Cutibacterium acnes KCTC 3314 Growth Reduction with the Combined Use of Bacteriophage PAP 1-1 and Nisin
Abstract
:1. Introduction
2. Results
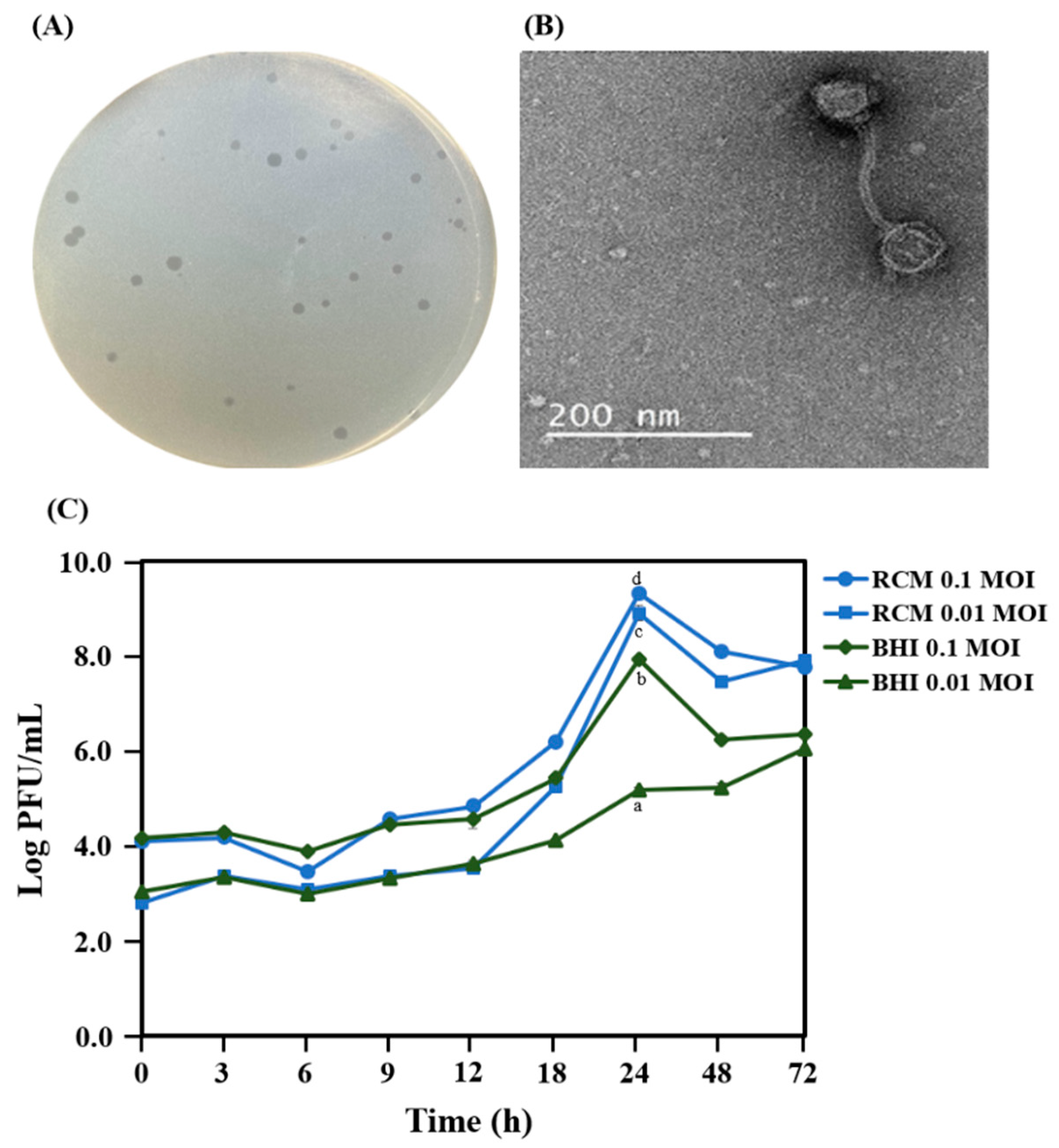
2.1. Isolation and Morphology of PAP 1-1
2.2. Genome Features
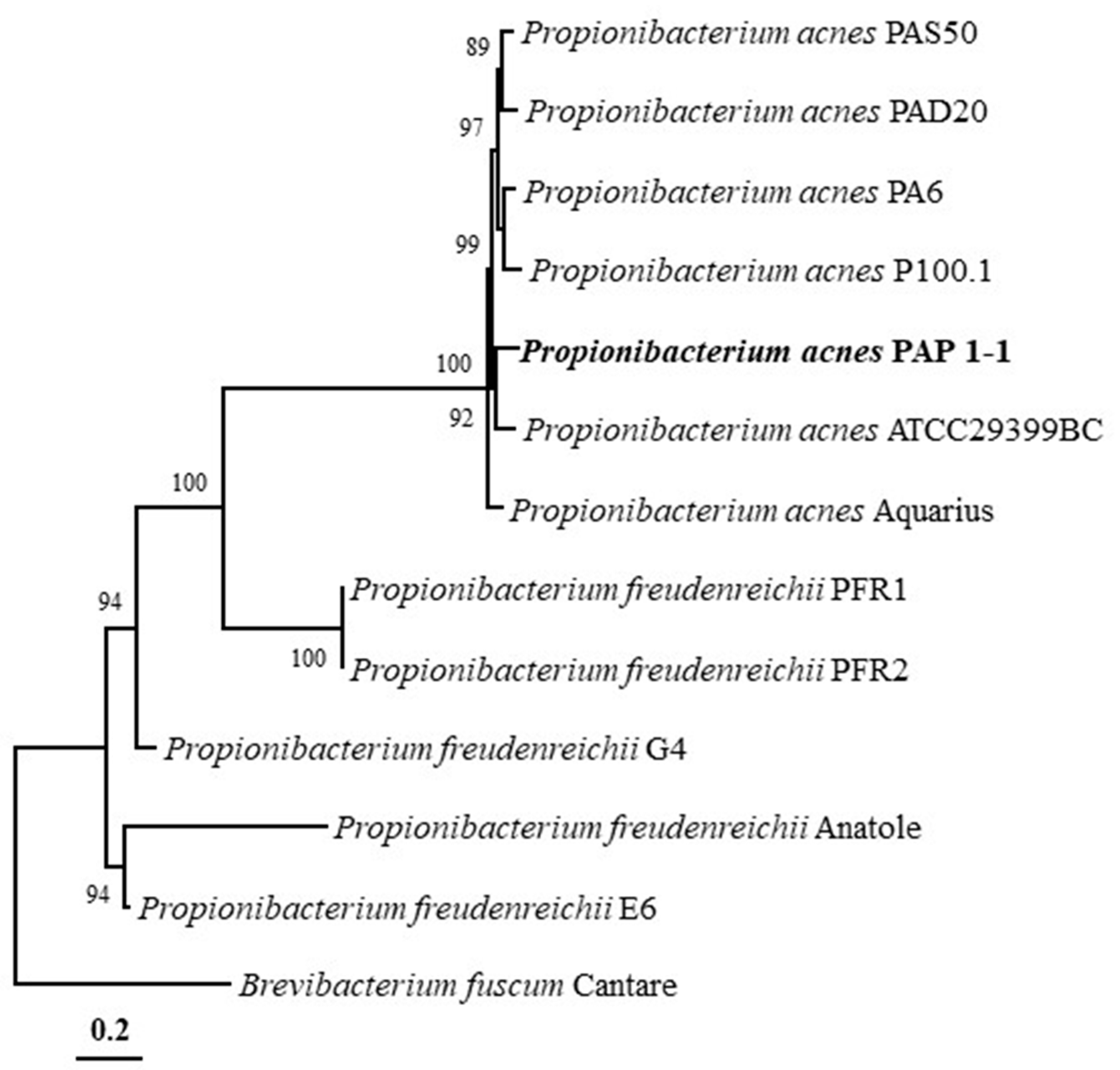
2.3. Phylogenetic Analysis and Genomic Comparison
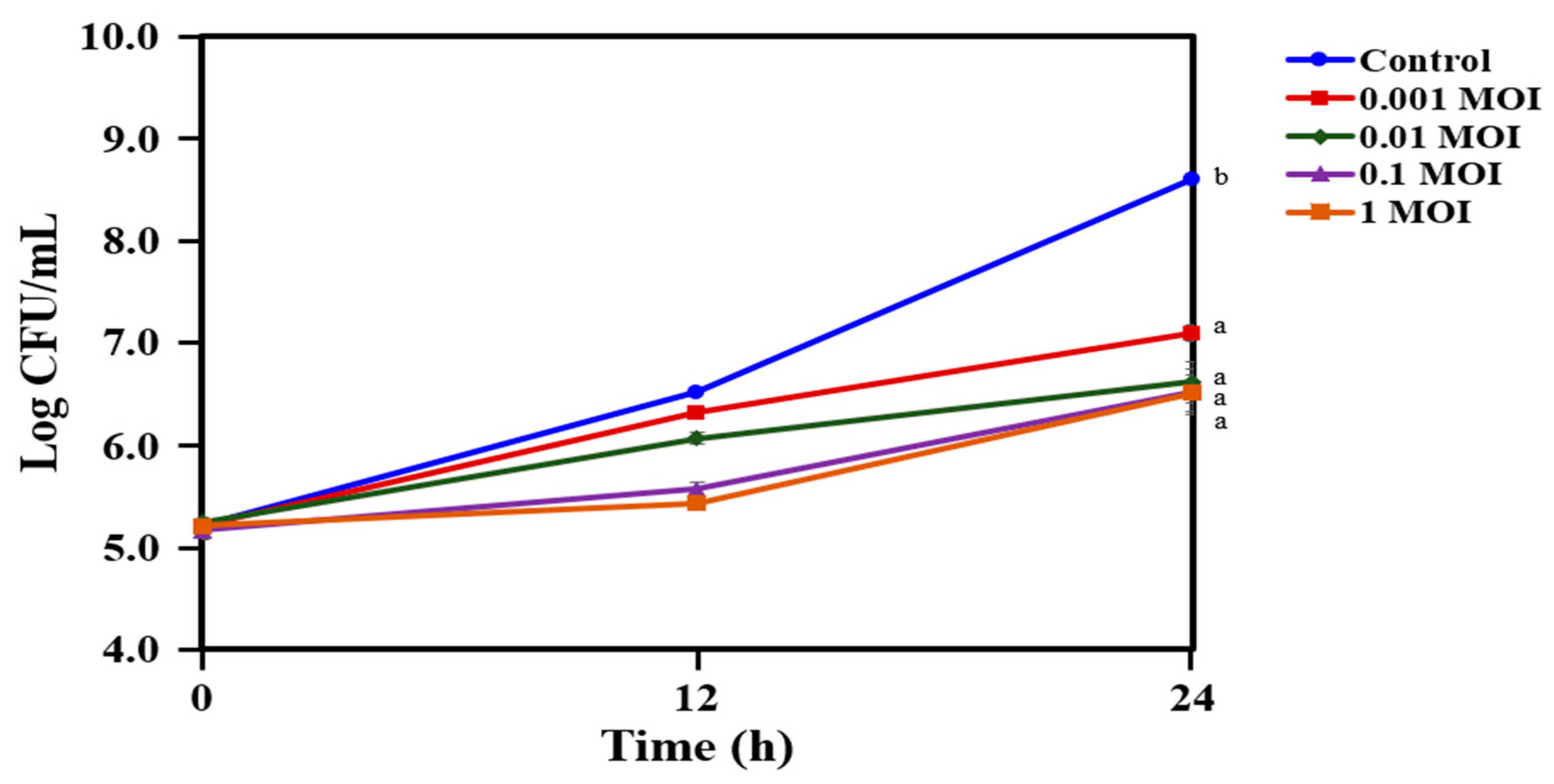
2.4. Reduction in the Growth of C. acnes KCTC 3314 with Bacteriophage PAP 1-1
2.5. Reduction in the Growth of C. acnes KCTC 3314 Followed by Co-treatment with Bacteriophage PAP 1-1 and Bacteriocin
2.6. Reduction in the Growth of C. acnes KCTC 3314 Followed by Co-treatment with Bacteriophage PAP 1-1 and Nisin
3. Discussion
4. Materials and Methods
4.1. Isolation of Bacteriophage
4.2. Transmission Electron Microscopy of PAP 1-1 Bacteriophage
4.3. Optimal Medium for Bacteriophage Propagation
4.4. Genomic DNA Extraction, Sequencing, and Annotation
4.5. Phylogenetic Analysis and Genomic Comparison
4.6. Phage Lytic Activity of Bacteriophage PAP 1-1
4.7. Bacteriocin Extraction and Purification
4.8. Combination Treatment with Bacteriophage PAP 1-1 and Crude Bacteriocin from Lactococcus Lactis and Nisin
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grice, E.A.; Segre, J.A. The Skin Microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Kao, H.J.; Wang, Y.H.; Keshari, S.; Yang, J.J.; Simbolon, S.; Chen, C.C.; Huang, C.M. Propionic acid produced by Cutibacterium acnes fermentation ameliorates ultraviolet B-induced melanin synthesis. Sci. Rep. 2021, 11, 11980. [Google Scholar] [CrossRef] [PubMed]
- Barnard, E.; Shi, B.; Kang, D.; Craft, N.; Li, H. The balance of metagenomic elements shapes the skin microbiome in acne and health. Sci. Rep. 2016, 6, 39491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szabó, K.; Erdei, L.; Bolla, B.S.; Tax, G.; Bíró, T.; Kemény, L. Factors shaping the composition of the cutaneous microbiota. Br. J. Dermatol. 2017, 176, 344–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Achermann, Y.; Goldstein, E.J.C.; Coenye, T.; Shirtliffa, M.E. Propionibacterium acnes: From Commensal to opportunistic biofilm-associated implant pathogen. Clin. Microbiol. Rev. 2014, 27, 419–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dessinioti, C.; Katsambas, A.D. The role of Propionibacterium acnes in acne pathogenesis: Facts and controversies. Clin. Dermatol. 2010, 28, 2–7. [Google Scholar] [CrossRef]
- Grange, P.A.; Raingeaud, J.; Morelle, W.; Marcelin, A.G.; Calvez, V.; Dupin, N. Characterization of a Propionibacterium acnes Surface Protein as a Fibrinogen-Binding Protein. Sci. Rep. 2017, 7, 6428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hay, R.J.; Johns, N.E.; Williams, H.C.; Bolliger, I.W.; Dellavalle, R.P.; Margolis, D.J.; Marks, R.; Naldi, L.; Weinstock, M.A.; Wulf, S.K.; et al. The global burden of skin disease in 2010: An analysis of the prevalence and impact of skin conditions. J. Investig. Dermatol. 2014, 134, 1527–1534. [Google Scholar] [CrossRef] [Green Version]
- Lynn, D.; Umari, T.; Dellavalle, R.; Dunnick, C. The epidemiology of acne vulgaris in late adolescence. Adolesc. Health Med. Ther. 2016, 7, 13–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Rosso, J. Topical and oral antibiotics for acne vulgaris. Semin. Cutan. Med. Surg. 2016, 35, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Dréno, B. Bacteriological resistance in acne: A call to action. Eur. J. Dermatol. 2016, 26, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Beylot, C.; Auffret, N.; Poli, F.; Claudel, J.P.; Leccia, M.T.; Del Giudice, P.; Dreno, B. Propionibacterium acnes: An update on its role in the pathogenesis of acne. J. Eur. Acad. Dermatol. Venereol. 2014, 28, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Eskenazi, A.; Lood, C.; Wubbolts, J.; Hites, M.; Balarjishvili, N.; Leshkasheli, L.; Askilashvili, L.; Kvachadze, L.; van Noort, V.; Wagemans, J.; et al. Combination of pre-adapted bacteriophage therapy and antibiotics for treatment of fracture-related infection due to pandrug-resistant Klebsiella pneumoniae. Nat. Comm. 2022, 13, 302. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Datta, S.; Chatterjee, S.; Dutta, M.; Samanta, J.; Vairale, M.G.; Gupta, R.; Veer, V.; Dwivedi, S.K. Isolation and characterization of a lytic bacteriophage against Pseudomonas aeruginosa. Sci. Rep. 2021, 11, 19393. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharm. Ther. 2017, 8, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Delwart, E.L. Viral metagenomics. Rev. Med. Virol. 2007, 17, 115–131. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V. The wonder world of microbial viruses. Expert Rev. Anti Infect. Ther. 2010, 8, 1097–1099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suttle, C. Viruses in the sea. Nature 2005, 437, 356–361. [Google Scholar] [CrossRef]
- Clokie, M.R.J.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef] [Green Version]
- Young, R.; Gill, J.J. Phage therapy redux—What is to be done? Science 2015, 350, 1163–1164. [Google Scholar] [CrossRef]
- Salmond, G.P.C.; Fineran, P.C. A century of the phage: Past, present and future. Nature Rev. Microbiol. 2015, 13, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Church, N.A.; McKillip, J.L. Antibiotic resistance crisis: Challenges and imperatives. Biologia 2021, 76, 1535–1550. [Google Scholar] [CrossRef]
- Hammami, R.; Fliss, I.; Corsetti, A. Editorial: Application of protective cultures and bacteriocins for food biopreservation. Front. Microbiol. 2019, 10, 1561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins-a viable alternative to antibiotics? Nature Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Yap, P.G.; Lai, Z.W.; Tan, J.S. Bacteriocins from lactic acid bacteria: Purification strategies and applications in food and medical industries: A review. Beni-Suef Univ. J. Basic Appl. Sci. 2022, 11, 51. [Google Scholar] [CrossRef]
- Jiang, S.; Cai, L.; Lv, L.; Li, L. Pediococcus pentosaceus, a future additive or probiotic candidate. Microb. Cell Factories 2021, 20, 45. [Google Scholar] [CrossRef]
- Cotter, P.D.; Hill, C.; Ross, R.P. Bacteriocins: Developing innate immunity for food. Nat. Rev. Microbiol. 2005, 3, 777–788. [Google Scholar] [CrossRef]
- Yang, R.; Johnson, M.C.; Ray, B. Novel method to extract large amounts of bacteriocins from lactic acid bacteria. Appl. Environ. Microbiol. 1992, 58, 3355–3359. [Google Scholar] [CrossRef] [Green Version]
- Muhammad, Z.; Ramzan, R.; Abdelazez, A.; Amjad, A.; Afzaal, M.; Zhang, S.; Pan, S. Assessment of the antimicrobial potentiality and functionality of Lactobacillus plantarum strains isolated from the conventional inner Mongolian fermented cheese against foodborne pathogens. Pathogens 2019, 8, 71. [Google Scholar] [CrossRef] [Green Version]
- da Costa, R.J.; Voloski, F.L.; Mondadori, R.G.; Duval, E.H.; Fiorentini, Â.M. Preservation of meat products with bacteriocins produced by lactic acid bacteria isolated from meat. J. Food Qual. 2019, 2019, 4726510. [Google Scholar] [CrossRef] [Green Version]
- Radaic, A.; Malone, E.; Kamarajan, P.; Kapila, Y.L. Solid Lipid Nanoparticles Loaded with Nisin (SLN-Nisin) are More Effective Than Free Nisin as Antimicrobial, Antibiofilm, and Anticancer Agents. J. Biomed. Nanotechnol. 2022, 18, 1227–1235. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, A.A.T.; de Melo, M.R.; da Silva, C.M.R.; Jain, S.; Dolabella, S.S. Nisin resistance in Gram-positive bacteria and approaches to circumvent resistance for successful therapeutic use. Crit. Rev. Microbiol. 2021, 47, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yan, R.; Zhong, Q.; Ngo, S.; Bangayan, N.J.; Nguyen, L.; Lui, T.; Liu, M.; Erfe, M.C.; Craft, N.; et al. The diversity and host interactions of Propionibacterium acnes bacteriophages on human skin. ISME J. 2015, 9, 2078–2093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dewanggana, M.N.; Evangeline, C.; Ketty, M.D.; Waturangi, D.E.; Yogiara; Magdalena, S. Isolation, characterization, molecular analysis and application of bacteriophage DW-EC to control Enterotoxigenic Escherichia coli on various foods. Sci. Rep. 2022, 12, 495. [Google Scholar] [CrossRef] [PubMed]
- Castillo, D.E.; Nanda, S.; Keri, J.E. Propionibacterium (Cutibacterium) acnes Bacteriophage Therapy in Acne: Current Evidence and Future Perspectives. Dermatol. Ther. 2019, 9, 19–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marinelli, L.J.; Fitz-Gibbon, S.; Hayes, C.; Bowman, C.; Inkeles, M.; Loncaric, A.; Russell, D.A.; Jacobs-Sera, D.; Cokus, S.; Pellegrini, M.; et al. Propionibacterium acnes bacteriophages display limited genetic diversity and broad killing activity against bacterial skin isolates. mBio 2012, 3, e00279-12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farrar, M.D.; Howson, K.M.; Bojar, R.A.; West, D.; Towler, J.C.; Parry, J.; Pelton, K.; Holland, K.T. Genome sequence and analysis of a Propionibacterium acnes bacteriophage. J. Bacteriol. 2007, 189, 4161–4167. [Google Scholar] [CrossRef] [Green Version]
- Schmelcher, M.; Donovan, D.M.; Loessner, M.J. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012, 7, 1147–1171. [Google Scholar] [CrossRef] [Green Version]
- Nelson, D.C.; Schmelcher, M.; Rodriguez-Rubio, L.; Klumpp, J.; Pritchard, D.G.; Dong, S.; Donovan, D.M. Endolysins as Antimicrobials. Adv. Virus Res. 2012, 83, 299–365. [Google Scholar]
- Jończyk-Matysiak, E.; Weber-Dabrowska, B.; Zaczek, M.; Miedzybrodzki, R.; Letkiewicz, S.; Łusiak-Szelchowska, M.; Górski, A. Prospects of phage application in the treatment of acne caused by Propionibacterium acnes. Front. Microbiol. 2017, 8, 164. [Google Scholar] [CrossRef] [Green Version]
- Rhoads, D.D.; Wolcott, R.D.; Kuskowski, M.A.; Wolcott, B.M.; Ward, L.S.; Sulakvelidze, A. Bacteriophage therapy of venous leg ulcers in humans: Results of a phase i safety trial. J. Wound Care 2009, 18, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Kutter, E.; De Vos, D.; Gvasalia, G.; Alavidze, Z.; Gogokhia, L.; Kuhl, S.; Abedon, S. Phage Therapy in Clinical Practice: Treatment of Human Infections. Curr. Pharm. Biotechnol. 2010, 11, 69–86. [Google Scholar] [CrossRef] [PubMed]
- Merabishvili, M.; de Vos, D.; Verbeken, G.; Kropinski, A.M.; Vandenheuvel, D.; Lavigne, R.; Wattiau, P.; Mast, J.; Ragimbeau, C.; Mossong, J.; et al. Selection and Characterization of a Candidate Therapeutic Bacteriophage That Lyses the Escherichia coli O104:H4 Strain from the 2011 Outbreak in Germany. PLoS ONE 2012, 7, e52709. [Google Scholar] [CrossRef] [Green Version]
- Pouillot, F.; Chomton, M.; Blois, H.; Courroux, C.; Noelig, J.; Bidet, P.; Bingen, E.; Bonacorsi, S. Efficacy of bacteriophage therapy in experimental sepsis and meningitis caused by a clone O25b: H4-ST131 Escherichia coli strain producing CTX-M-15. Antimicrob. Agents Chemother. 2012, 56, 3568–3575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chhibber, S.; Kaur, T.; Kaur, S. Co-Therapy Using Lytic Bacteriophage and Linezolid: Effective Treatment in Eliminating Methicillin Resistant Staphylococcus aureus (MRSA) from Diabetic Foot Infections. PLoS ONE 2013, 8, e56022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biswas, B.; Adhya, S.; Washart, P.; Paul, B.; Trostel, A.N.; Powell, B.; Carlton, R.; Merril, C.R. Erratum: Bacteriophage therapy rescues mice bacteremic from a clinical isolate of vancomycin-resistant Enterococcus faecium (Infection and Immunity (2002) 70:1 (204–210)). Infect. Immun. 2002, 70, 1664. [Google Scholar] [CrossRef] [Green Version]
- Wittebole, X.; De Roock, S.; Opal, S.M. A Historical Overview of Bacteriophage Therapy as an Alternative to Antibiotics for the Treatment of Bacterial Pathogens. Virulence 2014, 5, 226–235. [Google Scholar] [CrossRef]
- Brown, T.L.; Petrovski, S.; Dyson, Z.A.; Seviour, R.; Tucci, J. The Formulation of Bacteriophage in a Semi Solid Preparation for Control of Propionibacterium Acnes Growth. PLoS ONE 2016, 11, e0151184. [Google Scholar] [CrossRef] [Green Version]
- Golembo, M.; Puttagunta, S.; Rappo, U.; Weinstock, E.; Engelstein, R.; Gahali-Sass, I.; Moses, A.; Kario, E.; Ben-Dor Cohen, E.; Nicenboim, J.; et al. Development of a topical bacteriophage gel targeting Cutibacterium acnes for acne prone skin and results of a phase 1 cosmetic randomized clinical trial. Skin Health Dis. 2022, 2, e93. [Google Scholar] [CrossRef]
- de Sena Brandine, G.; Smith, A.D. Falco: High-Speed FastQC Emulation for Quality Control of Sequencing Data. F1000Res 2019, 8, 1874. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comp. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, D.M.; Kim, K.E.; Jeong, S.Y.; Park, C.S.; Ahn, K.H.; Kim, D.H.; Kang, D.O.; Chun, H.K.; Yoon, B.D.; Koh, H.B.; et al. Rapid concentration of some bacteriocin-like compounds using an organic solvent. Food Sci. Biotechnol. 2011, 20, 1457–1459. [Google Scholar] [CrossRef]
- Kim, S.G.; Lee, Y.D.; Park, J.H.; Moon, G.S. Synergistic inhibition by bacteriocin and bacteriophage against Staphylococcus aureus. Food Sci. Anim. Resour. 2019, 39, 1015–1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| CDs | Annotation | ||
|---|---|---|---|
| PAP1-1-1 | 47 | 373 | Terminase small subunit |
| PAP1-1-2 | 378 | 1889 | Gp2 (Propionibacterium phage PA6) |
| PAP1-1-3 | 1886 | 3211 | Capsid and scaffold protein |
| PAP1-1-4 | 3215 | 3970 | Capsid maturation protease |
| PAP1-1-5 | 4078 | 4638 | Putative head assembly scaffold |
| PAP1-1-6 | 4645 | 5592 | Major capsid protein |
| PAP1-1-7 | 5636 | 6097 | Head-to-tail adaptor |
| PAP1-1-8 | 6099 | 6446 | Gp8 |
| PAP1-1-9 | 6452 | 6742 | Gp9 (Propionibacterium phage PA6) |
| PAP1-1-10 | 6739 | 7110 | Gp10 (Propionibacterium phage PAS50) |
| PAP1-1-11 | 7162 | 7794 | Major tail protein (Propionibacterium phage P9.1) |
| PAP1-1-12 | 7822 | 8118 | Gp12 |
| PAP1-1-13 | 8217 | 8504 | Putative tail assembly chaperone |
| PAP1-1-14 | 8513 | 11,278 | Tape measure protein |
| PAP1-1-15 | 11,294 | 12,235 | Minor tail protein |
| PAP1-1-16 | 12,243 | 13,400 | Putative protease |
| PAP1-1-17 | 13,421 | 14,239 | Putative minor tail protein |
| PAP1-1-18 | 14,540 | 15,328 | Putative minor tail protein |
| PAP1-1-19 | 15,378 | 16,238 | Putative endolysin |
| PAP1-1-20 | 16,251 | 16,649 | Gp21 |
| PAP1-1-21 | 16,735 | 17,250 | Uncharacterized protein |
| PAP1-1-22 | 17,256 | 17,654 | Helix–turn–helix DNA-binding domain protein |
| PAP1-1-23 | 17,658 | 17,942 | Gp25 (Propionibacterium phage PAS50) |
| PAP1-1-24 | 17,954 | 18,274 | Helix–turn–helix DNA-binding domain protein |
| PAP1-1-25 | 18,284 | 19,330 | Gp27 |
| PAP1-1-26 | 19,327 | 19,521 | Gp28 (Propionibacterium phage PA6) |
| PAP1-1-27 | 19,505 | 20,062 | Gp29.1 (Propionibacterium phage PAS50) |
| PAP1-1-28 | 20,059 | 20,625 | Gp30 (Propionibacterium phage PA6) |
| PAP1-1-29 | 20,670 | 21,341 | DNA primase |
| PAP1-1-30 | 21,539 | 21,895 | Holliday junction resolvase |
| PAP1-1-31 | 21,895 | 22,758 | DNA primase/helicase |
| PAP1-1-32 | 22,804 | 23,268 | Gp35 (Propionibacterium phage PAS50) |
| PAP1-1-33 | 23,321 | 23,731 | Gp36 (Propionibacterium phage PAS50) |
| PAP1-1-34 | 23,728 | 24,675 | Cas4 family exonuclease (Propionibacterium phage Pirate) |
| PAP1-1-35 | 24,675 | 25,028 | Gp38 (Propionibacterium phage PAS50) |
| PAP1-1-36 | 25,134 | 25,337 | Gp39 |
| PAP1-1-37 | 25,334 | 25,561 | Gp40 (Propionibacterium phage PAS50) |
| PAP1-1-38 | 25,578 | 26,111 | Gp41 (Propionibacterium phage PAS50) |
| PAP1-1-39 | 26,232 | 26,543 | Gp43 (Propionibacterium phage PA6) |
| PAP1-1-40 | 26,578 | 26,886 | Gp43 (Propionibacterium phage PA6) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, M.-H.; Khan, S.A.; Moon, G.-S. Cutibacterium acnes KCTC 3314 Growth Reduction with the Combined Use of Bacteriophage PAP 1-1 and Nisin. Antibiotics 2023, 12, 1035. https://doi.org/10.3390/antibiotics12061035
Han M-H, Khan SA, Moon G-S. Cutibacterium acnes KCTC 3314 Growth Reduction with the Combined Use of Bacteriophage PAP 1-1 and Nisin. Antibiotics. 2023; 12(6):1035. https://doi.org/10.3390/antibiotics12061035
Chicago/Turabian StyleHan, Min-Hui, Shehzad Abid Khan, and Gi-Seong Moon. 2023. "Cutibacterium acnes KCTC 3314 Growth Reduction with the Combined Use of Bacteriophage PAP 1-1 and Nisin" Antibiotics 12, no. 6: 1035. https://doi.org/10.3390/antibiotics12061035







