P17722| DEFI_APIME | 53.5 | LTCDLLSFEAKGFAANHSLCAAHCLAIGRKGGACQNGVCVCRR | VTCDLLSFKGQVNDSACAANCLSLGKAGGHCEKGVCICRK |
| 01213| Hymenoptaecin | sp|Q10416| HYTA_APIME | 100.0 | RGSIVIQGTKEGKSRPSLDIDYKQRVYDKNGMTGDAYGGLNIRPGQPSRQHAGFEFGKEYKNGFIKGQSEVQRGPGGRLSPYFGINGGFRF | RGSIVIQGTKEGKSRPSLDIDYKQRVYDKNGMTGDAYGGLNIRPGQPSRQHAGFEFGKEYKNGFIKGQSEVQRGPGGRLSPYFGINGGFRF |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Łyskowski, A.; Miłek, M.; Dżugan, M. Assessing the Antimicrobial Properties of Honey Protein Components through In Silico Comparative Peptide Composition and Distribution Analysis. Antibiotics 2023, 12, 830. https://doi.org/10.3390/antibiotics12050830
Łyskowski A, Miłek M, Dżugan M. Assessing the Antimicrobial Properties of Honey Protein Components through In Silico Comparative Peptide Composition and Distribution Analysis. Antibiotics. 2023; 12(5):830. https://doi.org/10.3390/antibiotics12050830
Chicago/Turabian StyleŁyskowski, Andrzej, Michał Miłek, and Małgorzata Dżugan. 2023. "Assessing the Antimicrobial Properties of Honey Protein Components through In Silico Comparative Peptide Composition and Distribution Analysis" Antibiotics 12, no. 5: 830. https://doi.org/10.3390/antibiotics12050830
APA StyleŁyskowski, A., Miłek, M., & Dżugan, M. (2023). Assessing the Antimicrobial Properties of Honey Protein Components through In Silico Comparative Peptide Composition and Distribution Analysis. Antibiotics, 12(5), 830. https://doi.org/10.3390/antibiotics12050830