Co-Harboring of Beta-Lactamases and mcr-1 Genes in Escherichia coli and Klebsiella pneumoniae from Healthy Carriers and Backyard Animals in Rural Communities in Ecuador
Abstract
1. Introduction
2. Results
2.1. Genomic Coexistence of β-Lactamase and Mcr-1 Resistance Genes
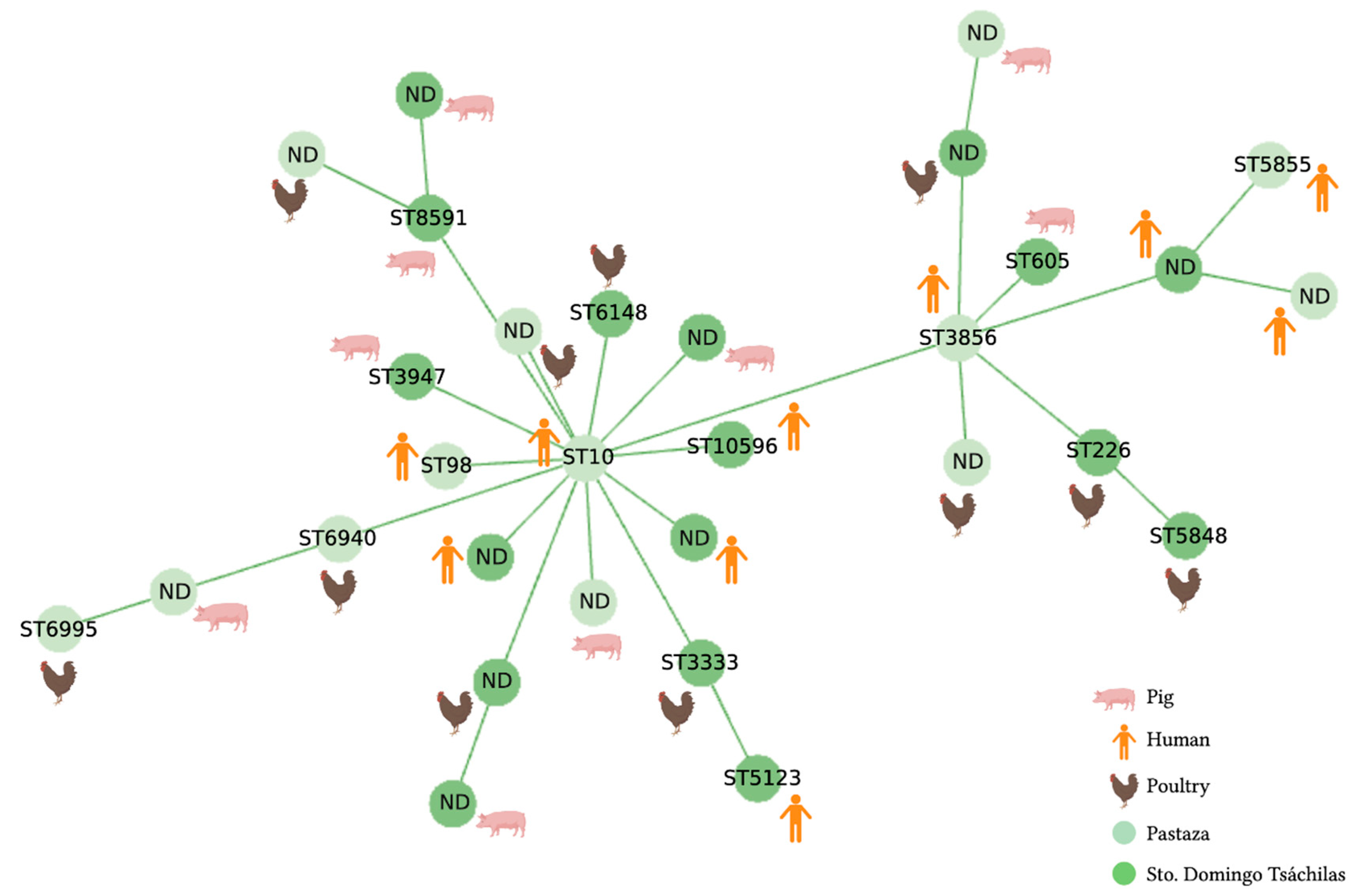
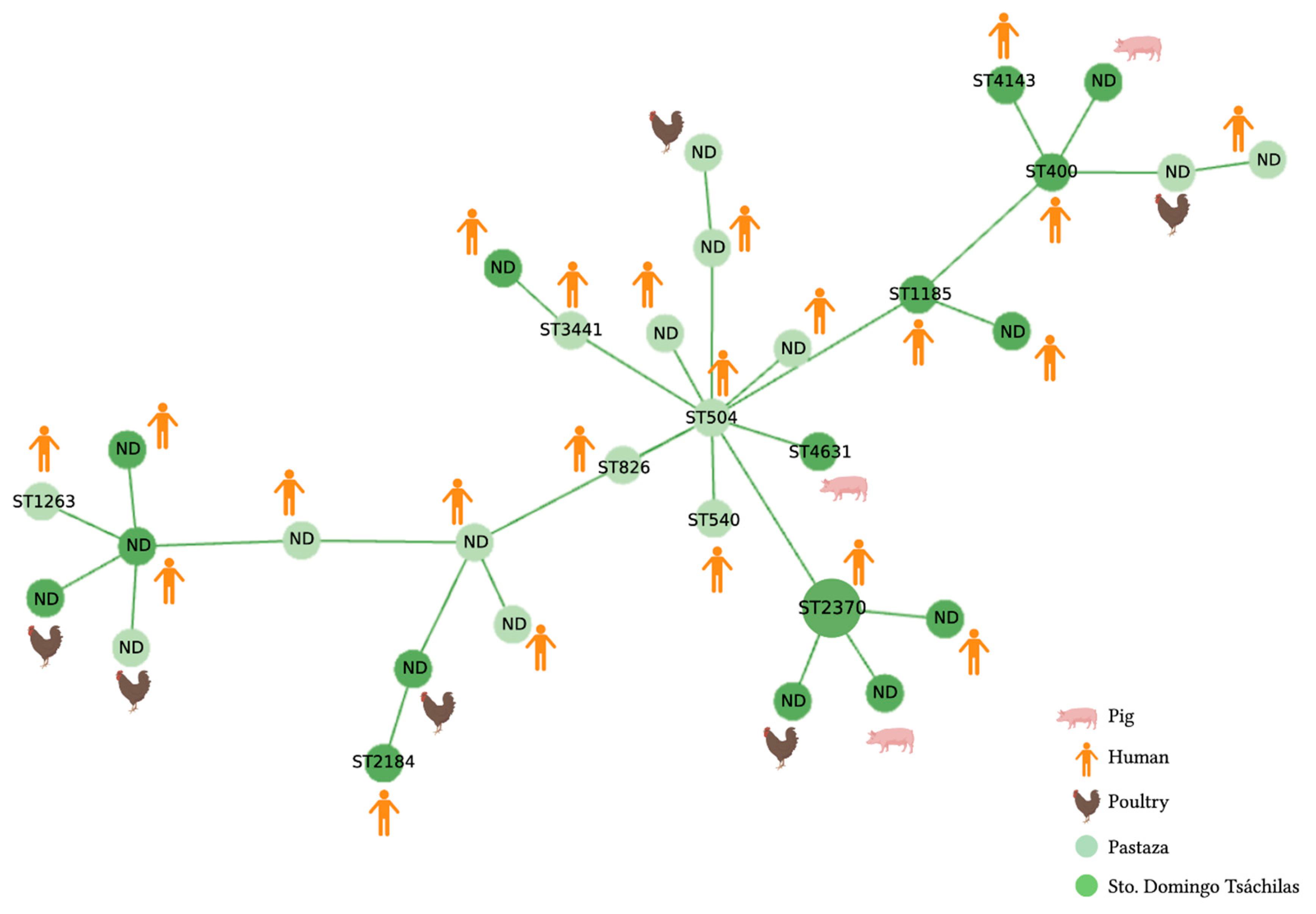
2.2. MLST Analysis
3. Discussion
3.1. Co-Harboring of Resistance Genes
3.2. MLST Analysis
4. Conclusions
5. Material and Methods
5.1. Bacterial Isolates
5.2. DNA Extraction
5.3. DNA Quantification
5.4. Molecular-Genetic Identification of β-Lactamase-Encoding Genes
5.5. Multilocus Sequence Typing (MLST)
5.6. Phylogenetic Analysis
6. Limitations of This Study
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO; Advisory Group on Integrated Surveillance (AGISAR). Critically Important Antimicrobials for Human Medicine 6th Revision 2018 Ranking of Medically Important Antimicrobials for Risk Management of Antimicrobial Resistance Due to Non-Human Use; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- McEwen, S.A.; Collignon, P.J.; Aarestrup, F.M.; Schwarz, S.; Shen, J.; Cavaco, L. Antimicrobial Resistance: A One Health Perspective. Microbiol. Spectr. 2018, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Samreen; Ahmad, I.; Malak, H.A.; Abulreesh, H.H. Environmental Antimicrobial Resistance and Its Drivers: A Potential Threat to Public Health. J. Glob. Antimicrob. Resist. 2021, 27, 101–111. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Calero-Cáceres, W.; Medina, J.; Ortuño-Gutiérrez, N.; Sunyoto, T.; Bastidas-Caldes, C.; Ramírez, M.S.; Harries, A.D. Genomic Insights of Mcr-1 Harboring Escherichia coli by Geographical Region and a One-Health Perspective. Front. Microbiol. 2023, 13, 1032753. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Calvopina, M.; Izurieta, R.; Villacres, I.; Kawahara, R.; Sasaki, M.; Yamamoto, M. Colistin-Resistant Escherichia coli with Mcr Genes in the Livestock of Rural Small-Scale Farms in Ecuador. BMC Res. Notes 2019, 12, 121. [Google Scholar] [CrossRef]
- Nang, S.C.; Li, J.; Velkov, T. The Rise and Spread of Mcr Plasmid-Mediated Polymyxin Resistance. Crit. Rev. Microbiol. 2019, 45, 131–161. [Google Scholar] [CrossRef]
- Li, Q.; Wang, H.; Xu, Y.; Bai, X.; Wang, J.; Zhang, Z.; Liu, X.; Miao, Y.; Zhang, L.; Li, X.; et al. Multidrug-Resistant Escherichia Albertii: Co-Occurrence of β-Lactamase and MCR-1 Encoding Genes. Front. Microbiol. 2018, 9, 258. [Google Scholar] [CrossRef]
- Kong, L.H.; Lei, C.W.; Ma, S.Z.; Jiang, W.; Liu, B.H.; Wang, Y.X.; Guan, R.; Men, S.; Yuan, Q.W.; Cheng, G.Y.; et al. Various Sequence Types of Escherichia coli Isolates Coharboring BlaNDM-5 and Mcr-1 Genes from a Commercial Swine Farm in China. Antimicrob. Agents Chemother. 2017, 61, e02167-16. [Google Scholar] [CrossRef]
- Gupta, C.L.; Blum, S.E.; Kattusamy, K.; Daniel, T.; Druyan, S.; Shapira, R.; Krifucks, O.; Zhu, Y.G.; Zhou, X.Y.; Su, J.Q.; et al. Longitudinal Study on the Effects of Growth-Promoting and Therapeutic Antibiotics on the Dynamics of Chicken Cloacal and Litter Microbiomes and Resistomes. Microbiome 2021, 9, 178. [Google Scholar] [CrossRef]
- Plata, G.; Baxter, N.T.; Susanti, D.; Volland-Munson, A.; Gangaiah, D.; Nagireddy, A.; Mane, S.P.; Balakuntla, J.; Hawkins, T.B.; Kumar Mahajan, A. Growth Promotion and Antibiotic Induced Metabolic Shifts in the Chicken Gut Microbiome. Commun. Biol. 2022, 5, 293. [Google Scholar] [CrossRef]
- Collignon, P.; McEwen, S. One Health—Its Importance in Helping to Better Control Antimicrobial Resistance. Trop. Med. Infect. Dis. 2019, 4, 22. [Google Scholar] [CrossRef] [PubMed]
- Thakuria, B.; Lahon, K. The Beta Lactam Antibiotics as an Empirical Therapy in a Developing Country: An Update on Their Current Status and Recommendations to Counter the Resistance against Them. J. Clin. Diagn. Res. 2013, 7, 1207–1214. [Google Scholar] [CrossRef]
- Alegría, Á.; Arias-Temprano, M.; Fernández-Natal, I.; Rodríguez-Calleja, J.M.; García-López, M.L.; Santos, J.A. Molecular Diversity of ESBL-Producing Escherichia coli from Foods of Animal Origin and Human Patients. Int. J. Environ. Res. Public Health 2020, 17, 1312. [Google Scholar] [CrossRef]
- Doi, Y.; Iovleva, A.; Bonomo, R.A. The Ecology of Extended-Spectrum β-Lactamases (ESBLs) in the Developed World. J. Travel Med. 2017, 24, S44–S51. [Google Scholar] [CrossRef] [PubMed]
- Woerther, P.L.; Burdet, C.; Chachaty, E.; Andremont, A. Trends in Human Fecal Carriage of Extended-Spectrum β-Lactamases in the Community: Toward the Globalization of CTX-M. Clin. Microbiol. Rev. 2013, 26, 744–758. [Google Scholar] [CrossRef]
- Bastidas-Caldes, C.; Ochoa, J.; Guerrero-Latorre, L.; Moyota-Tello, C.; Tapia, W.; Rey-Pérez, J.M.; Baroja, M.I. Removal of Extended-Spectrum Beta-Lactamase-Producing Escherichia coli, ST98, in Water for Human Consumption by Black Ceramic Water Filters in Low-Income Ecuadorian Highlands. Int. J. Environ. Res. Public Health 2022, 19, 4736. [Google Scholar] [CrossRef]
- Peirano, G.; Richardson, D.; Nigrin, J.; McGeer, A.; Loo, V.; Toye, B.; Alfa, M.; Pienaar, C.; Kibsey, P.; Pitout, J.D.D. High Prevalence of ST131 Isolates Producing CTX-M-15 and CTX-M-14 among Extended-Spectrum-β-Lactamase-Producing Escherichia coli Isolates from Canada. Antimicrob. Agents Chemother. 2010, 54, 1327–1330. [Google Scholar] [CrossRef]
- Carattoli, A. Animal Reservoirs for Extended Spectrum Beta-Lactamase Producers. Clin. Microbiol. Infect. 2008, 14 (Suppl. S1), 117–123. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-Spectrum β-Lactamases: An Update on Their Characteristics, Epidemiology and Detection. JAC-Antimicrob. Resist. 2021, 3, dlab092. [Google Scholar] [CrossRef]
- Soria Segarra, C.; Soria Baquero, E.; Cartelle Gestal, M. High Prevalence of CTX-M-1-Like Enzymes in Urinary Isolates of Escherichia coli in Guayaquil, Ecuador. Microb. Drug Resist. 2018, 24, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Bastidas-Caldes, C.; Romero-Alvarez, D.; Valdez-Vélez, V.; Morales, R.D.; Montalvo-Hernández, A.; Gomes-Dias, C.; Calvopiña, M. Extended-Spectrum Beta-Lactamases Producing Escherichia coli in South America: A Systematic Review with a One Health Perspective. Infect. Drug Resist. 2022, 15, 5759–5779. [Google Scholar] [CrossRef]
- Cantón, R.; Coque, T.M. The CTX-M β-Lactamase Pandemic. Curr. Opin. Microbiol. 2006, 9, 466–475. [Google Scholar] [CrossRef] [PubMed]
- Peirano, G.; Pitout, J.D.D. Molecular Epidemiology of Escherichia coli Producing CTX-M β-Lactamases: The Worldwide Emergence of Clone ST131 O25:H4. Int. J. Antimicrob. Agents 2010, 35, 316–321. [Google Scholar] [CrossRef]
- Mitsuda, T.; Muto, T.; Yamada, M.; Kobayashi, N.; Toba, M.; Aihara, Y.; Ito, A.; Yokota, S. Epidemiological Study of a Food-Borne Outbreak of Enterotoxigenic Escherichia coli O25:NM by Pulsed-Field Gel Electrophoresis and Randomly Amplified Polymorphic DNA Analysis. J. Clin. Microbiol. 1998, 36, 652–656. [Google Scholar] [CrossRef] [PubMed]
- Vinueza-Burgos, C.; Ortega-Paredes, D.; Narvaéz, C.; De Zutter, L.; Zurita, J. Characterization of Cefotaxime Resistant Escherichia coli Isolated from Broiler Farms in Ecuador. PLoS ONE 2019, 14, e0207567. [Google Scholar] [CrossRef]
- Villegas, M.V.; Pallares, C.J.; Escandón-Vargas, K.; Hernández-Gómez, C.; Correa, A.; Álvarez, C.; Rosso, F.; Matta, L.; Luna, C.; Zurita, J.; et al. Characterization and Clinical Impact of Bloodstream Infection Caused by Carbapenemase-Producing Enterobacteriaceae in Seven Latin American Countries. PLoS ONE 2016, 11, 1–13. [Google Scholar] [CrossRef]
- Zurita, J.; Yánez, F.; Sevillano, G.; Ortega-Paredes, D.; Miño, A.P.Y. Ready-to-Eat Street Food: A Potential Source for Dissemination of Multidrug-Resistant. Lett. Appl. Microbiol. 2020, 70, 203–209. [Google Scholar] [CrossRef]
- Ortega-Paredes, D.; Barba, P.; Mena-López, S.; Espinel, N.; Zurita, J. Escherichia coli Hyperepidemic Clone ST410-A Harboring blaCTX-M-15 Isolated from Fresh Vegetables in a Municipal Market in Quito-Ecuador. Int. J. Food Microbiol. 2018, 280, 41–45. [Google Scholar] [CrossRef]
- Villacís, J.E.; Reyes, J.A.; Castelán-Sánchez, H.G.; Dávila-Ramos, S.; Lazo, M.A.; Wali, A.; Bodero, L.A.; Toapanta, Y.; Naranjo, C.; Montero, L.; et al. OXA-48 Carbapenemase in Klebsiella pneumoniae Sequence Type 307 in Ecuador. Microorganisms 2020, 8, 435. [Google Scholar] [CrossRef]
- Nuñez Quezada, T.; Rodríguez, C.H.; Castro Cañarte, G.; Nastro, M.; Balderrama Yarhui, N.; Dabos, L.; Acosta Mosquera, Y.; Plaza Moreira, N.; Famiglietti, A. Outbreak of BlaOXA-72-Producing Acinetobacter Baumannii in South America. J. Chemother. 2017, 29, 321–324. [Google Scholar] [CrossRef] [PubMed]
- Reyes, J.A.; Villavicencio, F.; Villacís, J.E.; Pavón, E.; Campoverde, N.; Espinel, M.; Núñez, B.; Trueba, G. First Report of a Clinical Isolate of BlaOXA-48- Carbapenemase Producing Raoultella Ornithinolytica in South America. Rev. Argent. Microbiol. 2020, 52, 82–83. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Paredes, D.; de Janon, S.; Villavicencio, F.; Ruales, K.J.; De La Torre, K.; Villacís, J.E.; Wagenaar, J.A.; Matheu, J.; Bravo-Vallejo, C.; Fernández-Moreira, E.; et al. Broiler Farms and Carcasses Are an Important Reservoir of Multi-Drug Resistant Escherichia coli in Ecuador. Front. Vet. Sci. 2020, 7, 1–8. [Google Scholar] [CrossRef]
- Hoepers, P.G.; Silva, P.L.; Rossi, D.A.; Valadares Júnior, E.C.; Ferreira, B.C.; Zuffo, J.P.; Koerich, P.K.; Fonseca, B.B. The Association between Extended Spectrum Beta-Lactamase (ESBL) and Ampicillin C (AmpC) Beta-Lactamase Genes with Multidrug Resistance in Escherichia coli Isolates Recovered from Turkeys in Brazil. Br. Poult. Sci. 2018, 59, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Crecencio, R.B.; Brisola, M.C.; Bitner, D.; Frigo, A.; Rampazzo, L.; Borges, K.A.; Furian, T.Q.; Salle, C.T.P.; Moraes, H.L.S.; Faria, G.A.; et al. Antimicrobial Susceptibility, Biofilm Formation and Genetic Profiles of Escherichia coli Isolated from Retail Chicken Meat. Infect. Genet. Evol. 2020, 84, 104355. [Google Scholar] [CrossRef]
- Jena, J.; Sahoo, R.K.; Debata, N.K.; Subudhi, E. Prevalence of TEM, SHV, and CTX-M Genes of Extended-Spectrum β-Lactamase-Producing Escherichia coli Strains Isolated from Urinary Tract Infections in Adults. 3 Biotech 2017, 7, 1–17. [Google Scholar] [CrossRef]
- Abujnah, A.A.; Zorgani, A.; Sabri, M.A.M.; El-Mohammady, H.; Khalek, R.A.; Ghenghesh, K.S. Multidrug Resistance and Extended-Spectrum β-Lactamases Genes among Escherichia coli from Patients with Urinary Tract Infections in Northwestern Libya. Libyan J. Med. 2015, 10, 1–7. [Google Scholar] [CrossRef]
- Wu, C.; Wang, Y.; Shi, X.; Wang, S.; Ren, H.; Shen, Z.; Wang, Y.; Lin, J.; Wang, S. Rapid Rise of the ESBL and Mcr-1 Genes in Escherichia coli of Chicken Origin in China, 2008–2014. Emerg. Microbes Infect. 2018, 7, 1–10. [Google Scholar] [CrossRef]
- Moghanni, M.; Ghazvini, K.; Farsiani, H.; Namaei, M.H.; Derakhshan, M.; Yousefi, M.; Maragheh, A.; Jamehdar, S.A. High Prevalence of Sequence Type 131 Isolates Producing CTX-M-15 among Extended-Spectrum β-Lactamase-Producing Escherichia coli Strains in Northeast Iran. J. Glob. Antimicrob. Resist. 2018, 15, 74–78. [Google Scholar] [CrossRef]
- Amin, M.B.; Sraboni, A.S.; Hossain, M.I.; Roy, S.; Mozmader, T.A.U.; Unicomb, L.; Rousham, E.K.; Islam, M.A. Occurrence and Genetic Characteristics of Mcr-1-Positive Colistin-Resistant E. coli from Poultry Environments in Bangladesh. J. Glob. Antimicrob. Resist. 2020, 22, 546–552. [Google Scholar] [CrossRef]
- Shafiq, M.; Huang, J.; Ur Rahman, S.; Shah, J.M.; Chen, L.; Gao, Y.; Wang, M.; Wang, L. High Incidence of Multidrug-Resistant Escherichia coli Coharboring Mcr-1 and blaCTX-M-15 Recovered from Pigs. Infect. Drug Resist. 2019, 12, 2135–2149. [Google Scholar] [CrossRef]
- D’Andrea, M.M.; Arena, F.; Pallecchi, L.; Rossolini, G.M. CTX-M-Type β-Lactamases: A Successful Story of Antibiotic Resistance. Int. J. Med. Microbiol. 2013, 303, 305–317. [Google Scholar] [CrossRef]
- Ur Rahman, S.; Ali, T.; Ali, I.; Khan, N.A.; Han, B.; Gao, J. The Growing Genetic and Functional Diversity of Extended Spectrum Beta-Lactamases. BioMed Res. Int. 2018, 2018, 9519718. [Google Scholar] [CrossRef] [PubMed]
- Araque, M.; Labrador, I. Prevalence of Fecal Carriage of CTX-M-15 Beta-Lactamase-Producing Escherichia coli in Healthy Children from a Rural Andean Village in Venezuela. Osong Public Health Res. Perspect. 2018, 9, 9–15. [Google Scholar] [CrossRef]
- Ryoo, N.H.; Kim, E.C.; Hong, S.G.; Park, Y.J.; Lee, K.; Bae, I.K.; Song, E.H.; Jeong, S.H. Dissemination of SHV-12 and CTX-M-Type Extended-Spectrum β-Lactamases among Clinical Isolates of Escherichia coli and Klebsiella pneumoniae and Emergence of GES-3 in Korea. J. Antimicrob. Chemother. 2005, 56, 698–702. [Google Scholar] [CrossRef]
- Komijani, M.; Bouzari, M.; Rahimi, F. Detection of TEM, SHV and CTX-M Antibiotic Resistance Genes in Escherichia coli Isolates from Infected Wounds. Med. Lab. J. 2017, 11, 30–35. [Google Scholar]
- Hülter, N.; Ilhan, J.; Wein, T.; Kadibalban, A.S.; Hammerschmidt, K.; Dagan, T. An Evolutionary Perspective on Plasmid Lifestyle Modes. Curr. Opin. Microbiol. 2017, 38, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Tsang, J. Bacterial Plasmid Addiction Systems and Their Implications for Antibiotic Drug Development. In Postdoc Journal: A Journal of Postdoctoral Research and Postdoctoral Affairs; NIH: Washington, DC, USA, 2017. [Google Scholar]
- Touati, A.; Mairi, A. Plasmid-Determined Colistin Resistance in the North African Countries: A Systematic Review. Microb. Drug Resist. 2021, 27, 121–133. [Google Scholar] [CrossRef]
- Millan, A.S.; Escudero, J.A.; Gifford, D.R.; Mazel, D.; MacLean, R.C. Multicopy Plasmids Potentiate the Evolution of Antibiotic Resistance in Bacteria. Nat. Ecol. Evol. 2016, 1, 0010. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wang, J.; Wang, X.; Bai, X.; Ma, J.; Dang, R.; Xiong, Y.; Fanning, S.; Bai, L.; Yang, Z. Characterization of Five Escherichia coli Isolates Co-Expressing ESBL and Mcr-1 Resistance Mechanisms from Different Origins in China. Front. Microbiol. 2019, 10, 1994. [Google Scholar] [CrossRef]
- Díaz-Jiménez, D.; García-Meniño, I.; Herrera, A.; García, V.; López-Beceiro, A.M.; Alonso, M.P.; Blanco, J.; Mora, A. Genomic Characterization of Escherichia coli Isolates Belonging to a New Hybrid Aepec/Expec Pathotype O153:H10-a-St10 Eae-Beta1 Occurred in Meat, Poultry, Wildlife and Human Diarrheagenic Samples. Antibiotics 2020, 9, 192. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, Y.; Hagiya, H.; Akeda, Y.; Takeuchi, D.; Sakamoto, N.; Matsumoto, Y.; Motooka, D.; Nishi, I.; Tomono, K.; Hamada, S. Community Spread and Acquisition of Clinically Relevant Escherichia coli Harbouring BlaNDMamong Healthy Japanese Residents of Yangon, Myanmar. J. Antimicrob. Chemother. 2021, 76, 1448–1454. [Google Scholar] [CrossRef]
- Cheng, P.; Yang, Y.; Cao, S.; Liu, H.; Li, X.; Sun, J.; Li, F.; Ishfaq, M.; Zhang, X. Prevalence and Characteristic of Swine-Origin Mcr-1-Positive Escherichia coli in Northeastern China. Front. Microbiol. 2021, 12, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ćwiek, K.; Woźniak-Biel, A.; Karwańska, M.; Siedlecka, M.; Lammens, C.; Rebelo, A.R.; Hendriksen, R.S.; Kuczkowski, M.; Chmielewska-Władyka, M.; Wieliczko, A. Phenotypic and Genotypic Characterization of Mcr-1-Positive Multidrug-Resistant Escherichia coli ST93, ST117, ST156, ST10, and ST744 Isolated from Poultry in Poland. Brazilian J. Microbiol. 2021, 52, 1597–1609. [Google Scholar] [CrossRef]
- He, W.Y.; Zhang, X.X.; Gao, G.L.; Gao, M.Y.; Zhong, F.G.; Lv, L.C.; Cai, Z.P.; Si, X.F.; Yang, J.; Liu, J.H. Clonal Spread of Escherichia coli O101:H9-St10 and O101:H9-St167 Strains Carrying fosA3 and blaCTX-M-14 among Diarrheal Calves in a Chinese Farm, with Australian Chroicocephalus as the Possible Origin of E. coli O101:H9-St10. Zool. Res. 2021, 42, 461–468. [Google Scholar] [CrossRef]
- Blaak, H.; De Kruijf, P.; Hamidjaja, R.A.; Van Hoek, A.H.A.M.; De Roda Husman, A.M.; Schets, F.M. Prevalence and Characteristics of ESBL-Producing E. coli in Dutch Recreational Waters Influenced by Wastewater Treatment Plants. Vet. Microbiol. 2014, 171, 448–459. [Google Scholar] [CrossRef] [PubMed]
- MK, A.; JKP, K.; RS, H.; EC, O.; Thakur, S. Genetic Relatedness of Multidrug Resistant Escherichia coli Isolated from Humans, Chickens and Poultry Environments. Antimicrob. Resist. Infect. Control. 2021, 10, 58. [Google Scholar]
- Matamoros, S.; Van Hattem, J.M.; Arcilla, M.S.; Willemse, N.; Melles, D.C.; Penders, J.; Vinh, T.N.; Thi Hoa, N.; Bootsma, M.C.J.; Van Genderen, P.J.; et al. Global Phylogenetic Analysis of Escherichia coli and Plasmids Carrying the Mcr-1 Gene Indicates Bacterial Diversity but Plasmid Restriction. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Chen, X.; Liu, W.; Li, H.; Yan, S.; Jiang, F.; Cai, W.; Li, G. Whole Genome Sequencing Analysis of Avian Pathogenic Escherichia coli from China. Vet. Microbiol. 2021, 259, 109158. [Google Scholar] [CrossRef]
- Geyer, C.N.; Fowler, R.C.; Johnson, J.R.; Johnston, B.; Weissman, S.J.; Hawkey, P.; Hanson, N.D. Evaluation of CTX-M Steady-State MRNA, MRNA Half-Life and Protein Production in Various STs of Escherichia coli. J. Antimicrob. Chemother. 2016, 71, 607–616. [Google Scholar] [CrossRef]
- Chen, B.; Berglund, B.; Wang, S.; Börjesson, S.; Bi, Z.; Nilsson, M.; Yin, H.; Zheng, B.; Xiao, Y.; Bi, Z.; et al. Rapid Increase in Occurrence of Carbapenem-Resistant Enterobacteriaceae in Healthy Rural Residents in Shandong Province, China, from 2015 to 2017. J. Glob. Antimicrob. Resist. 2022, 28, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Truong, D.T.Q.; Hounmanou, Y.M.G.; Dang, S.T.T.; Olsen, J.E.; Truong, G.T.H.; Tran, N.T.; Scheutz, F.; Dalsgaard, A. Genetic Comparison of Esbl-Producing Escherichia coli from Workers and Pigs at Vietnamese Pig Farms. Antibiotics 2021, 10, 1165. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, Y.; Xi, W.; Liu, S.; Liu, J.; Mu, H.; Chen, B.; He, H.; Fan, Y.; Ma, W.; et al. Genetic Features of Plasmid- and Chromosome-Mediated Mcr-1 in Escherichia coli Isolates From Animal Organs With Lesions. Front. Microbiol. 2021, 12, 707332. [Google Scholar] [CrossRef]
- Novovic, K.; Vasiljevic, Z.; Kuzmanovic, M.; Lozo, J.; Begovic, J.; Kojic, M.; Jovcic, B.; Novel, E. Coli ST5123 Containing BlaNDM-1 Carried by IncF Plasmid Isolated from a Pediatric Patient in Serbia. Microb. Drug Resist. 2016, 22, 707–711. [Google Scholar] [CrossRef]
- Abril, D.; Vergara, E.; Palacios, D.; Leal, A.L.; Marquez-Ortiz, R.A.; Madroñero, J.; Corredor Rozo, Z.L.; De La Rosa, Z.; Nieto, C.A.; Vanegas, N.; et al. Within Patient Genetic Diversity of Bla KPC Harboring Klebsiella pneumoniae in a Colombian Hospital and Identification of a New NTEKPC Platform. Sci. Rep. 2021, 11, 1–16. [Google Scholar] [CrossRef]
- Lu, B.; Zhou, H.; Zhang, X.; Qu, M.; Huang, Y.; Wang, Q. Molecular Characterization of Klebsiella pneumoniae Isolates from Stool Specimens of Outpatients in Sentinel Hospitals Beijing, China, 2010-2015. Gut Pathog. 2017, 9, 1–5. [Google Scholar] [CrossRef]
- Yin, L.; He, L.; Miao, J.; Yang, W.; Wang, X.; Ma, J.; Wu, N.; Cao, Y.; Wang, L.; Lu, G.; et al. Active Surveillance and Appropriate Patient Placement in Contact Isolation Dramatically Decreased Carbapenem-Resistant Enterobacterales Infection and Colonization in Paediatric Patients in China. J. Hosp. Infect. 2020, 105. [Google Scholar] [CrossRef] [PubMed]
- Compain, F.; Decré, D.; Fulgencio, J.P.; Berraho, S.; Arlet, G.; Verdet, C. Molecular Characterization of DHA-1-Producing Klebsiella pneumoniae Isolates Collected during a 4-Year Period in an Intensive Care Unit. Diagn. Microbiol. Infect. Dis. 2014, 80, 159–161. [Google Scholar] [CrossRef]
- Zhang, D.F.; Zhang, Z.F.; Li, P.D.; Qu, P.H. Characterization of Carbapenem-Resistant Acinetobacter Baumannii ST540 and Klebsiella pneumoniae ST2237 Isolates in a Pneumonia Case from China. J. Appl. Microbiol. 2022, 133, 1434–1445. [Google Scholar] [CrossRef]
- Rakotondrasoa, A.; Andrianonimiadana, L.M.; Rahajandraibe, S.; Razafimahatratra, S.; Andrianaivoarimanana, V.; Rahelinirina, S.; Crucitti, T.; Brisse, S.; Jeannoda, V.; Rajerison, M.; et al. Characterization of Klebsiella pneumoniae Isolated from Patients Suspected of Pulmonary or Bubonic Plague during the Madagascar Epidemic in 2017. Sci. Rep. 2022, 12, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Saxenborn, P.; Baxter, J.; Tilevik, A.; Fagerlind, M.; Dyrkell, F.; Pernestig, A.K.; Enroth, H.; Tilevik, D. Genotypic Characterization of Clinical Klebsiella Spp. Isolates Collected From Patients With Suspected Community-Onset Sepsis, Sweden. Front. Microbiol. 2021, 12, 1091. [Google Scholar] [CrossRef] [PubMed]
- Bastidas-Caldes, C.; Guerrero-Freire, S.; Ortuño-Gutiérrez, N.; Sunyoto, T.; Gomes-Dias, C.A.; Ramírez, M.S.; Calero-Cáceres, W.; Harries, A.D.; Rey, J.; de Waard, J.H.; et al. Colistin resistance in Escherichia coli and Klebsiella pneumoniae in humans and backyard animals in Ecuador. Revista Panamericana Salud Pública 2023, 47, e48. [Google Scholar] [CrossRef] [PubMed]
- Suenaga, E.; Nakamura, H. Evaluation of Three Methods for Effective Extraction of DNA from Human Hair. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2005, 820, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Le, Q.P.; Ueda, S.; Nguyen, T.N.H.; Dao, T.V.K.; Van Hoang, T.A.; Tran, T.T.N.; Hirai, I.; Nakayama, T.; Kawahara, R.; Do, T.H.; et al. Characteristics of Extended-Spectrum β-Lactamase-Producing Escherichia coli in Retail Meats and Shrimp at a Local Market in Vietnam. Foodborne Pathog. Dis. 2015, 12, 719–725. [Google Scholar] [CrossRef]
- Poirel, L.; Walsh, T.R.; Cuvillier, V.; Nordmann, P. Multiplex PCR for Detection of Acquired Carbapenemase Genes. Diagn. Microbiol. Infect. Dis. 2011, 70, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Feil, E.J.; Li, B.C.; Aanensen, D.M.; Hanage, W.P.; Spratt, B.G. EBURST: Inferring Patterns of Evolutionary Descent among Clusters of Related Bacterial Genotypes from Multilocus Sequence Typing Data. J. Bacteriol. 2004, 186, 1518–1530. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Rebelo, A.R.; Bortolaia, V.; Leekitcharoenphon, P.; Hansen, D.S.; Nielsen, H.L.; Ellermann-Eriksen, S.; Kemp, M.; Røder, B.L.; Frimodt-Møller, N.; Søndergaard, T.S.; et al. One Day in Denmark: Comparison of Phenotypic and Genotypic Antimicrobial Susceptibility Testing in Bacterial Isolates From Clinical Settings. Front. Microbiol. 2022, 13, 804627. [Google Scholar] [CrossRef]


| Host | blaSHV | blaTEM | blaCTXM-1 | blaCTXM-9 | blaCTXM-8/25 | blaOXA48 | blaNDM | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Healthy humans (n = 10) | 1 * (10) | [0–34.4] | 9 (90) | [65.5–100] | 3 (30) | [0–67] | ||||||||
| Pigs (n = 9) | 1 (11.1) | [0–38] | 8 (89) | [62.1–100] | 2 (22.2) | [0–57] | 2 (22.2) | [0–57] | 1 (11.1) | [0–38] | ||||
| Chickens (n = 11) | 7 (63.6) | [26.2–100] | 1 (9.1) | [0–31.4] | 2 (18.1) | [11.8–48] | ||||||||
| Total isolates (N = 30) | 2 (6.6) | [0–18.2] | 24 (80) | [61–98] | 5 (16.6) | [1–34.1] | 6 (20) | [1.19–39] | 1 (3.3) | [0–11.7] | 2 (6.6) | [0–18.2] | 1 (3.3) | [0–11.7] |
| Host | blaSHV | blaTEM | blaCTXM-1 | blaCTXM-9 | blaCTXM-8/25 | blaNDM | blaKPC | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Healthy humans (n = 23) | 19 * (82.6) | [62.2–102] | 12 (37.5) | [11.5–63.5] | 2 (8.7) | [0–23.8] | 3 (13) | [0–17.3] | 1 (5.3) | [0–17.3] | 1 (5.3) | [0–17.3] | 1 (5.3) | [0–17.3] |
| Pigs (n = 3) | 3 (100) | [-] | 3 (100) | [-] | ||||||||||
| Chickens (n = 6) | 5 (83) | [43.5–100] | 4 (66.6) | [17–100] | 1 (16.6) | [0–55.7] | 1 (16.6) | [0–55.7] | ||||||
| Total of isolates (N = 32) | 27 (84.4) | [67.8–100] | 19 (59.4) | [37–81.7] | 3 (9.4) | [0–22.7] | 4 (13) | [0–17.4] | 1 (3.1) | [0–17.36] | 1 (3.1) | [0–17.36] | 1 (3.1) | [0–17.36] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bastidas-Caldes, C.; Cisneros-Vásquez, E.; Zambrano, A.; Mosquera-Maza, A.; Calero-Cáceres, W.; Rey, J.; Yamamoto, Y.; Yamamoto, M.; Calvopiña, M.; de Waard, J.H. Co-Harboring of Beta-Lactamases and mcr-1 Genes in Escherichia coli and Klebsiella pneumoniae from Healthy Carriers and Backyard Animals in Rural Communities in Ecuador. Antibiotics 2023, 12, 856. https://doi.org/10.3390/antibiotics12050856
Bastidas-Caldes C, Cisneros-Vásquez E, Zambrano A, Mosquera-Maza A, Calero-Cáceres W, Rey J, Yamamoto Y, Yamamoto M, Calvopiña M, de Waard JH. Co-Harboring of Beta-Lactamases and mcr-1 Genes in Escherichia coli and Klebsiella pneumoniae from Healthy Carriers and Backyard Animals in Rural Communities in Ecuador. Antibiotics. 2023; 12(5):856. https://doi.org/10.3390/antibiotics12050856
Chicago/Turabian StyleBastidas-Caldes, Carlos, Emily Cisneros-Vásquez, Antonella Zambrano, Andrea Mosquera-Maza, William Calero-Cáceres, Joaquín Rey, Yoshimasa Yamamoto, Mayumi Yamamoto, Manuel Calvopiña, and Jacobus H. de Waard. 2023. "Co-Harboring of Beta-Lactamases and mcr-1 Genes in Escherichia coli and Klebsiella pneumoniae from Healthy Carriers and Backyard Animals in Rural Communities in Ecuador" Antibiotics 12, no. 5: 856. https://doi.org/10.3390/antibiotics12050856
APA StyleBastidas-Caldes, C., Cisneros-Vásquez, E., Zambrano, A., Mosquera-Maza, A., Calero-Cáceres, W., Rey, J., Yamamoto, Y., Yamamoto, M., Calvopiña, M., & de Waard, J. H. (2023). Co-Harboring of Beta-Lactamases and mcr-1 Genes in Escherichia coli and Klebsiella pneumoniae from Healthy Carriers and Backyard Animals in Rural Communities in Ecuador. Antibiotics, 12(5), 856. https://doi.org/10.3390/antibiotics12050856











