Extended-Spectrum β-Lactamase-Producing Escherichia coli Isolated from Food-Producing Animals in Tamaulipas, Mexico
Abstract
:1. Introduction
2. Results
2.1. Identification of ESBL-EC
2.2. Antimicrobial Susceptibility
2.3. Detection of Virulence Factors
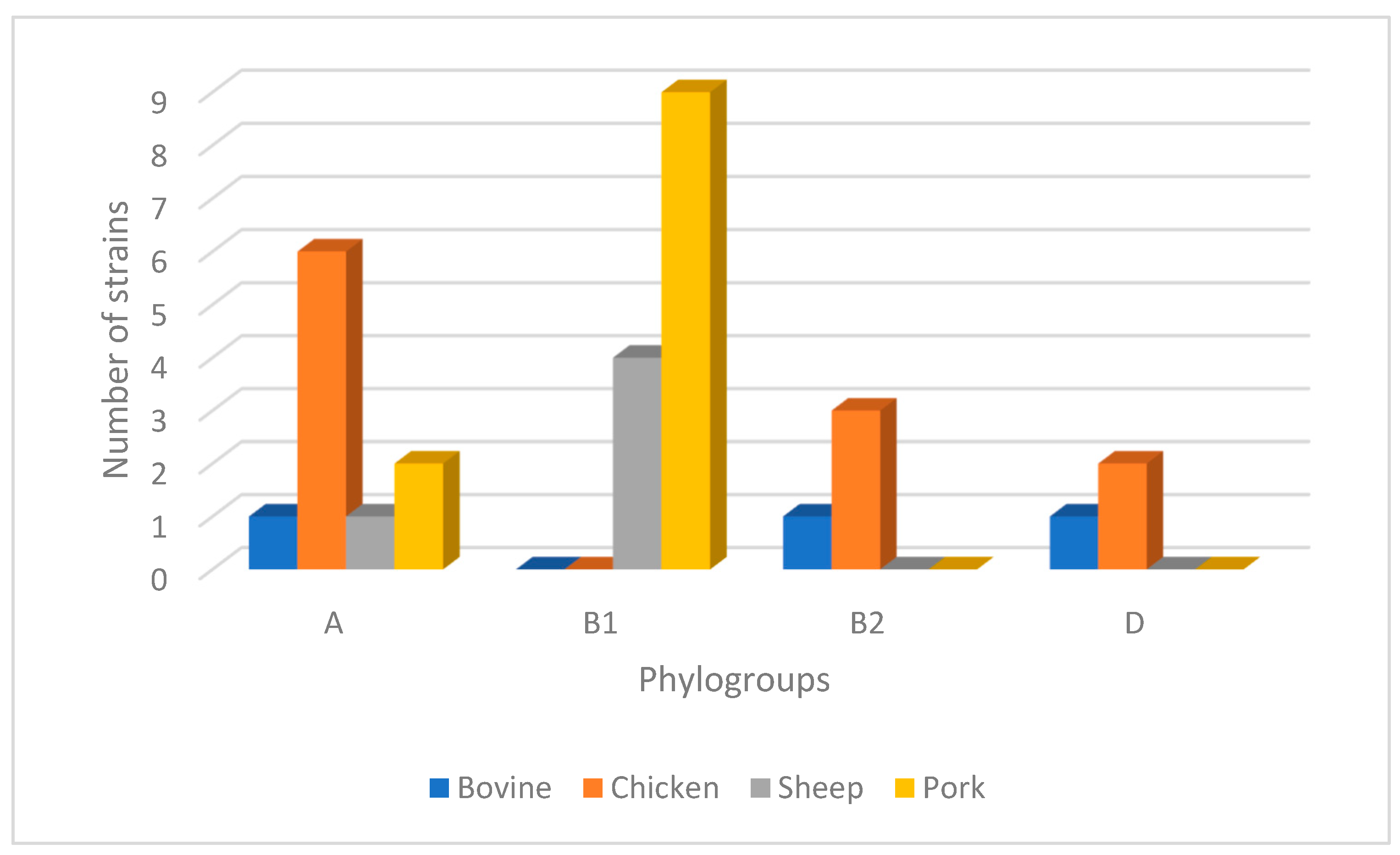
2.4. Phylogenetic Groups
3. Discussion
4. Materials and Methods
4.1. Identification of ESBL-EC
4.2. Antimicrobial Susceptibility
4.3. Detection of Virulence Factors
4.4. Phylogenetic Groups
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rosini, R.; Nicchi, S.; Pizza, M.; Rappouli, R. Vaccines Against Antimicrobial Resistance. Front. Inmunol. 2020, 11, 1048. [Google Scholar] [CrossRef]
- Nossair, M.A.; Abd El Baqy, F.A.; Rizk, M.S.Y.; Elaadli, H.; Mansour, A.M.; El-Aziz, A.H.A.; Alkhedaide, A.; Soliman, M.M.; Ramadan, H.; Shukry, M. Prevalence and Molecular Characterization of Extended-Spectrum β-Lactamases and AmpC β-lactamase-Producing Enterobacteriaceae among Human, Cattle, and Poultry. Pathogens 2022, 11, 852. [Google Scholar] [CrossRef]
- Alsamawi, M.; Joudeh, A.I.; Eldeeb, Y.; Al-Dahshan, A.; Khan, F.; Ghadban, W.; Almaslamani, M.; Alkhal, A. Epidemiology of Extended-Spectrum Beta-Lactamase Producing Enterobacteriaceae in Qatar: A 3-year Hospital-Based Study. Front. Antibiot. 2022, 1, 980686. [Google Scholar] [CrossRef]
- Adler, A.; Katz, D.E.; Marchaim, D. The Continuing Plague of Extended-Spectrum β-Lactamase Producing Enterobacterales Infections: An Update. Infect. Dis. Clin. 2020, 34, 677–708. [Google Scholar] [CrossRef] [PubMed]
- Benavides, J.A.; Salgado-Caxito, M.; Opazo-Capurro, A.; González Muñoz, P.; Piñeiro, A.; Otto Medina, M.; Rivas, L.; Munita, J.; Millán, J. ESBL-Producing Escherichia coli Carrying CTX-M Genes Circulating among Livestock, Dogs, and Wild Mammals in Small-Scale Farms of Central Chile. Antibiotics 2021, 10, 510. [Google Scholar] [CrossRef]
- Ajuga, M.U.; Otokunefor, K.; Agbagwa, O.E. Antibiotic Resistance and ESBL Production in Escherichia coli From Various Sources in Aba Metropolis, Nigeria. Bull. Natl. Res. Cent. 2021, 45, 173. [Google Scholar] [CrossRef]
- Kimera, Z.I.; Mgaya, F.X.; Mshana, S.E.; Karimuribo, E.D.; Matee, M.I.N. Occurrence of Extended Spectrum Beta Lactamase (ESBL) Producers, Quinolone and Carbapenem Resistant Enterobacteriaceae Isolated from Environmental Samples along Msimbazi River Basin Ecosystem in Tanzania. Int. J. Environ. Res. Public Health 2021, 18, 8264. [Google Scholar] [CrossRef] [PubMed]
- Soncini, J.G.M.; Cerdeira, L.; Sano, E.; Koga, V.L.; Tizura, A.T.; Tano, Z.N.; Nakazato, G.; Kobayashi, R.K.T.; Aires, C.A.M.; Lincopan, N.; et al. Genomic Insights of High-Risk Clones of ESBL-Producing Escherichia coli Isolated from Community Infections and Commercial Meat in Southern Brazil. Sci. Rep. 2022, 12, 9354. [Google Scholar] [CrossRef] [PubMed]
- Barrios, E.; Cortés, G.; Lozano, P.; Romero, S.; Lara, N.; Estepa, V.; Somalo, S.; Torres, C.; Rocha, R.C. Characterization of Extended-Spectrum and CMY-2 ß-Lactamases, and Associated Virulence Genes in Escherichia coli from Food of Animal Origin in México. Br. Food J. 2018, 120, 1457–1473. [Google Scholar] [CrossRef]
- Martínez-Vázquez, A.V.; Mandujano, A.; Cruz, E.; Guerrero, A.; Vazquez, J.; Cruz, W.L.; Rivera, G.; Bocanegra-García, V. Evaluation of Retail Meat as a Source of ESBL Escherichia coli in Tamaulipas, Mexico. Antibiotics 2022, 11, 1795. [Google Scholar] [CrossRef]
- Vega, V.; Talavera Rojas, M.; Barba, J.; Zepeda, A.P.; Reyes, N.E. Antimicrobial Resistance of Escherichia coli Isolated from Cattle Carcasses and Feces in Center of Mexico. Rev. Mex. Cienc. Pecu. 2020, 11, 991–1003. [Google Scholar]
- Delgado, E.J.; Palós, T.; Ruíz, F.A.; Hernández, C.F.; Ballesteros, N.E.; Soberanis, O.; Mendez, R.D.; Allard, M.W.; Rubio, M.S. Genomic Surveillance of Antimicrobial Resistance Shows Cattle and Poultry are a Moderate source of multi-drug resistant non-typhoidal Salmonella in Mexico. PLoS ONE 2021, 16, e0243681. [Google Scholar]
- Navarro, C.L.; Ibarra, L.M.; Diosdado, J.D.; Madriz, A.L.; Cardona, M.A.; Varela, J.J.; Silva, J.; Arvizu, S.M.; Padilla, J.J. Frequency, Territorial Distribution and Antimicrobial Resistance of Salmonella spp. on Bovine Cattle Feces from the Altos Sur Region of Jalisco State, Mexico. Biotecnia 2021, 23, 5–13. [Google Scholar]
- Martínez-Vázquez, A.V.; Vázquez, J.; Leyva, L.M.; Barrios, H.; Rivera, G.; Bocanegra, V. Multidrug Resistance of Escherichia coli Strains Isolated from Bovine Feces and Carcasses in Northeast Mexico. Front. Vet. Sci. 2021, 8, 643802. [Google Scholar] [CrossRef]
- Ramos, S.; Silva, V.; Dapkevicius, M.L.E.; Caniça, M.; Tejedor-Junco, M.T.; Igrejas, G.; Poeta, P. Escherichia coli as Commensal and Pathogenic Bacteria among Food-producing Animals: Health Implications of Extended Spectrum β-lactamase (ESBL) Production. Animals 2020, 10, 2239. [Google Scholar] [CrossRef] [PubMed]
- Jalil, A.; Gul, S.; Bhatti, M.F.; Siddiqui, M.F.; Adnan, F. High Occurrence of Multidrug-Resistant Escherichia coli Strains in Bovine Fecal Samples from Healthy Cows Serves as Rich Reservoir for AMR Transmission. Antibiotics 2023, 12, 37. [Google Scholar] [CrossRef]
- Sanou, S.; Salam, A.; Lounnas, M.; Zougmore, A.; Pooda, A.; Zoungrana, J.; Anicet, G.; Traore-Ouedraogo, R.; Ouchar, O.; Jean-Pierre, H.; et al. Epidemiology and Molecular Characterization of Enterobacteriaceae Producing Extended-Spectrum β-lactamase in Intensive and Extensive Breeding Animals in Burkina Faso. PAMJ One Health 2022, 8, 4. [Google Scholar] [CrossRef]
- Li, C.; Chen, X.; Ju, Z.; Li, C.; Xu, Y.; Ding, J.; Wang, Y.; Ma, P.; Gu, K.; Lei, C.; et al. Comparative Analysis of Phylogenetic Relationships and Virulence Factor Characteristics Between Extended-Spectrum β-Lactamase-Producing Escherichia coli Isolates Derived from Clinical Sites and Chicken Farm. Microbiol. Spectr. 2022, 10, e02557-22. [Google Scholar] [CrossRef]
- Shafiq, M.; Huang, J.; Shah, J.M.; Ali, I.; Rahman, S.U.; Wang, L. Characterization and Resistant Determinants Linked to Mobile Elements of ESBL-Producing and mcr-1-Positive Escherichia coli Recovered from the Chicken Origin. Microb. Pathog. 2021, 150, 104722. [Google Scholar] [CrossRef]
- Giufrè, M.; Mazzolini, E.; Cerquetti, M.; Brusaferro, S. Extended-Spectrum β-lactamase-Producing Escherichia coli from Extraintestinal Infections in Humans and from Food-Producing Animals in Italy: A ‘One Health’ Study. Int. J. Antimicrob. Agents 2021, 58, 106433. [Google Scholar] [CrossRef]
- Sghaier, S.; Abbassi, M.S.; Pascual, A.; Serrano, L.; Díaz-De-Alba, P.; Said, M.B.; Hassen, B.; Ibrahim, C.; Hassen, A.; López-Cerero, L. Extended-Spectrum β-lactamase-Producing Enterobacteriaceae from Animal Origin and Wastewater in Tunisia: First Detection of O25b-B23-CTX-M-27-ST131 Escherichia coli and CTX-M-15/OXA-204-Producing Citrobacter freundii from Wastewater. J. Glob. Antimicrob. Resist. 2019, 17, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Miltgen, G.; Martak, D.; Valot, B.; Kamus, L.; Garrigos, T.; Verchere, G.; Gbaguidi-Haore, H.; Ben Cimon, C.; Ramiandrisoa, M.; Picot, S.; et al. One Health Compartmental Analysis of ESBL-Producing Escherichia coli on Reunion Island Reveals Partitioning Between Humans and Livestock. J. Antimicrob. Chemother. 2022, 77, 1254–1262. [Google Scholar] [CrossRef] [PubMed]
- Rhouma, M.; Soufi, L.; Cenatus, S.; Archambault, M.; Butaye, P. Current Insights Regarding the Role of Farm Animals in the Spread of Antimicrobial Resistance from a One Health Perspective. Vet. Sci. 2022, 9, 480. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, N.A.; Gonçalves, B.L.; Lee, S.H.; Oliveira, C.A.F.; Corassin, C.H. Use of Antibiotics in Animal Production and its Impact on Human Health. J. Food Chem. Nanotechnol. 2020, 6, 40–47. [Google Scholar] [CrossRef] [Green Version]
- Tamma, P.D.; Aitken, S.L.; Bonomo, R.A.; Mathers, A.J.; Van Duin, D.; Clancy, C.J. Infectious Diseases Society of America Guidance on the Treatment of AmpC β-Lactamase-Producing Enterobacterales, Carbapenem-Resistant Acinetobacter baumannii, and Stenotrophomonas maltophilia Infections; Version 2.0; Infectious Diseases Society of America: Chicago, IL, USA, 2022; Available online: https://www.idsociety.org/practice-guideline/amr-guidance-2.0/ (accessed on 17 January 2023).
- Karaiskos, I.; Giamarellou, H. Carbapenem-Sparing Strategies for ESBL Producers: When and How. Antibiotics 2020, 9, 61. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez, B.; Rodríguez, J. Current Options for the Treatment of Infections due to Extended-Spectrum Beta-Lactamase-Producing Enterobacteriaceae in Different Groups of Patients. Clin. Microbiol. Infect. 2019, 25, 932–942. [Google Scholar] [CrossRef]
- Pana, Z.D.; Zaoutis, T. Treatment of Extended-Spectrum β-lactamase-Producing Enterobacteriaceae (ESBLs) Infections: What Have we Learned Until Now? F1000Research 2018, 7, 1347. [Google Scholar] [CrossRef] [Green Version]
- Michaelis, C.; Grohmann, E. Horizontal Gene Transfer of Antibiotic Resistance Genes in Biofilms. Antibiotics 2023, 12, 328. [Google Scholar] [CrossRef]
- Afunwa, R.A.; Ezeanyinka, J.; Afunwa, E.C.; Udeh, A.S.; Oli, N.A.; Unachukwu, M. Multiple Antibiotic Resistant Index of Gram-negative Bacteria from Bird Droppings in two Commercial Poultries in Enugu, Nigeria. Open J. Med. Microbiol. 2020, 10, 171–181. [Google Scholar] [CrossRef]
- Tiantian, T.; Shiting, D.; Dejun, L.; Yang, W.; Wei, Q.; Min, Y.; Yu, Z. Occurrence and Transfer Characteristics of blaCTX-M Genes among Escherichia coli in Anaerobic Digestion Systems Treating Swine Waste. Sci. Total Environ. 2022, 834, 155321. [Google Scholar] [CrossRef] [PubMed]
- Salah, F.D.; Soubeiga, S.T.; Ouattara, A.K.; Sadji, A.Y.; Metuor-Dabire, A.; Obiri-Yeboah, D.; Banla-Kere, A.; Karou, S.; Simpore, S. Distribution of Quinolone Resistance Gene (qnr) in ESBL-Producing Escherichia coli and Klebsiella spp. in Lomé, Togo. Antimicrob. Resist. Infect. Control 2019, 8, 104. [Google Scholar] [CrossRef] [Green Version]
- Kammili, N.; Rani, M.; Styczynski, A.; Latha, M.; Pavuluri, P.R.; Reddy, V.; Alsan, M. Plasmid Mediated Antibiotic Resistance among Uropathogens in Primigravid Women—Hyderabad, India. PLoS ONE 2020, 15, e0232710. [Google Scholar] [CrossRef]
- Ghaly, T.M.; Geoghegan, J.L.; Tetu, S.G.; Gillings, M.R. The Peril and Promise of Integrons: Beyond Antibiotic Resistance. Trends Microbiol. 2020, 28, 455–464. [Google Scholar] [CrossRef]
- Karimi Dehkordi, M.; Halaji, M.; Nouri, S. Prevalence of Class 1 Integron in Escherichia coli Isolated from Animal Sources in Iran: A Systematic Review and Meta-Analysis. Trop. Med. Health 2020, 48, 16. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and Simple Determination of the Escherichia coli Phylogenetic Group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef] [Green Version]
- Bourne, J.A.; Chong, W.L.; Gordon, D.M. Genetic Structure, Antimicrobial Resistance and Frequency of Human Associated Escherichia coli Sequence Types among Faecal Isolates from Healthy Dogs and Cats Living in Canberra, Australia. PLoS ONE 2019, 14, e0212867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguirre-Sánchez, J.R.; Valdez-Torres, J.B.; Castro del Campo, N.; Martínez-Urtaza, J.; Castro del Campo, N.; Lee, B.G.; Quiñones, B.; Chaidez-Quiroz, C. Phylogenetic Group and Virulence Profile Classification in Escherichia coli from Distinct Isolation Sources in Mexico. Infect. Genet. Evol. 2022, 106, 105380. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Gordon, D.; Denamur, E. Guide to the Various Phylogenetic Classification Schemes for Escherichia coli and the Correspondence among Schemes. Microbiology 2015, 161, 980–988. [Google Scholar] [CrossRef]
- Barzan, M.; Rad, M.; Hashemi, G.R.; Azizzadeh, M. Phylogenetic Analysis of Escherichia coli Isolates from Healthy and Diarrheic Calves in Mashhad, Iran. Bulg. J. Vet. Med. 2017, 20, 11–18. [Google Scholar] [CrossRef]
- Alfinete, N.W.; Bolukaoto, J.Y.; Heine, L.; Potgieter, N.; Barnard, T.G. Virulence and Phylogenetic Analysis of enteric pathogenic Escherichia coli Isolated from Children with Diarrhea in South Africa. Int. J. Infect. Dis. 2022, 114, 226–232. [Google Scholar] [CrossRef]
- Karakaya, E.; Aydin, F.; Kayman, T.; Abay, S. Escherichia coli in Different Animal Feces: Phylotypes and Virulence Genes. World J. Microbiol. Biotechnol. 2023, 39, 14. [Google Scholar] [CrossRef]
- Vázquez-Villanueva, J.; Vázquez, K.; Martínez-Vázquez, A.V.; Wong-González, A.; Hernández-Escareño, J.; Cabrero-Martínez, O.; Cruz-Pulido, W.L.; Guerrero, A.; Rivera, G.; Bocanegra-García, V. Molecular and Antimicrobial Susceptibility Characterization of Escherichia coli Isolates from Bovine slaughterhouse Process. Antibiotics 2023, 12, 291. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing: 30th Informational Supplement; CLSI Document M100-Ed31; CLSI: Wayne, PA, USA, 2021. [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters; Version 11.0; EUCAST: Växjö, Sweden, 2021. [Google Scholar]
- Kozak, G.K.; Boerlin, P.; Janecko, N.; Reid-Smith, R.J.; Jardine, C. Antimicrobial Resistance in Escherichia coli Isolates from Swine and Wild Small Mammals in the Proximity of Swine Farms and in Natural Environments in Ontario, Canada. Appl. Environ. Microbiol. 2009, 75, 559–566. [Google Scholar] [CrossRef] [Green Version]
- Ng, L.K.; Martin, I.; Alfa, M.; Mulvey, M. Multiplex PCR for the Detection of Tetracycline Resistant Genes. Mol. Cell. Probes. 2001, 15, 209–215. [Google Scholar] [CrossRef]
- Yu, T.; Jiang, X.; Fu, K.; Liu, B.; Xu, D.; Ji, S.; Zhou, L. Detection of Extended-Spectrum β-Lactamase and Plasmid-mediated Quinolone Resistance Determinants in Escherichia coli Isolates from Retail Meat in China. J. Food Sci. 2015, 80, M1039–M1043. [Google Scholar] [CrossRef] [PubMed]
- Kargar, M.; Mohammadalipour, Z.; Doosti, A.; Lorzadeh, S.; Japoni, A. High Prevalence of Class 1 to 3 Integrons among Multidrug-Resistant Diarrheagenic Escherichia coli in Sothwest of Iran. Osong Public Health Res. Perspt. 2014, 5, 193–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canizalez, A.; González, E.; Vidal, J.E.; Flores, H.; León, N. Prevalence and Antibiotic Resistance Profiles of Diarrheagenic Escherichia coli Strains Isolated from Food Items in Northwestern Mexico. Int. J. Food Microbiol. 2013, 164, 36–45. [Google Scholar] [CrossRef]
| Type of Sample | Number of Samples | ESBL-Positive Samples | Number of Strains of E. coli | CTX-Resistant or Intermediate | ESBL-Positive Strains |
|---|---|---|---|---|---|
| Bovines | 50 | 6.0% (3/50) | 150 | 36.6% (55/150) | 2.0% (3/150) |
| Chickens | 50 | 16.0% (8/50) | 150 | 35.3% (53/150) | 7.3% (11/150) |
| Pigs | 50 | 10.0% (5/50) | 150 | 37.3% (56/150) | 7.3% (11/150) |
| Sheep | 50 | 14.0% (7/50) | 150 | 44.6% (67/150) | 3.3% (5/150) |
| Total | 200 | 11.5% (23/200) | 600 | 38.5% (231/600) | 5.0% (30/600) |
| Sample | No. of Isolates | Resistance Patterns | No. of Antibiotics | MARI |
|---|---|---|---|---|
| Bovines | 1 | GE-NET-S-C-AM-FEP-STX-TE | 8 | 0.471 |
| 2 | AN-GE-NET-S-CF-FEP-CTX-TE | 8 | 0.471 | |
| 3 | AN-GE-NET-S-CTX-CRO-CIP-TE | 8 | 0.471 | |
| Chickens | 1 | AN-GE-CF-CTX-CRO | 5 | 0.294 |
| 2 | AN-GE-CTX-CRO-TE | 5 | 0.294 | |
| 3 | S-C-FEP-CIP-STX-TE-LV | 7 | 0.412 | |
| 4 | AN-GE-C-CTX-CRO-CIP-STX-TE-LV | 9 | 0.529 | |
| 5 | AN-GE-NET-S-C-AM-CRO-STX-TE | 9 | 0.529 | |
| 6 | AN-GE-S-CF-FEP-CTX-CRO-AmC-TE | 9 | 0.529 | |
| 7 | AN-GE-S-CF-CTX-CRO-AmC-STX-TE | 9 | 0.529 | |
| 8 | AN-GE-AM-CF-CTX-CRO-AmC-STX-TE | 9 | 0.529 | |
| 9 | AN-GE-S-C-AM-FEP-CTX-CAZ-CRO-STX | 10 | 0.588 | |
| 10 | AN-G-CF-CTX-CRO-AmC-CIP-STX-TE-LV | 10 | 0.588 | |
| 11 | AN-GE-S-C-CF-FEP-CTX-CRO-CIP-STX-TE-LV | 12 | 0.706 | |
| Sheep | 1 | GE-S-AM-CF-TE | 5 | 0.294 |
| 2 | AN-GE-S-C-AM | 5 | 0.294 | |
| 3 | GE-S-CF-CTX-TE | 5 | 0.294 | |
| 4 | AN-GE-S-AM-CF-CTX | 6 | 0.353 | |
| 5 | GE-S-C-AM-CF-CTX-TE | 7 | 0.412 | |
| Pigs | 1 | AN-GE-S-C-AM-CF-AmC-STX-TE | 9 | 0.529 |
| 2 | AN-GE-C-CF-FEP-CAZ-CRO-STX-TE-LV | 10 | 0.588 | |
| 3 | GE-S-C-AM-CF-FEP-CTX-CRO-CIP-STX-TE-LV | 12 | 0.706 | |
| 4 | GE-S-C-AM-CF-FEP-CTX-CRO-AmC-CIP-STX-TE-LV | 13 | 0.765 | |
| 5 | AN-GE-NET-S-C-CF-FEP-CRO-AmC-CIP-STX-TE-LV | 13 | 0.765 | |
| 6 | GE-S-C-AM-CF-FEP-CTX-CAZ-CRO-CIP-STX-TE-LV | 13 | 0.765 | |
| 7 | GE-S-C-AM-CF-FEP-CTX-CAZ-CRO-CIP-STX-TE-LV | 13 | 0.765 | |
| 8 | GE-S-C-AM-CF-FEP-CTX-CAZ-CRO-CIP-STX-TE-LV | 13 | 0.765 | |
| 9 | GE-S-C-AM-CF-FEP-CTX-CAZ-CRO-CIP-STX-TE-LV | 13 | 0.765 | |
| 10 | AN-GE-S-C-AM-CF-FEP-CTX-CAZ-CRO-CIP-STX-TE-LV | 14 | 0.824 | |
| 11 | AN-GE-S-C-AM-CF-CTX-CAZ-CRO-AmC-CIP-STX-TE-LV | 14 | 0.824 |
| Antibiotic Group | Phenotype | Genotype | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Aminoglycoside | AN | GM | NET | S | acc(3)-VI | aadA1 | strA | strB | |||
| 63.3% (19/30) | 93.3% (28/30) | 16.6% (5/30) | 83.3% (25/30) | 0% (0/30) | 60.0% (18/30) | 13.3% (4/30) | 33.3% (10/30) | ||||
| β-lactam | AM | CF | FEP | CTX | CAZ | CRO | blaCTX-M | blaCTX-M2 | blaCTX-M3 | blaTEM | blaSHV |
| 56.6% (17/30) | 73.3% (22/30) | 50.0% (15/30) | 73.3% (22/30) | 26.6% (8/30) | 70.0% (21/30) | 23.3% (7/30) | 0% (0/30) | 0% (0/30) | 33.3% (10/30) | 0% (0/30) | |
| Sulfonamide | STX | sul1 | sul2 | sul3 | |||||||
| 66.6% (20/30) | 23.3% (7/30) | 43.3% (13/30) | 13.3% (4/30) | ||||||||
| Tetracycline | TET | tetA | tetB | ||||||||
| 86.6% (26/30) | 73.3% (22/30) | 13.3% (4/30) | |||||||||
| Quinolone | LVX | qnrA | qnrB | ||||||||
| 50% (15/30) | 0% (0/30) | 20% (6/30) | |||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mandujano, A.; Cortés-Espinosa, D.V.; Vásquez-Villanueva, J.; Guel, P.; Rivera, G.; Juárez-Rendón, K.; Cruz-Pulido, W.L.; Aguilera-Arreola, G.; Guerrero, A.; Bocanegra-García, V.; et al. Extended-Spectrum β-Lactamase-Producing Escherichia coli Isolated from Food-Producing Animals in Tamaulipas, Mexico. Antibiotics 2023, 12, 1010. https://doi.org/10.3390/antibiotics12061010
Mandujano A, Cortés-Espinosa DV, Vásquez-Villanueva J, Guel P, Rivera G, Juárez-Rendón K, Cruz-Pulido WL, Aguilera-Arreola G, Guerrero A, Bocanegra-García V, et al. Extended-Spectrum β-Lactamase-Producing Escherichia coli Isolated from Food-Producing Animals in Tamaulipas, Mexico. Antibiotics. 2023; 12(6):1010. https://doi.org/10.3390/antibiotics12061010
Chicago/Turabian StyleMandujano, Antonio, Diana Verónica Cortés-Espinosa, José Vásquez-Villanueva, Paulina Guel, Gildardo Rivera, Karina Juárez-Rendón, Wendy Lizeth Cruz-Pulido, Guadalupe Aguilera-Arreola, Abraham Guerrero, Virgilio Bocanegra-García, and et al. 2023. "Extended-Spectrum β-Lactamase-Producing Escherichia coli Isolated from Food-Producing Animals in Tamaulipas, Mexico" Antibiotics 12, no. 6: 1010. https://doi.org/10.3390/antibiotics12061010
APA StyleMandujano, A., Cortés-Espinosa, D. V., Vásquez-Villanueva, J., Guel, P., Rivera, G., Juárez-Rendón, K., Cruz-Pulido, W. L., Aguilera-Arreola, G., Guerrero, A., Bocanegra-García, V., & Martínez-Vázquez, A. V. (2023). Extended-Spectrum β-Lactamase-Producing Escherichia coli Isolated from Food-Producing Animals in Tamaulipas, Mexico. Antibiotics, 12(6), 1010. https://doi.org/10.3390/antibiotics12061010











