Comparison of Fecal Antimicrobial Resistance Genes in Captive and Wild Asian Elephants
Abstract
1. Introduction
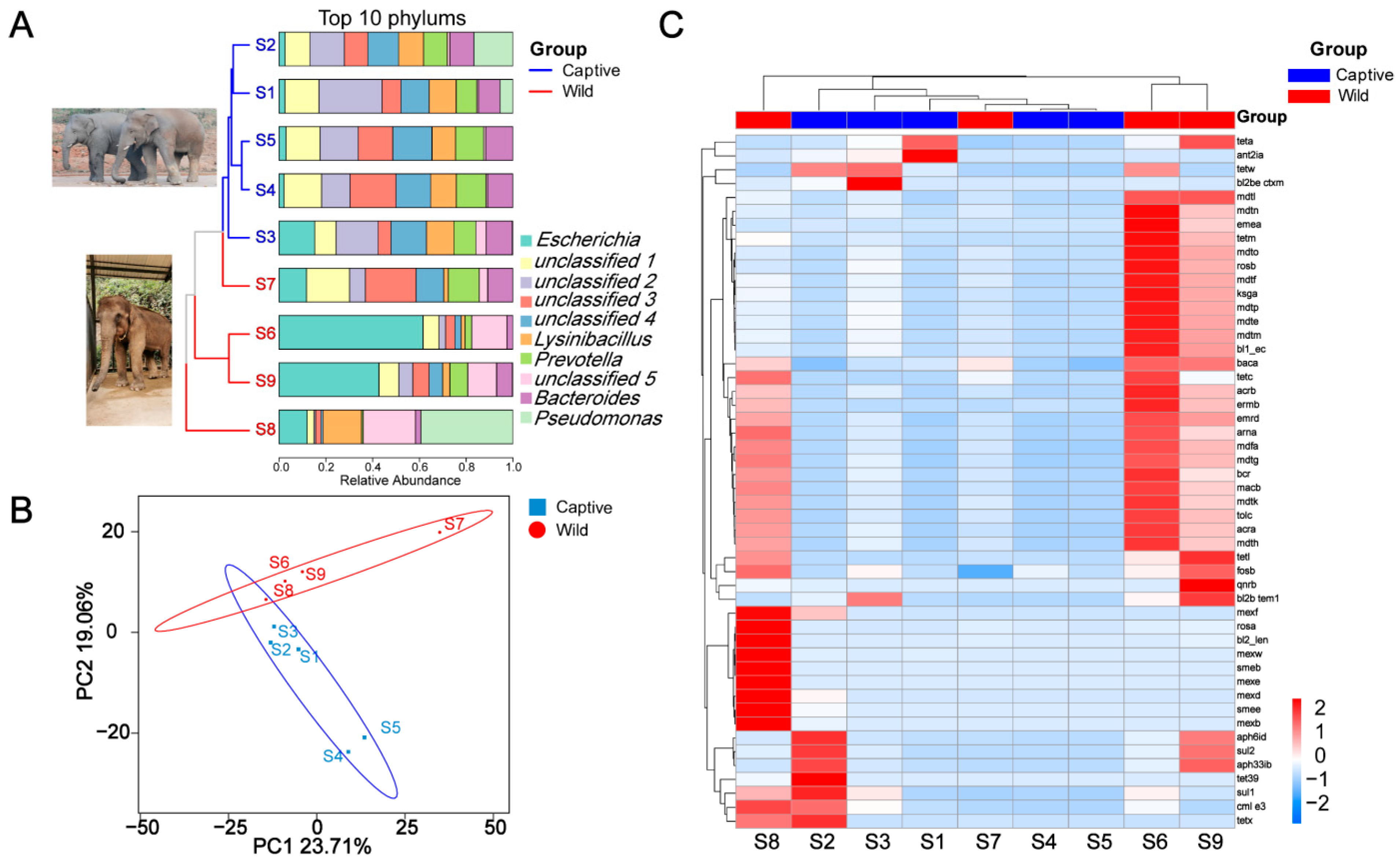
2. Results
2.1. Fecal Metabolomics Profile
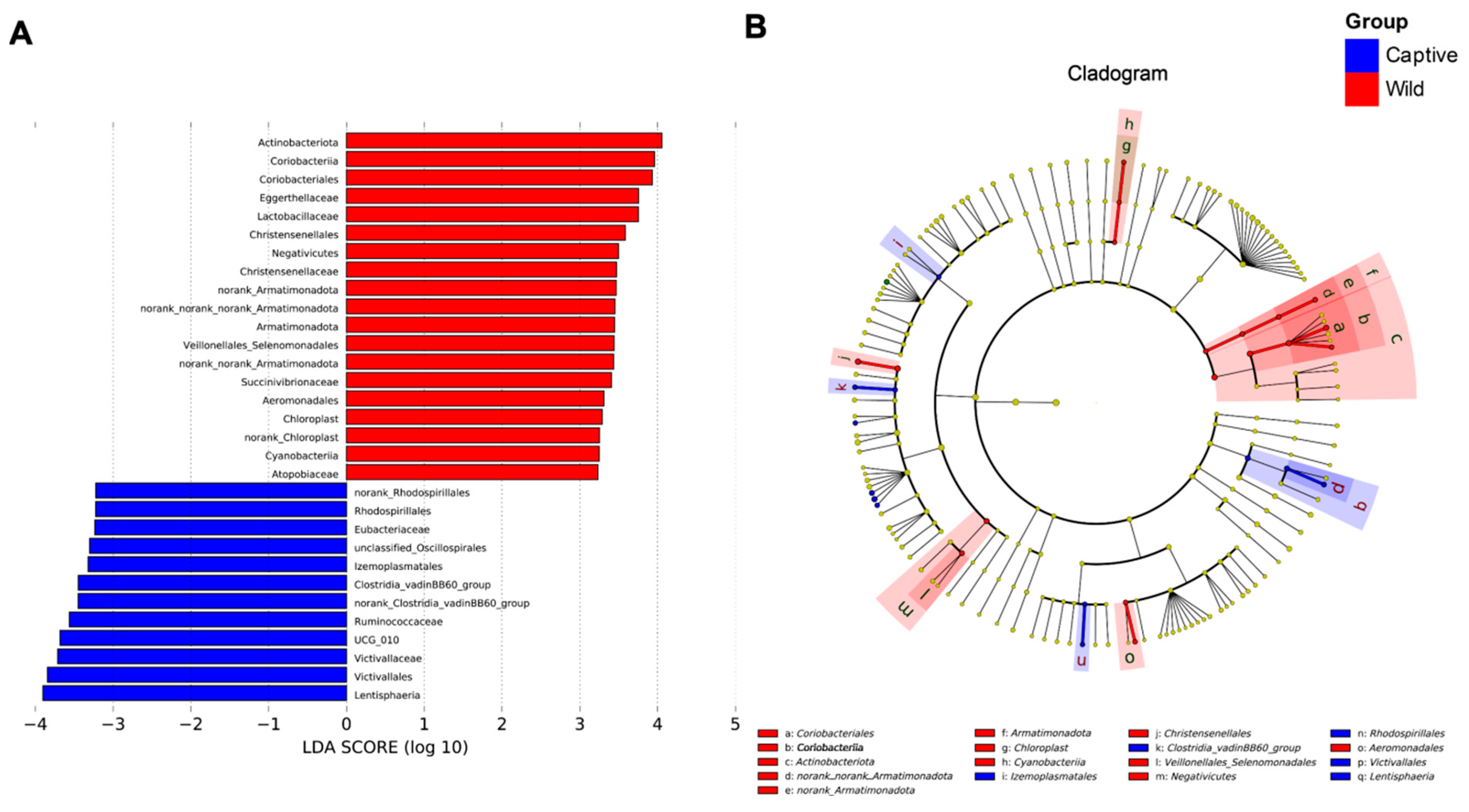
2.2. Linear Discriminant Analysis Effect Size (LEfSe) Analysis
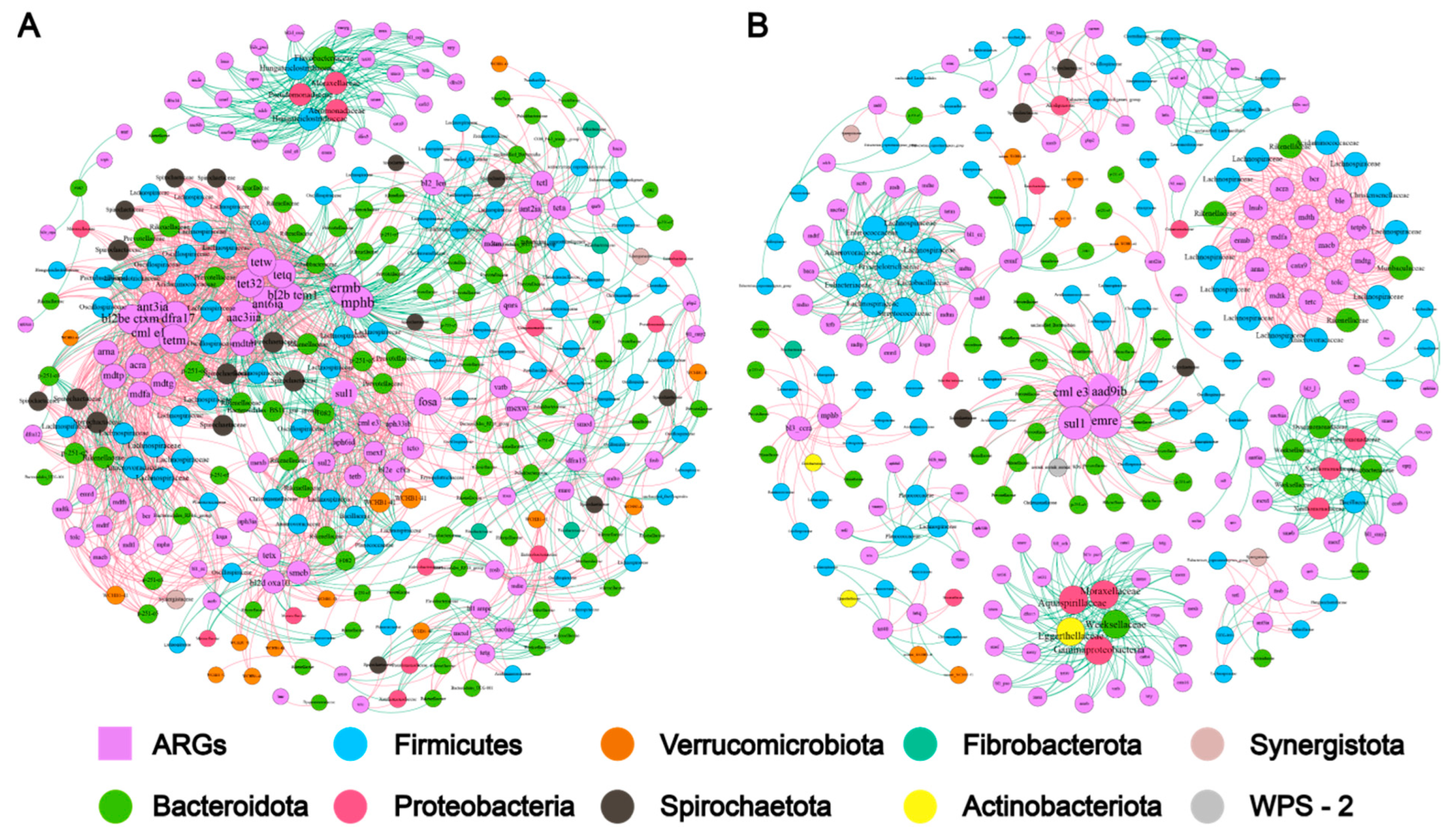
2.3. Microbiome-Metabolome Associations
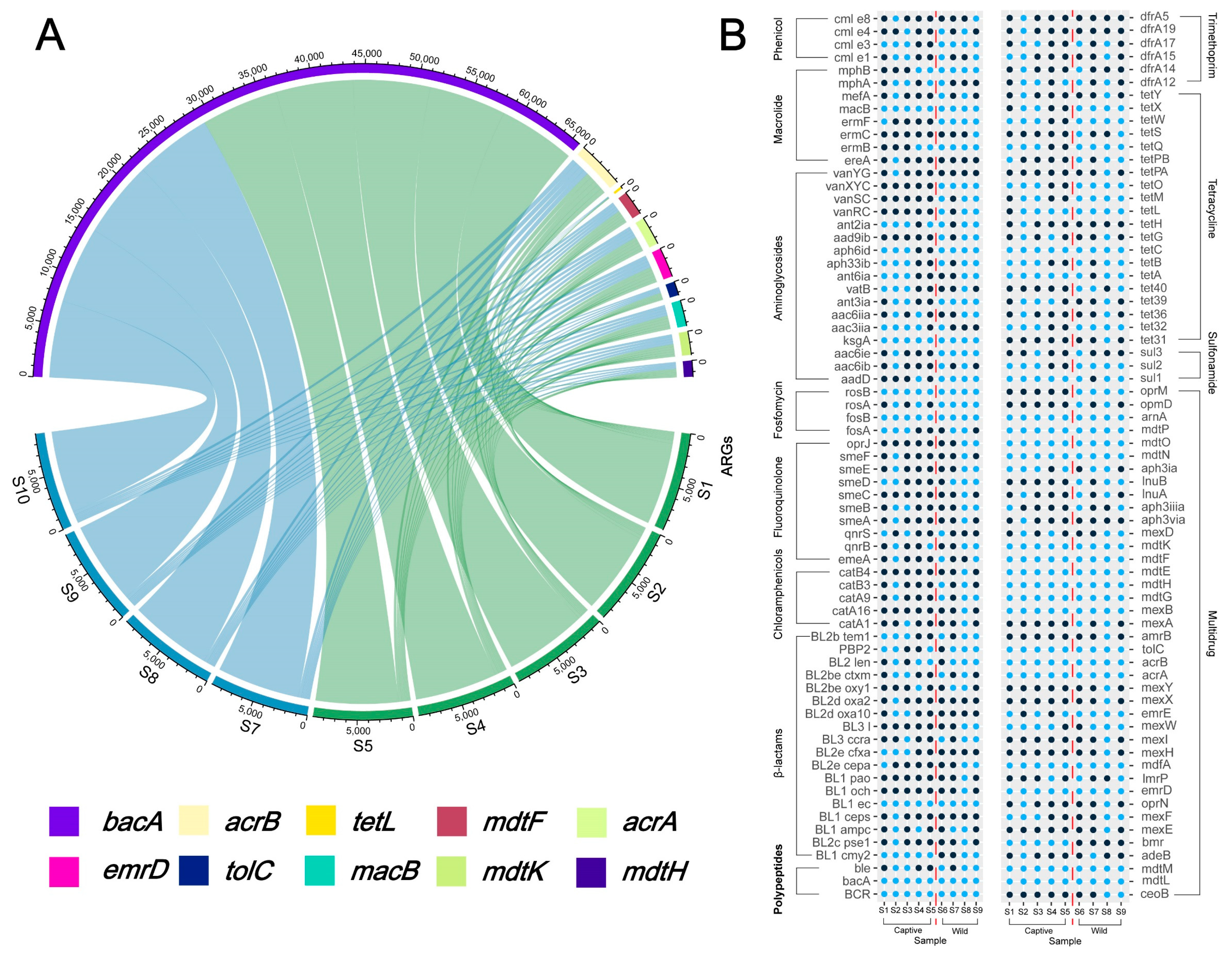
2.4. Overview of Resistance Gene Abundance
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Sample Collection and Pretreatment
5.2. DNA Extraction and PCR Amplification
5.3. Bioinformatics and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- IUCN. International Union for Conservation of Nature Red List. Available online: http://www.iucnredlist.org. (accessed on 1 February 2022).
- Wang, H.; Wang, P.; Zhao, X.; Zhang, W.; Li, J.; Xu, C.; Xie, P. What triggered the Asian elephant′s northward migration across southwestern Yunnan? Innovation 2021, 2. [Google Scholar] [CrossRef]
- Zhang, L. Research on Asian Elephant Conservation in China; China Science Publishing & Media Ltd.: Beijing, China, 2018; pp. 10–15. [Google Scholar]
- Jin, Y.; Fan, H. Land use/land cover change and its impacts on protected areas in Mengla County, Xishuangbanna, Southwest China. Environ. Monit. Assess. 2018, 190, 509. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, P.; Guo, X.; Wang, L.; Wang, Q.; Yu, Y.; Dai, Y.; Li, L.; Zhang, L. Human-elephant conflict in Xishuangbanna Prefecture, China: Distribution, diffusion, and mitigation. Glob. Ecol. Conserv. 2018, 16, e00462. [Google Scholar] [CrossRef]
- Rahman, M.M.; Alam Tumpa, M.A.; Zehravi, M.; Sarker, M.T.; Yamin, M.; Islam, M.R.; Harun-Or-Rashid, M.; Ahmed, M.; Ramproshad, S.; Mondal, B.; et al. An Overview of Antimicrobial Stewardship Optimization: The Use of Antibiotics in Humans and Animals to Prevent Resistance. Antibiotics 2022, 11, 667. [Google Scholar] [CrossRef] [PubMed]
- Tzialla, C.; Civardi, E.; Pozzi, M.; Stronati, M. Antibiotics and multi-resistant organisms. Ital. J. Pediatr. 2015, 41, A45. [Google Scholar] [CrossRef]
- Beyene, A.M.; Gezachew, M.; Mengesha, D.; Yousef, A.; Gelaw, B. Prevalence and drug resistance patterns of Gram-negative enteric bacterial pathogens from diarrheic patients in Ethiopia: A systematic review and meta-analysis. PLoS ONE 2022, 17, e0265271. [Google Scholar] [CrossRef]
- Vargas, J.; Máttar, S.; Monsalve, S. Captive animals at Barranquilla s zoo are reservoirs of high resistance bacterial pathogens. Infectio 2010, 14, 6–19. [Google Scholar] [CrossRef]
- Kümmerer, K. The presence of pharmaceuticals in the environment due to human use–present knowledge and future challenges. J. Environ. Manag. 2009, 90, 2354–2366. [Google Scholar] [CrossRef]
- Szmolka, A.; Nagy, B. Multidrug resistant commensal Escherichia coli in animals and its impact for public health. Front. Microbiol. 2013, 4, 258. [Google Scholar] [CrossRef]
- Roose-Amsaleg, C.; Laverman, A.M. Do antibiotics have environmental side-effects? Impact of synthetic antibiotics on biogeochemical processes. Environ. Sci. Pollut. Res. 2016, 23, 4000–4012. [Google Scholar] [CrossRef]
- Wichmann, F.; Udikovic-Kolic, N.; Andrew, S.; Handelsman, J. Diverse Antibiotic Resistance Genes in Dairy Cow Manure. mBio 2014, 5, e01017-13. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; He, J. Effect of antibiotics in the environment on microbial populations. Appl. Microbiol. Biotechnol. 2010, 87, 925–941. [Google Scholar] [CrossRef] [PubMed]
- Grenni, P.; Ancona, V.; Caracciolo, A.B. Ecological effects of antibiotics on natural ecosystems: A review. Microchem. J. 2018, 136, 25–39. [Google Scholar] [CrossRef]
- Ben, Y.; Fu, C.; Hu, M.; Liu, L.; Wong, M.H.; Zheng, C. Human health risk assessment of antibiotic resistance associated with antibiotic residues in the environment: A review. Environ. Res. 2019, 169, 483–493. [Google Scholar] [CrossRef]
- Wang, L.; Xu, Y.; Wang, Z. Identification of Novel Mutations Associated with Bedaquiline Resistance in Mycobacterium Marinum. Zoonoses 2023, 3. [Google Scholar] [CrossRef]
- Cao, J.; Hu, Y.; Liu, F.; Wang, Y.; Bi, Y.; Lv, N.; Li, J.; Zhu, B.; Gao, G.F. Metagenomic analysis reveals the microbiome and resistome in migratory birds. Microbiome 2020, 8, 26. [Google Scholar] [CrossRef]
- Nieto-Claudin, A.; Deem, S.L.; Rodríguez, C.; Cano, S.; Moity, N.; Cabrera, F.; Esperón, F. Antimicrobial resistance in Galapagos tortoises as an indicator of the growing human footprint. Environ. Pollut. 2021, 284, 117453. [Google Scholar] [CrossRef]
- Lagerstrom, K.M.; Hadly, E.A. The under-investigated wild side of Escherichia coli: Genetic diversity, pathogenicity and antimicrobial resistance in wild animals. Proc. R. Soc. B 2021, 288, 20210399. [Google Scholar] [CrossRef] [PubMed]
- Dolejska, M. Antibiotic-resistant bacteria in wildlife. In Antibiotic Resistance in the Environment: A Worldwide Overview; Springer Nature: Berlin/Heidelberg, Germany, 2020; pp. 19–70. [Google Scholar]
- Manaia, C.M. Assessing the risk of antibiotic resistance transmission from the environment to humans: Non-direct proportionality between abundance and risk. Trends Microbiol. 2017, 25, 173–181. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, X.; Xu, T.; Yin, D. Effects of carbamazepine on gut microbiota, ARGs and intestinal health in zebrafish. Ecotoxicol. Environ. Saf. 2023, 249, 114473. [Google Scholar] [CrossRef]
- Francino, M. Antibiotics and the human gut microbiome: Dysbioses and accumulation of resistances. Front. Microbiol. 2016, 6, 1543. [Google Scholar] [CrossRef] [PubMed]
- Hyde, E.R.; Navas-Molina, J.A.; Song, S.J.; Kueneman, J.G.; Ackermann, G.; Cardona, C.; Humphrey, G.; Boyer, D.; Weaver, T.; Mendelson, J.R.; et al. The Oral and Skin Microbiomes of Captive Komodo Dragons Are Significantly Shared with Their Habitat. mSystems 2016, 1, e00046-16. [Google Scholar] [CrossRef]
- Trevelline, B.K.; Fontaine, S.S.; Hartup, B.K.; Kohl, K.D. Conservation biology needs a microbial renaissance: A call for the consideration of host-associated microbiota in wildlife management practices. Proc. R. Soc. B 2019, 286, 20182448. [Google Scholar] [CrossRef] [PubMed]
- Patterson, E.; Cryan, J.F.; Fitzgerald, G.F.; Ross, R.P.; Dinan, T.G.; Stanton, C. Gut microbiota, the pharmabiotics they produce and host health. Proc. Nutr. Soc. 2014, 73, 477–489. [Google Scholar] [CrossRef]
- Li, G.; Jiang, Y.; Li, Q.; An, D.; Bao, M.; Lang, L.; Han, L.; Huang, X.; Jiang, C. Comparative and functional analyses of fecal microbiome in Asian elephants. Antonie Leeuwenhoek 2022, 115, 1187–1202. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef]
- Skurnik, D.; Ruimy, R.; Andremont, A.; Amorin, C.; Rouquet, P.; Picard, B.; Denamur, E. Effect of human vicinity on antimicrobial resistance and integrons in animal faecal Escherichia coli. J. Antimicrob. Chemother. 2006, 57, 1215–1219. [Google Scholar] [CrossRef]
- Wu, K.; Xu, Y.; Zhang, W.; Mao, H.; Chen, B.; Zheng, Y.; Hu, X. Differences in Fecal Microbiome and Antimicrobial Resistance between Captive and Free-Range Sika Deer under the Same Exposure of Antibiotic Anthelmintics. Microbiol. Spectr. 2021, 9, e01918–e01921. [Google Scholar] [CrossRef]
- Hu, T.; Dai, Q.; Chen, H.; Zhang, Z.; Dai, Q.; Gu, X.; Yang, X.; Yang, Z.; Zhu, L. Geographic pattern of antibiotic resistance genes in the metagenomes of the giant panda. Microb. Biotechnol. 2021, 14, 186–197. [Google Scholar] [CrossRef]
- Campbell, T.P.; Sun, X.; Patel, V.H.; Sanz, C.; Morgan, D.; Dantas, G. The microbiome and resistome of chimpanzees, gorillas, and humans across host lifestyle and geography. ISME J. 2020, 14, 1584–1599. [Google Scholar] [CrossRef]
- Larsson, D.J.; Flach, C.-F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef]
- Lee, S.; Fan, P.; Liu, T.; Yang, A.; Boughton, R.K.; Pepin, K.M.; Miller, R.S.; Jeong, K.C. Transmission of antibiotic resistance at the wildlife-livestock interface. Commun. Biol. 2022, 5, 585. [Google Scholar] [CrossRef]
- Gibson, K.M.; Nguyen, B.N.; Neumann, L.M.; Miller, M.; Buss, P.; Daniels, S.; Ahn, M.J.; Crandall, K.A.; Pukazhenthi, B. Gut microbiome differences between wild and captive black rhinoceros–implications for rhino health. Sci. Rep. 2019, 9, 7570. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, S.B.; Martínez-Mota, R.; Stapleton, T.E.; Klure, D.M.; Greenhalgh, R.; Orr, T.J.; Dale, C.; Kohl, K.D.; Dearing, M.D. Microbiome stability and structure is governed by host phylogeny over diet and geography in woodrats (Neotoma spp.). Proc. Natl. Acad. Sci. USA 2021, 118, e2108787118. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Mishra, S.; Wang, C.; Zhang, H.; Ning, R.; Kong, F.; Zeng, B.; Zhao, J.; Li, Y. Comparative study of gut microbiota in wild and captive giant pandas (Ailuropoda melanoleuca). Genes 2019, 10, 827. [Google Scholar] [CrossRef] [PubMed]
- Paz, H.A.; Hales, K.E.; Wells, J.E.; Kuehn, L.A.; Freetly, H.C.; Berry, E.D.; Flythe, M.D.; Spangler, M.L.; Fernando, S.C. Rumen bacterial community structure impacts feed efficiency in beef cattle. J. Anim. Sci. 2018, 96, 1045–1058. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Nanda, A.; Khadka, B. Novel molecular, structural and evolutionary characteristics of the phosphoketolases from bifidobacteria and Coriobacteriales. PLoS ONE 2017, 12, e0172176. [Google Scholar] [CrossRef]
- Liew, K.J.; Liang, C.H.; Lau, Y.T.; Yaakop, A.S.; Chan, K.-G.; Shahar, S.; Shamsir, M.S.; Goh, K.M. Thermophiles and carbohydrate-active enzymes (CAZymes) in biofilm microbial consortia that decompose lignocellulosic plant litters at high temperatures. Sci. Rep. 2022, 12, 2850. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X.; Li, K.; Huang, Y.; Yang, H.; Zhu, P.; Chi, Z.; Xu, Y.; Li, Q. Signature of dissolved organic matter and microbial communities based on different oxygen levels response during distillers dried grains with solubles plus sugarcane pith co-fermentations. Bioresour. Technol. 2022, 349, 126868. [Google Scholar] [CrossRef]
- Ning, J.; Huang, S.-Y.; Chen, S.-D.; Zhang, Y.-R.; Huang, Y.-Y.; Yu, J.-T. Investigating Casual Associations among Gut Microbiota, Metabolites, and Neurodegenerative Diseases: A Mendelian Randomization Study. J. Alzheimer’s Dis. 2022, 87, 211–222. [Google Scholar] [CrossRef]
- Juckel, G.; Manitz, M.-P.; Freund, N.; Gatermann, S. Impact of Poly I: C induced maternal immune activation on offspring′s gut microbiome diversity–implications for schizophrenia. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2021, 110, 110306. [Google Scholar] [CrossRef]
- Ratto, D.; Roda, E.; Romeo, M.; Venuti, M.T.; Desiderio, A.; Lupo, G.; Capelli, E.; Sandionigi, A.; Rossi, P. The Many Ages of Microbiome-Gut-Brain Axis. Nutrients 2022, 14, 2937. [Google Scholar] [CrossRef]
- Sun, S.; Wang, H.; Howard, A.G.; Zhang, J.; Su, C.; Wang, Z.; Du, S.; Fodor, A.A.; Gordon-Larsen, P.; Zhang, B. Loss of Novel Diversity in Human Gut Microbiota Associated with Ongoing Urbanization in China. mSystems 2022, 7, e0020022. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, R.; Scharch, C.; Sandvang, D. The link between broiler flock heterogeneity and cecal microbiome composition. Anim. Microbiome 2021, 3, 54. [Google Scholar] [CrossRef]
- Zhou, Z.-C.; Zheng, J.; Wei, Y.-Y.; Chen, T.; Dahlgren, R.A.; Shang, X.; Chen, H. Antibiotic resistance genes in an urban river as impacted by bacterial community and physicochemical parameters. Environ. Sci. Pollut. Res. 2017, 24, 23753–23762. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Guan, Y.; Zhao, R.; Feng, J.; Huang, J.; Ma, L.; Li, B. Metagenomic and network analyses decipher profiles and co-occurrence patterns of antibiotic resistome and bacterial taxa in the reclaimed wastewater distribution system. J. Hazard. Mater. 2020, 400, 123170. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Ge, F.; Yao, X.; Guo, X.; Bao, P.; Ma, X.; Wu, X.; Chu, M.; Yan, P.; Liang, C. Microbiome and metabolomics reveal the effects of different feeding systems on the growth and ruminal development of yaks. Front. Microbiol. 2021, 12, 682989. [Google Scholar] [CrossRef]
- Zang, X.-W.; Sun, H.-Z.; Xue, M.-Y.; Zhang, Z.; Plastow, G.; Yang, T.; Guan, L.L.; Liu, J.-X. Heritable and Nonheritable Rumen Bacteria Are Associated with Different Characters of Lactation Performance of Dairy Cows. mSystems 2022, 7, e0042222. [Google Scholar] [CrossRef]
- Theelen, M.J.; Luiken, R.E.; Wagenaar, J.A.; Sloet van Oldruitenborgh-Oosterbaan, M.M.; Rossen, J.W.; Zomer, A.L. The equine faecal microbiota of healthy horses and ponies in the Netherlands: Impact of host and environmental factors. Animals 2021, 11, 1762. [Google Scholar] [CrossRef]
- Mu, Y.; Qi, W.; Zhang, T.; Zhang, J.; Mao, S. Multi-omics Analysis Revealed Coordinated Responses of Rumen Microbiome and Epithelium to High-Grain-Induced Subacute Rumen Acidosis in Lactating Dairy Cows. mSystems 2022, 7, e0149021. [Google Scholar] [CrossRef]
- Wang, J.; Fan, H.; Han, Y.; Zhao, J.; Zhou, Z. Characterization of the microbial communities along the gastrointestinal tract of sheep by 454 pyrosequencing analysis. Asian-Australas J. Anim. Sci. 2017, 30, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Radolf, J.D. Medical Microbiology. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996; Chapter 36. [Google Scholar]
- Stamm, L.V. Global challenge of antibiotic-resistant Treponema pallidum. Antimicrob. Agents Chemother. 2010, 54, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Wang, W.; Xing, Y.; Chen, H.; Luo, X.; Chen, W.; Huang, Q. Niche overlap is a predictor of the interspecies correlations detected by microbial network analysis in soil micro-aggregates. J. Soils Sediments 2022, 22, 1521–1529. [Google Scholar] [CrossRef]
- Yuan, K.; Yu, K.; Yang, R.; Zhang, Q.; Yang, Y.; Chen, E.; Lin, L.; Luan, T.; Chen, W.; Chen, B. Metagenomic characterization of antibiotic resistance genes in Antarctic soils. Ecotoxicol. Environ. Saf. 2019, 176, 300–308. [Google Scholar] [CrossRef]
- Liu, S.; Wang, P.; Wang, C.; Wang, X.; Chen, J. Anthropogenic disturbances on antibiotic resistome along the Yarlung Tsangpo River on the Tibetan Plateau: Ecological dissemination mechanisms of antibiotic resistance genes to bacterial pathogens. Water Res. 2021, 202, 117447. [Google Scholar] [CrossRef]
- Chen, B.; Yang, Y.; Liang, X.; Yu, K.; Zhang, T.; Li, X. Metagenomic Profiles of Antibiotic Resistance Genes (ARGs) between Human Impacted Estuary and Deep Ocean Sediments. Environ. Sci. Technol. 2013, 47, 12753–12760. [Google Scholar] [CrossRef]
- Tikhonova, E.B.; Zgurskaya, H.I. AcrA, AcrB, and TolC of Escherichia coli form a stable intermembrane multidrug efflux complex. J. Biol. Chem. 2004, 279, 32116–32124. [Google Scholar] [CrossRef]
- Murakami, S.; Nakashima, R.; Yamashita, E.; Yamaguchi, A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 2002, 419, 587–593. [Google Scholar] [CrossRef]
- McMurry, L.M.; Park, B.H.; Burdett, V.; Levy, S.B. Energy-dependent efflux mediated by class L (tetL) tetracycline resistance determinant from streptococci. Antimicrob. Agents Chemother. 1987, 31, 1648–1650. [Google Scholar] [CrossRef]
- Horiyama, T.; Yamaguchi, A.; Nishino, K. TolC dependency of multidrug efflux systems in Salmonella enterica serovar Typhimurium. J. Antimicrob. Chemother. 2010, 65, 1372–1376. [Google Scholar] [CrossRef]
- Schuster, S.; Vavra, M.; Greim, L.; Kern, W.V. Exploring the Contribution of the AcrB Homolog MdtF to Drug Resistance and Dye Efflux in a Multidrug Resistant E. coli Isolate. Antibiotics 2021, 10, 503. [Google Scholar] [CrossRef] [PubMed]
- Massella, E.; Reid, C.J.; Cummins, M.L.; Anantanawat, K.; Zingali, T.; Serraino, A.; Piva, S.; Giacometti, F.; Djordjevic, S.P. Snapshot Study of Whole Genome Sequences of Escherichia coli from Healthy Companion Animals, Livestock, Wildlife, Humans and Food in Italy. Antibiotics 2020, 9, 782. [Google Scholar] [CrossRef] [PubMed]
- Kang, Q.; Wang, X.; Zhao, J.; Liu, Z.; Ji, F.; Chang, H.; Yang, J.; Hu, S.; Jia, T.; Wang, X. Multidrug-resistant Proteus mirabilis isolates carrying blaOXA-1 and blaNDM-1 from wildlife in China: Increasing public health risk. Integr. Zool. 2021, 16, 798–809. [Google Scholar] [CrossRef]
- Campos-Arceiz, A.; Larrinaga, A.R.; Weerasinghe, U.R.; Takatsuki, S.; Pastorini, J.; Leimgruber, P.; Fernando, P.; Santamaría, L. Behavior rather than diet mediates seasonal differences in seed dispersal by Asian elephants. Ecology 2008, 89, 2684–2691. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Goebel, B.M. Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 1994, 44, 846–849. [Google Scholar] [CrossRef]
- Ma, Z.; Li, L. Measuring metagenome diversity and similarity with Hill numbers. Mol. Ecol. Resour. 2018, 18, 1339–1355. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, K.; Wang, Y.; Bai, X.; Wang, J.; Zhang, L.; Tang, Y.; Thuku, R.C.; Hou, W.; Mo, G.; Chen, F.; et al. Comparison of Fecal Antimicrobial Resistance Genes in Captive and Wild Asian Elephants. Antibiotics 2023, 12, 859. https://doi.org/10.3390/antibiotics12050859
Cao K, Wang Y, Bai X, Wang J, Zhang L, Tang Y, Thuku RC, Hou W, Mo G, Chen F, et al. Comparison of Fecal Antimicrobial Resistance Genes in Captive and Wild Asian Elephants. Antibiotics. 2023; 12(5):859. https://doi.org/10.3390/antibiotics12050859
Chicago/Turabian StyleCao, Kaixun, Yepeng Wang, Xuewei Bai, Jishan Wang, Liting Zhang, Yongjing Tang, Rebecca Caroline Thuku, Wei Hou, Guoxiang Mo, Fei Chen, and et al. 2023. "Comparison of Fecal Antimicrobial Resistance Genes in Captive and Wild Asian Elephants" Antibiotics 12, no. 5: 859. https://doi.org/10.3390/antibiotics12050859
APA StyleCao, K., Wang, Y., Bai, X., Wang, J., Zhang, L., Tang, Y., Thuku, R. C., Hou, W., Mo, G., Chen, F., & Jin, L. (2023). Comparison of Fecal Antimicrobial Resistance Genes in Captive and Wild Asian Elephants. Antibiotics, 12(5), 859. https://doi.org/10.3390/antibiotics12050859





