Insect Antimicrobial Peptides: Advancements, Enhancements and New Challenges
Abstract
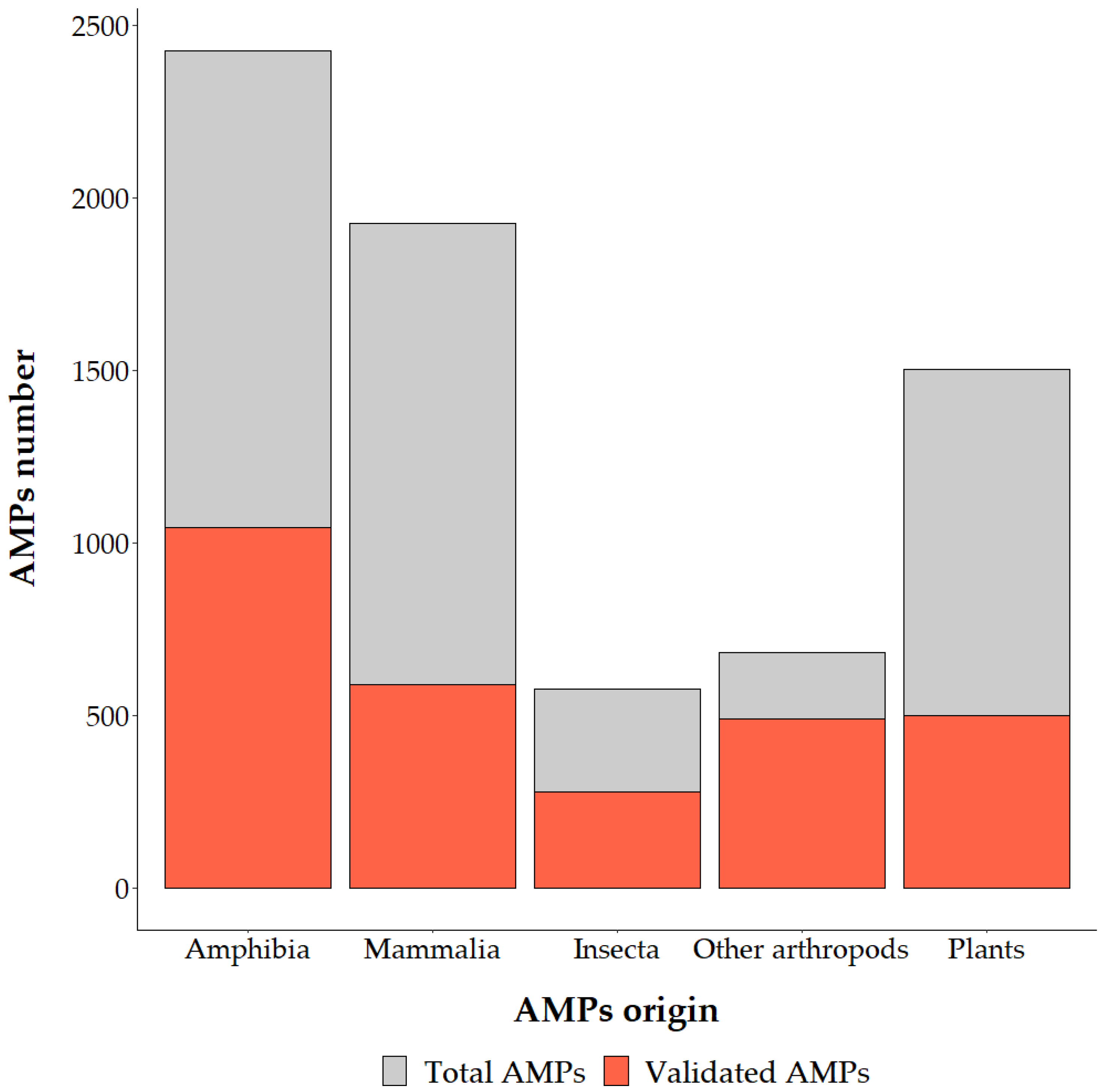
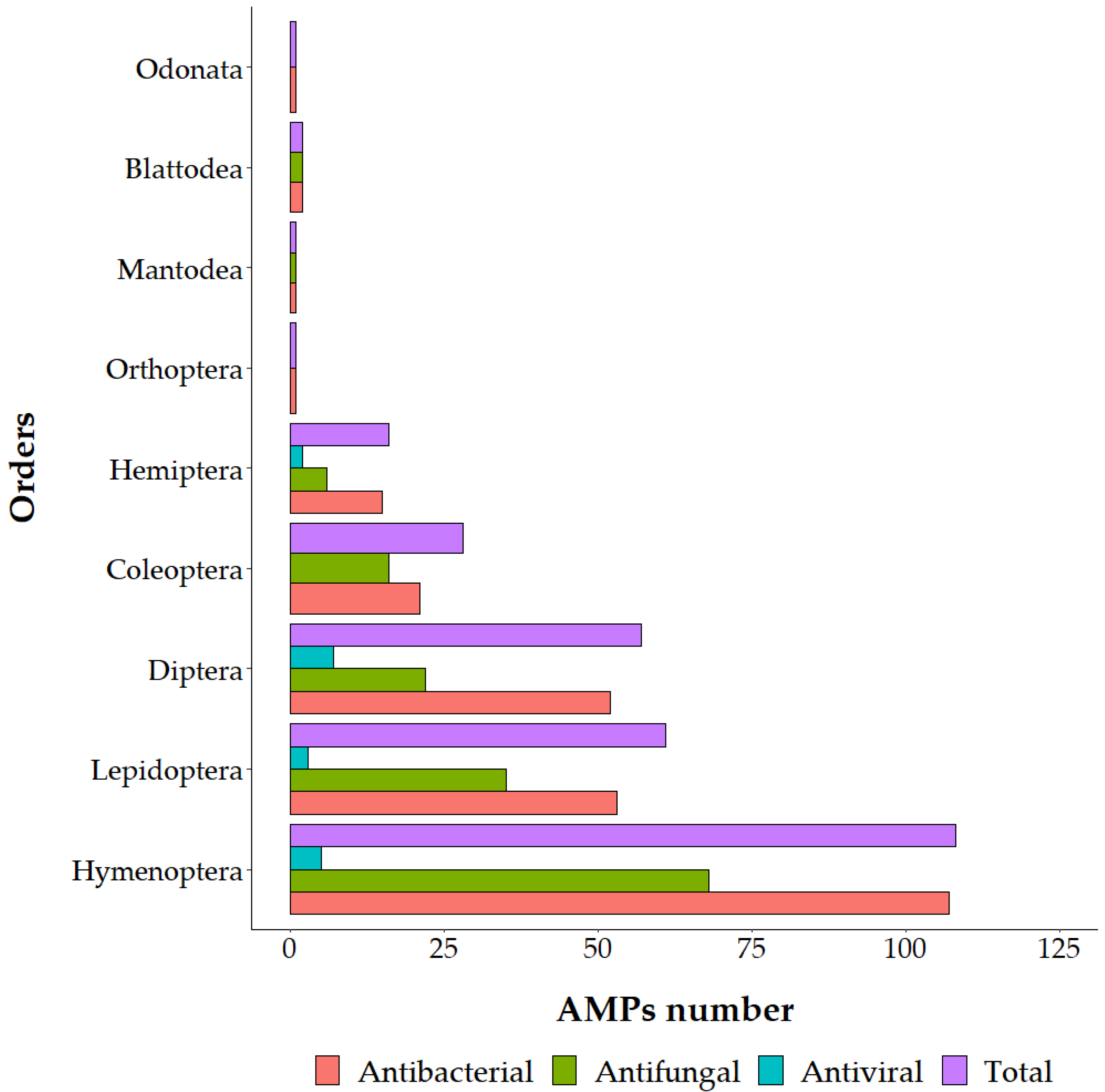
:1. Introduction
2. Mechanism of Action of AMPs
2.1. Pore Formation
2.2. Other Mechanisms of Action
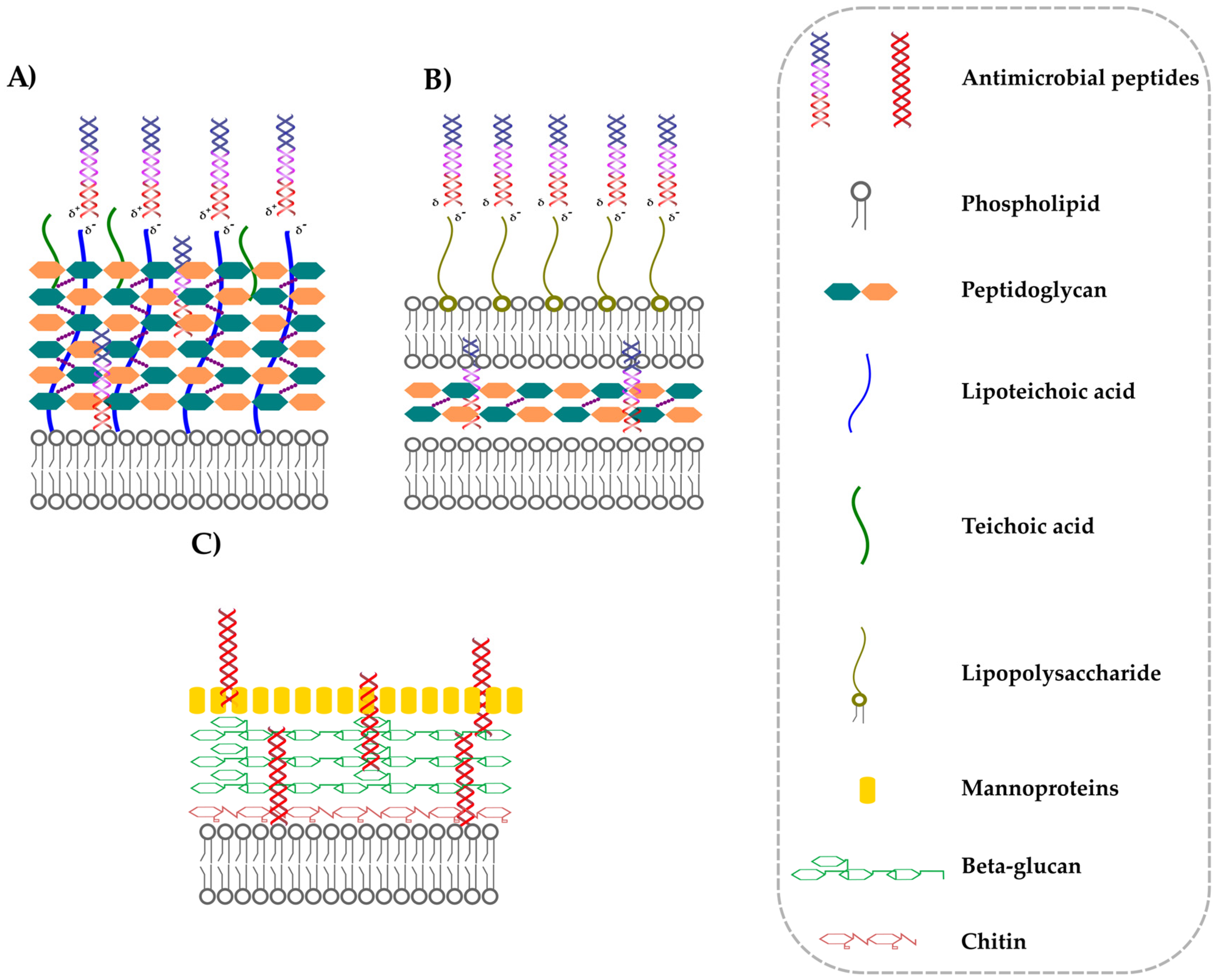
3. Interactions between AMPs and Cell Wall
4. Proposed Applications of Insect AMPs
4.1. Proposed Applications of AMPs in the Medical Field
4.2. Proposed Applications of Insect AMPs in Agriculture
4.3. Proposed Applications of Insect AMPs in the Food Chain
5. Effect of Insect Diet on AMP Translation and Synthesis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reddy, K.; Yedery, R.; Aranha, C. Antimicrobial peptides: Premises and promises. Int. J. Antimicrob. Agents 2004, 24, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.; Kjos, M.; Nes, I.; Diep, D.; Lotfipour, F. Natural antimicrobial peptides from bacteria: Characteristics and potential applications to fight against antibiotic resistance. J. Appl. Microbiol. 2012, 113, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Manniello, M.; Moretta, A.; Salvia, R.; Scieuzo, C.; Lucchetti, D.; Vogel, H.; Sgambato, A.; Falabella, P. Insect antimicrobial peptides: Potential weapons to counteract the antibiotic resistance. Cell. Mol. Life Sci. 2021, 78, 4259–4282. [Google Scholar] [CrossRef]
- Jhong, J.-H.; Yao, L.; Pang, Y.; Li, Z.; Chung, C.-R.; Wang, R.; Li, S.; Li, W.; Luo, M.; Ma, R. dbAMP 2.0: Updated resource for antimicrobial peptides with an enhanced scanning method for genomic and proteomic data. Nucleic Acids Res. 2022, 50, D460–D470. [Google Scholar] [CrossRef]
- Yi, H.-Y.; Chowdhury, M.; Huang, Y.-D.; Yu, X.-Q. Insect antimicrobial peptides and their applications. Appl. Microbiol. Biotechnol. 2014, 98, 5807. [Google Scholar] [CrossRef] [PubMed]
- Hillyer, J.F. Insect immunology and hematopoiesis. Dev. Comp. Immunol. 2016, 58, 102–118. [Google Scholar] [CrossRef] [PubMed]
- Buonocore, F.; Fausto, A.M.; Della Pelle, G.; Roncevic, T.; Gerdol, M.; Picchietti, S. Attacins: A promising class of insect antimicrobial peptides. Antibiotics 2021, 10, 212. [Google Scholar] [CrossRef]
- Vilcinskas, A.; Schmidtberg, H.; Estoup, A.; Tayeh, A.; Facon, B.; Vogel, H. Evolutionary ecology of microsporidia associated with the invasive ladybird Harmonia axyridis. Insect Sci. 2015, 22, 313–324. [Google Scholar] [CrossRef]
- Moretta, A.; Salvia, R.; Scieuzo, C.; Di Somma, A.; Vogel, H.; Pucci, P.; Sgambato, A.; Wolff, M.; Falabella, P. A bioinformatic study of antimicrobial peptides identified in the Black Soldier Fly (BSF) Hermetia illucens (Diptera: Stratiomyidae). Sci. Rep. 2020, 10, 16875. [Google Scholar] [CrossRef]
- Kim, I.-W.; Lee, J.H.; Subramaniyam, S.; Yun, E.-Y.; Kim, I.; Park, J.; Hwang, J.S. De novo transcriptome analysis and detection of antimicrobial peptides of the American cockroach Periplaneta americana (Linnaeus). PLoS ONE 2016, 11, e0155304. [Google Scholar] [CrossRef]
- Waghu, F.H.; Barai, R.S.; Gurung, P.; Idicula-Thomas, S. CAMPR3: A database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res. 2015, 44, D1094–D1097. [Google Scholar] [CrossRef] [PubMed]
- Vilcinskas, A.; Mukherjee, K.; Vogel, H. Expansion of the antimicrobial peptide repertoire in the invasive ladybird Harmonia axyridis. Proc. R. Soc. B Biol. Sci. 2013, 280, 20122113. [Google Scholar] [CrossRef] [PubMed]
- Orivel, J.; Redeker, V.; Le Caer, J.-P.; Krier, F.; Revol-Junelles, A.-M.; Longeon, A.; Chaffotte, A.; Dejean, A.; Rossier, J. Ponericins, new antibacterial and insecticidal peptides from the venom of the ant Pachycondyla goeldii. J. Biol. Chem. 2001, 276, 17823–17829. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.-W.; Markkandan, K.; Lee, J.H.; Subramaniyam, S.; Yoo, S.; Park, J.; Hwang, J.S. Transcriptome profiling and in silico analysis of the antimicrobial peptides of the grasshopper Oxya chinensis sinuosa. J. Microbiol 2016, 26, 1863–1870. [Google Scholar] [CrossRef] [PubMed]
- Remington, J.M.; Liao, C.; Sharafi, M.; Ste. Marie, E.J.; Ferrell, J.B.; Hondal, R.J.; Wargo, M.J.; Schneebeli, S.T.; Li, J. Aggregation state of synergistic antimicrobial peptides. J. Phys. Chem. Lett. 2020, 11, 9501–9506. [Google Scholar] [CrossRef] [PubMed]
- Mylonakis, E.; Podsiadlowski, L.; Muhammed, M.; Vilcinskas, A. Diversity, evolution and medical applications of insect antimicrobial peptides. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150290. [Google Scholar] [CrossRef]
- Misof, B.; Liu, S.; Meusemann, K.; Peters, R.S.; Donath, A.; Mayer, C.; Frandsen, P.B.; Ware, J.; Flouri, T.; Beutel, R.G. Phylogenomics resolves the timing and pattern of insect evolution. Science 2014, 346, 763–767. [Google Scholar] [CrossRef] [PubMed]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial peptides: Classification, design, application and research progress in multiple fields. Front. Microbiol. 2020, 11, 2559. [Google Scholar] [CrossRef]
- Morton, J.T.; Freed, S.D.; Lee, S.W.; Friedberg, I. A large scale prediction of bacteriocin gene blocks suggests a wide functional spectrum for bacteriocins. BMC Bioinform. 2015, 16, 381. [Google Scholar] [CrossRef]
- Bíliková, K.; Mirgorodskaya, E.; Bukovská, G.; Gobom, J.; Lehrach, H.; Šimúth, J. Towards functional proteomics of minority component of honeybee royal jelly: The effect of post-translational modifications on the antimicrobial activity of apalbumin2. Proteomics 2009, 9, 2131–2138. [Google Scholar] [CrossRef]
- Koehbach, J. Structure-activity relationships of insect defensins. Front. Chem. 2017, 5, 45. [Google Scholar] [CrossRef] [PubMed]
- Darveau, R.P.; Blake, J.; Seachord, C.L.; Cosand, W.L.; Cunningham, M.D.; Cassiano-Clough, L.; Maloney, G. Peptides related to the carboxyl terminus of human platelet factor IV with antibacterial activity. J. Clin. Investig. 1992, 90, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Hultmark, D.; Steiner, H.; Rasmuson, T.; Boman, H.G. Insect immunity. Purification and properties of three inducible bactericidal proteins from hemolymph of immunized pupae of Hyalophora cecropia. Eur. J. Biochem. 1980, 106, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, K.; Natori, S. Purification of three antibacterial proteins from the culture medium of NIH-Sape-4, an embryonic cell line of Sarcophaga peregrina. J. Biol. Chem. 1988, 263, 17112–17116. [Google Scholar] [CrossRef] [PubMed]
- Laver, D. The barrel-stave model as applied to alamethicin and its analogs reevaluated. Biophys. J. 1994, 66, 355–359. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Liu, S.; Wang, W.; Deng, L.; Ming, J.; Yao, S.; Zeng, K. Control of sour rot in citrus fruit by three insect antimicrobial peptides. Postharvest Biol. Technol. 2019, 149, 200–208. [Google Scholar] [CrossRef]
- Neshani, A.; Sedighian, H.; Mirhosseini, S.A.; Ghazvini, K.; Zare, H.; Jahangiri, A. Antimicrobial peptides as a promising treatment option against Acinetobacter baumannii infections. Microb. Pathog. 2020, 146, 104238. [Google Scholar] [CrossRef]
- Aumer, T.; Voisin, S.b.N.; Knobloch, T.; Landon, C.; Bulet, P. Impact of an antifungal insect defensin on the proteome of the phytopathogenic fungus Botrytis cinerea. J. Proteome Res. 2020, 19, 1131–1146. [Google Scholar] [CrossRef]
- Vogel, H.; MŘller, A.; Heckel, D.G.; Gutzeit, H.; Vilcinskas, A. Nutritional immunology: Diversification and diet-dependent expression of antimicrobial peptides in the black soldier fly Hermetia illucens. Dev. Comp. Immunol. 2018, 78, 141–148. [Google Scholar] [CrossRef]
- Candian, V.; Savio, C.; Meneguz, M.; Gasco, L.; Tedeschi, R. Effect of the rearing diet on gene expression of antimicrobial peptides in Hermetia illucens (Diptera: Stratiomyidae). Insect Sci. 2023. [Google Scholar] [CrossRef] [PubMed]
- Candian, V.; Meneguz, M.; Tedeschi, R. Immune responses of the Black Soldier Fly Hermetia illucens (L.) (Diptera: Stratiomyidae) reared on catering waste. Life 2023, 13, 213. [Google Scholar] [CrossRef] [PubMed]
- Candian, V.; Tedeschi, R. Impact of the diet on the mortality and on gene expression of the antimicrobial peptide tenecin 3 in Tenebrio molitor larvae infected by Beauveria bassiana. Insects 2023, 14, 359. [Google Scholar] [CrossRef]
- Dang, X.; Zheng, X.; Wang, Y.; Wang, L.; Ye, L.; Jiang, J. Antimicrobial peptides from the edible insect Musca domestica and their preservation effect on chilled pork. J. Food Process. Preserv. 2020, 44, e14369. [Google Scholar] [CrossRef]
- Sultana, A.; Luo, H.; Ramakrishna, S. Harvesting of antimicrobial peptides from insect (Hermetia illucens) and its applications in the food packaging. Appl. Sci. 2021, 11, 6991. [Google Scholar] [CrossRef]
- Bin Hafeez, A.; Jiang, X.; Bergen, P.J.; Zhu, Y. Antimicrobial peptides: An update on classifications and databases. Int. J. Mol. Sci. 2021, 22, 11691. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.-H.; Hofferek, V.; Separovic, F.; Reid, G.E.; Aguilar, M.-I. The role of bacterial lipid diversity and membrane properties in modulating antimicrobial peptide activity and drug resistance. Curr. Opin. Struct. Biol. 2019, 52, 85–92. [Google Scholar] [CrossRef]
- Rajamuthiah, R.; Jayamani, E.; Conery, A.L.; Fuchs, B.B.; Kim, W.; Johnston, T.; Vilcinskas, A.; Ausubel, F.M.; Mylonakis, E. A defensin from the model beetle Tribolium castaneum acts synergistically with telavancin and daptomycin against multidrug resistant Staphylococcus aureus. PLoS ONE 2015, 10, e0128576. [Google Scholar] [CrossRef]
- Hitchner, M.A.; Santiago-Ortiz, L.E.; Necelis, M.R.; Shirley, D.J.; Palmer, T.J.; Tarnawsky, K.E.; Vaden, T.D.; Caputo, G.A. Activity and characterization of a pH-sensitive antimicrobial peptide. Biochim. Biophys. Acta Biomembr. 2019, 1861, 182984. [Google Scholar] [CrossRef]
- Melo, M.N.; Ferre, R.; Castanho, M.A. Antimicrobial peptides: Linking partition, activity and high membrane-bound concentrations. Nat. Rev. Microbiol. 2009, 7, 245–250. [Google Scholar] [CrossRef]
- Hollmann, A.; Martinez, M.; Maturana, P.; Semorile, L.C.; Maffia, P.C. Antimicrobial peptides: Interaction with model and biological membranes and synergism with chemical antibiotics. Front. Chem. 2018, 6, 204. [Google Scholar] [CrossRef] [PubMed]
- Pfalzgraff, A.; Brandenburg, K.; Weindl, G. Antimicrobial peptides and their therapeutic potential for bacterial skin infections and wounds. Front. Pharmacol. 2018, 9, 281. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.; Chapple, D.S. Peptide antibiotics. Antimicrob. Agents Chemother. 1999, 43, 1317–1323. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Feix, J.B. Peptide–membrane interactions and mechanisms of membrane destruction by amphipathic α-helical antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 2006, 1758, 1245–1256. [Google Scholar] [CrossRef]
- Dagan, A.; Efron, L.; Gaidukov, L.; Mor, A.; Ginsburg, H. In vitro antiplasmodium effects of dermaseptin S4 derivatives. Antimicrob. Agents Chemother. 2002, 46, 1059–1066. [Google Scholar] [CrossRef]
- Ludtke, S.J.; He, K.; Heller, W.T.; Harroun, T.A.; Yang, L.; Huang, H.W. Membrane pores induced by magainin. Biochem. 1996, 35, 13723–13728. [Google Scholar] [CrossRef]
- Leontiadou, H.; Mark, A.E.; Marrink, S.J. Antimicrobial Peptides in Action. J. Am. Chem. Soc. 2006, 128, 12156–12161. [Google Scholar] [CrossRef]
- Patrzykat, A.; Friedrich, C.L.; Zhang, L.; Mendoza, V.; Hancock, R.E. Sublethal concentrations of pleurocidin-derived antimicrobial peptides inhibit macromolecular synthesis in Escherichia coli. Antimicrob. Agents Chemother. 2002, 46, 605–614. [Google Scholar] [CrossRef]
- Otvos, L.O.I.; Rogers, M.E.; Consolvo, P.J.; Condie, B.A.; Lovas, S.; Bulet, P.; Blaszczyk-Thurin, M. Interaction between heat shock proteins and antimicrobial peptides. Biochemistry 2000, 39, 14150–14159. [Google Scholar] [CrossRef]
- Castle, M.; Nazarian, A.; Tempst, P. Lethal effects of apidaecin on Escherichia coli involve sequential molecular interactions with diverse targets. J. Biol. Chem. 1999, 274, 32555–32564. [Google Scholar] [CrossRef]
- Di Somma, A.; Avitabile, C.; Cirillo, A.; Moretta, A.; Merlino, A.; Paduano, L.; Duilio, A.; Romanelli, A. The antimicrobial peptide Temporin L impairs E. coli cell division by interacting with FtsZ and the divisome complex. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129606. [Google Scholar] [CrossRef]
- Yadavalli, S.S.; Carey, J.N.; Leibman, R.S.; Chen, A.I.; Stern, A.M.; Roggiani, M.; Lippa, A.M.; Goulian, M. Antimicrobial peptides trigger a division block in Escherichia coli through stimulation of a signalling system. Nat. Commun. 2016, 7, 12340. [Google Scholar] [CrossRef] [PubMed]
- Florin, T.; Maracci, C.; Graf, M.; Karki, P.; Klepacki, D.; Berninghausen, O.; Beckmann, R.; Vázquez-Laslop, N.; Wilson, D.N.; Rodnina, M.V. An antimicrobial peptide that inhibits translation by trapping release factors on the ribosome. Nat. Struct. Biol. 2017, 24, 752–757. [Google Scholar] [CrossRef] [PubMed]
- Seefeldt, A.C.; Graf, M.; Pérébaskine, N.; Nguyen, F.; Arenz, S.; Mardirossian, M.; Scocchi, M.; Wilson, D.N.; Innis, C.A. Structure of the mammalian antimicrobial peptide Bac7 (1–16) bound within the exit tunnel of a bacterial ribosome. Nucleic Acids Res. 2016, 44, 2429–2438. [Google Scholar] [CrossRef]
- Seefeldt, A.C.; Nguyen, F.; Antunes, S.; Pérébaskine, N.; Graf, M.; Arenz, S.; Inampudi, K.K.; Douat, C.; Guichard, G.; Wilson, D.N. The proline-rich antimicrobial peptide Onc112 inhibits translation by blocking and destabilizing the initiation complex. Nat. Struct. Biol. 2015, 22, 470–475. [Google Scholar] [CrossRef]
- Roy, R.N.; Lomakin, I.B.; Gagnon, M.G.; Steitz, T.A. The mechanism of inhibition of protein synthesis by the proline-rich peptide oncocin. Nat. Struct. Biol. 2015, 22, 466–469. [Google Scholar] [CrossRef] [PubMed]
- Schneider, T.; Kruse, T.; Wimmer, R.; Wiedemann, I.; Sass, V.; Pag, U.; Jansen, A.; Nielsen, A.K.; Mygind, P.H.; Raventós, D.S. Plectasin, a fungal defensin, targets the bacterial cell wall precursor Lipid II. Science 2010, 328, 1168–1172. [Google Scholar] [CrossRef]
- Schmitt, P.; Wilmes, M.; Pugnière, M.; Aumelas, A.; Bachère, E.; Sahl, H.-G.; Schneider, T.; Destoumieux-Garzón, D. Insight into invertebrate defensin mechanism of action: Oyster defensins inhibit peptidoglycan biosynthesis by binding to lipid II. J. Biol. Chem. 2010, 285, 29208–29216. [Google Scholar] [CrossRef]
- Essig, A.; Hofmann, D.; Münch, D.; Gayathri, S.; Künzler, M.; Kallio, P.T.; Sahl, H.-G.; Wider, G.; Schneider, T.; Aebi, M. Copsin, a novel peptide-based fungal antibiotic interfering with the peptidoglycan synthesis. J. Biol. Chem. 2014, 289, 34953–34964. [Google Scholar] [CrossRef]
- Yount, N.Y.; Yeaman, M.R. Peptide antimicrobials: Cell wall as a bacterial target. Ann. N. Y. Acad. Sci. 2013, 1277, 127–138. [Google Scholar] [CrossRef]
- Yun, J.; Lee, D.G. Cecropin A-induced apoptosis is regulated by ion balance and glutathione antioxidant system in Candida albicans. IUBMB Life 2016, 68, 652–662. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, D.G. Melittin triggers apoptosis in Candida albicans through the reactive oxygen species-mediated mitochondria/caspase-dependent pathway. FEMS Microbiol. Lett. 2014, 355, 36–42. [Google Scholar] [CrossRef]
- Moghaddam, M.-R.B.; Gross, T.; Becker, A.; Vilcinskas, A.; Rahnamaeian, M. The selective antifungal activity of Drosophila melanogaster metchnikowin reflects the species-dependent inhibition of succinate–coenzyme Q reductase. Scie. Rep. 2017, 7, 8192. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Hwang, J.S.; Lee, D.G. Periplanetasin-4, a novel antimicrobial peptide from the cockroach, inhibits communications between mitochondria and vacuoles. Biochem. J. 2019, 476, 1267–1284. [Google Scholar] [CrossRef] [PubMed]
- Buda De Cesare, G.; Cristy, S.A.; Garsin, D.A.; Lorenz, M.C. Antimicrobial peptides: A new frontier in antifungal therapy. Mbio 2020, 11, e02123-20. [Google Scholar] [CrossRef]
- He, M.; Zhang, H.; Li, Y.; Wang, G.; Tang, B.; Zhao, J.; Huang, Y.; Zheng, J. Cathelicidin-derived antimicrobial peptides inhibit Zika virus through direct inactivation and interferon pathway. Front. Immunol. 2018, 9, 722. [Google Scholar] [CrossRef]
- Wachinger, M.; Kleinschmidt, A.; Winder, D.; von Pechmann, N.; Ludvigsen, A.; Neumann, M.; Holle, R.; Salmons, B.; Erfle, V.; Brack-Werner, R. Antimicrobial peptides Melittin and Cecropin inhibit replication of human immunodeficiency virus 1 by suppressing viral gene expression. J. Gen. Virol. 1998, 79, 731–740. [Google Scholar] [CrossRef]
- Liu, X.; Guo, C.; Huang, Y.; Zhang, X.; Chen, Y. Inhibition of porcine reproductive and respiratory syndrome virus by Cecropin D in vitro. Infect. Genet. Evol. 2015, 34, 7–16. [Google Scholar] [CrossRef]
- Guo, C.; Huang, Y.; Cong, P.; Liu, X.; Chen, Y.; He, Z. Cecropin P1 inhibits porcine reproductive and respiratory syndrome virus by blocking attachment. BMC Microbiol. 2014, 14, 273. [Google Scholar] [CrossRef]
- de Kruijff, B.; van Dam, V.; Breukink, E. Lipid II: A central component in bacterial cell wall synthesis and a target for antibiotics. Prostaglandins Leukot. Essent. Fat. Acids 2008, 79, 117–121. [Google Scholar] [CrossRef]
- Scott, M.G.; Gold, M.R.; Hancock, R.E. Interaction of cationic peptides with lipoteichoic acid and gram-positive bacteria. Infect. Immun. 1999, 67, 6445–6453. [Google Scholar] [CrossRef] [PubMed]
- Saar-Dover, R.; Bitler, A.; Nezer, R.; Shmuel-Galia, L.; Firon, A.; Shimoni, E.; Trieu-Cuot, P.; Shai, Y. D-alanylation of lipoteichoic acids confers resistance to cationic peptides in group B Streptococcus by increasing the cell wall density. PLoS Pathog. 2012, 8, e1002891. [Google Scholar] [CrossRef]
- Ebbensgaard, A.; Mordhorst, H.; Aarestrup, F.M.; Hansen, E.B. The role of outer membrane proteins and lipopolysaccharides for the sensitivity of Escherichia coli to antimicrobial peptides. Front. Microbiol. 2018, 9, 2153. [Google Scholar] [CrossRef]
- Van Der Weerden, N.L.; Hancock, R.E.W.; Anderson, M.A. Permeabilization of fungal hyphae by the plant defensin NaD1 occurs through a cell wall-dependent process. J. Biol. Chem. 2010, 285, 37513–37520. [Google Scholar] [CrossRef] [PubMed]
- Ibeas, J.I.; Lee, H.; Damsz, B.; Prasad, D.T.; Pardo, J.M.; Hasegawa, P.M.; Bressan, R.A.; Narasimhan, M.L. Fungal cell wall phosphomannans facilitate the toxic activity of a plant PR-5 protein. Plant J. 2000, 23, 375–383. [Google Scholar] [CrossRef]
- Thevissen, K.; de Mello Tavares, P.; Xu, D.; Blankenship, J.; Vandenbosch, D.; Idkowiak-Baldys, J.; Govaert, G.; Bink, A.; Rozental, S.; De Groot, P.W. The plant defensin RsAFP2 induces cell wall stress, septin mislocalization and accumulation of ceramides in Candida albicans. Mol. Microbiol. 2012, 84, 166–180. [Google Scholar] [CrossRef]
- Thevissen, K.; Warnecke, D.C.; François, I.E.; Leipelt, M.; Heinz, E.; Ott, C.; Zähringer, U.; Thomma, B.P.; Ferket, K.K.; Cammue, B.P. Defensins from insects and plants interact with fungal glucosylceramides. J. Biol. Chem. 2004, 279, 3900–3905. [Google Scholar] [CrossRef]
- Nakajima, Y.; Ishibashi, J.; Yukuhiro, F.; Asaoka, A.; Taylor, D.; Yamakawa, M. Antibacterial activity and mechanism of action of tick defensin against Gram-positive bacteria. Biochim. Biophys. Acta Gen. Subj. 2003, 1624, 125–130. [Google Scholar] [CrossRef]
- Bulet, P.; Stocklin, R. Insect antimicrobial peptides: Structures, properties and gene regulation. Protein Pept. Lett. 2005, 12, 3–11. [Google Scholar] [CrossRef]
- Hirsch, R.; Wiesner, J.; Marker, A.; Bauer, A.; Hammann, P.E.; Vilcinskas, A. Biological profiling of coleoptericins and coleoptericin-like antimicrobial peptides from the invasive harlequin ladybird Harmonia axyridis. In Advances in Microbiology, Infectious Diseases and Public Health; Springer: Berlin/Heidelberg, Germany, 2018; pp. 43–59. [Google Scholar]
- Terreni, M.; Taccani, M.; Pregnolato, M. New antibiotics for multidrug-resistant bacterial strains: Latest research developments and future perspectives. Molecules 2021, 26, 2671. [Google Scholar] [CrossRef]
- Park, S.-C.; Park, Y.; Hahm, K.-S. The role of antimicrobial peptides in preventing multidrug-resistant bacterial infections and biofilm formation. Int. J. Mol. Sci. 2011, 12, 5971–5992. [Google Scholar] [CrossRef] [PubMed]
- Simons, A.; Alhanout, K.; Duval, R.E. Bacteriocins, antimicrobial peptides from bacterial origin: Overview of their biology and their impact against multidrug-resistant bacteria. Microorganisms 2020, 8, 639. [Google Scholar] [CrossRef] [PubMed]
- Deslouches, B.; Montelaro, R.C.; Urish, K.L.; Di, Y.P. Engineered cationic antimicrobial peptides (eCAPs) to combat multidrug-resistant bacteria. Pharmaceutics 2020, 12, 501. [Google Scholar] [CrossRef] [PubMed]
- Lindhauer, N.S.; Bertrams, W.; Pöppel, A.; Herkt, C.E.; Wesener, A.; Hoffmann, K.; Greene, B.; Van Der Linden, M.; Vilcinskas, A.; Seidel, K. Antibacterial activity of a Tribolium castaneum defensin in an in vitro infection model of Streptococcus pneumoniae. Virulence 2019, 10, 902–909. [Google Scholar] [CrossRef]
- Robles-Fort, A.; García-Robles, I.; Fernando, W.; Hoskin, D.W.; Rausell, C.; Real, M.D. Dual antimicrobial and antiproliferative activity of TcPaSK peptide derived from a Tribolium castaneum insect defensin. Microorganisms 2021, 9, 222. [Google Scholar] [CrossRef] [PubMed]
- Toro Segovia, L.J.; Téllez Ramírez, G.A.; Henao Arias, D.C.; Rivera Duran, J.D.; Bedoya, J.P.; Castaño Osorio, J.C. Identification and characterization of novel cecropins from the Oxysternon conspicillatum neotropic dung beetle. PLoS ONE 2017, 12, e0187914. [Google Scholar] [CrossRef]
- Romoli, O.; Mukherjee, S.; Mohid, S.A.; Dutta, A.; Montali, A.; Franzolin, E.; Brady, D.; Zito, F.; Bergantino, E.; Rampazzo, C. Enhanced silkworm cecropin B antimicrobial activity against Pseudomonas aeruginosa from single amino acid variation. ACS Infect. Dis. 2019, 5, 1200–1213. [Google Scholar] [CrossRef]
- Araki, M.; Kurihara, M.; Kinoshita, S.; Awane, R.; Sato, T.; Ohkawa, Y.; Inoue, Y.H. Anti-tumour effects of antimicrobial peptides, components of the innate immune system, against haematopoietic tumours in Drosophila mxc mutants. Dis. Model. Mech. 2019, 12, dmm037721. [Google Scholar] [CrossRef]
- Wu, Y.-L.; Xia, L.-J.; Li, J.-Y.; Zhang, F.-C. CecropinXJ inhibits the proliferation of human gastric cancer BGC823 cells and induces cell death in vitro and in vivo. Int. J. Oncol. 2015, 46, 2181–2193. [Google Scholar] [CrossRef]
- Xia, L.; Wu, Y.; Kang, S.; Ma, J.; Yang, J.; Zhang, F. CecropinXJ, a silkworm antimicrobial peptide, induces cytoskeleton disruption in esophageal carcinoma cells. Acta Biochim. Biophys. Sin. 2014, 46, 867–876. [Google Scholar] [CrossRef]
- Suttmann, H.; Retz, M.; Paulsen, F.; Harder, J.; Zwergel, U.; Kamradt, J.; Wullich, B.; Unteregger, G.; Stöckle, M.; Lehmann, J. Antimicrobial peptides of the Cecropin-family show potent antitumor activity against bladder cancer cells. BMC Urol. 2008, 8, 5. [Google Scholar] [CrossRef]
- Rady, I.; Siddiqui, I.A.; Rady, M.; Mukhtar, H. Melittin, a major peptide component of bee venom, and its conjugates in cancer therapy. Cancer Lett. 2017, 402, 16–31. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.Y.; Lee, M.K.; Kim, K.L.; HAHM, K.S. Structure-antitumor and hemolytic activity relationships of synthetic peptides derived from cecropin A-magainin 2 and cecropin A-melittin hybrid peptides. J. Pept. Res. 1997, 50, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Garrido, P.; Cárdenas-Guerra, R.E.; Martínez, I.; Poggio, S.; Rodríguez-Hernández, K.; Rivera-Santiago, L.; Ortega-López, J.; Sánchez-Esquivel, S.; Espinoza, B. Differential activity on trypanosomatid parasites of a novel recombinant defensin type 1 from the insect Triatoma (Meccus) pallidipennis. Insect Biochem. Mol. Biol. 2021, 139, 103673. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Cordero, J.J.; Lozano, J.M.; Cortés, J.; Delgado, G. Leishmanicidal activity of synthetic antimicrobial peptides in an infection model with human dendritic cells. Peptides 2011, 32, 683–690. [Google Scholar] [CrossRef]
- Hu, Y.; Aksoy, S. An antimicrobial peptide with trypanocidal activity characterized from Glossina morsitans morsitans. Insect Biochem. Mol. Biol. 2005, 35, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Tanphaichitr, N.; Srakaew, N.; Alonzi, R.; Kiattiburut, W.; Kongmanas, K.; Zhi, R.; Li, W.; Baker, M.; Wang, G.; Hickling, D. Potential use of antimicrobial peptides as vaginal spermicides/microbicides. Pharmaceuticals 2016, 9, 13. [Google Scholar] [CrossRef]
- Alengebawy, A.; Abdelkhalek, S.T.; Qureshi, S.R.; Wang, M.-Q. Heavy metals and pesticides toxicity in agricultural soil and plants: Ecological risks and human health implications. Toxics 2021, 9, 42. [Google Scholar] [CrossRef]
- Sharma, A.; Kumar, V.; Shahzad, B.; Tanveer, M.; Sidhu, G.P.S.; Handa, N.; Kohli, S.K.; Yadav, P.; Bali, A.S.; Parihar, R.D. Worldwide pesticide usage and its impacts on ecosystem. SN Appl. Sci. 2019, 1, 1446. [Google Scholar] [CrossRef]
- Ab Rahman, S.F.S.; Singh, E.; Pieterse, C.M.; Schenk, P.M. Emerging microbial biocontrol strategies for plant pathogens. Plant Sci. 2018, 267, 102–111. [Google Scholar] [CrossRef]
- Banzet, N.; Latorse, M.-P.; Bulet, P.; François, E.; Derpierre, C.; Dubald, M. Expression of insect cystein-rich antifungal peptides in transgenic tobacco enhances resistance to a fungal disease. Plant Sci. 2002, 162, 995–1006. [Google Scholar] [CrossRef]
- Rahnamaeian, M.; Langen, G.; Imani, J.; Khalifa, W.; Altincicek, B.; Von Wettstein, D.; Kogel, K.-H.; Vilcinskas, A. Insect peptide Metchnikowin confers on barley a selective capacity for resistance to fungal ascomycetes pathogens. J. Exp. Bot. 2009, 60, 4105–4114. [Google Scholar] [CrossRef] [PubMed]
- Boscariol, R.L.; Monteiro, M.; Takahashi, E.K.; Chabregas, S.M.; Vieira, M.L.C.; Vieira, L.G.; Pereira, L.F.; de AA Mourão Filho, F.; Cardoso, S.C.; Christiano, R.S. Attacin A gene from Tricloplusia ni reduces susceptibility to Xanthomonas axonopodis pv. citri in transgenic Citrus sinensis ‘Hamlin’. J. Am. Soc. Hortic. Sci. 2006, 131, 530–536. [Google Scholar] [CrossRef]
- Zakharchenko, N.; Buryanov, Y.I.; Lebedeva, A.; Pigoleva, S.; Vetoshkina, D.; Loktyushov, E.; Chepurnova, M.; Kreslavski, V.; Kosobryukhov, A. Physiological features of rapeseed plants expressing the gene for an antimicrobial peptide cecropin P1. Russ. J. Plant Physiol. 2013, 60, 411–419. [Google Scholar] [CrossRef]
- Shi, W.; Li, C.; Li, M.; Zong, X.; Han, D.; Chen, Y. Antimicrobial peptide melittin against Xanthomonas oryzae pv. oryzae, the bacterial leaf blight pathogen in rice. Appl. Microbiol. Biotechnol. 2016, 100, 5059–5067. [Google Scholar]
- Kamysz, W.; Królicka, A.; Bogucka, K.; Ossowski, T.; Lukasiak, J.; Lojkowska, E. Antibacterial activity of synthetic peptides against plant pathogenic Pectobacterium species. J. Phytopathol. 2005, 153, 313–317. [Google Scholar] [CrossRef]
- Józefiak, A.; Engberg, R.M. Insect proteins as a potential source of antimicrobial peptides in livestock production. A review. J. Anim. Feed Sci. 2017, 26, 87–99. [Google Scholar] [CrossRef]
- Wen, L.-F.; He, J.-G. Dose–response effects of an antimicrobial peptide, a cecropin hybrid, on growth performance, nutrient utilisation, bacterial counts in the digesta and intestinal morphology in broilers. Br. J. Nutr 2012, 108, 1756–1763. [Google Scholar] [CrossRef]
- Dai, J.; Zheng, J.; Ou, W.; Xu, W.; Ai, Q.; Zhang, W.; Niu, J.; Zhang, Y.; Mai, K. The effect of dietary cecropin AD on intestinal health, immune response and disease resistance of juvenile turbot (Scophthalmus maximus L.). Fish Shellfish Immunol. 2020, 100, 117–125. [Google Scholar] [CrossRef]
- Lin, X.; Chen, W.; Lin, S.; Luo, L. Effects of dietary cecropin on growth, non-specific immunity and disease resistance of tilapia (Oreochromis niloticus × O. aureus). Aquac. Res. 2015, 46, 2999–3007. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, F.; Huang, Z.; Liu, H.; Xie, C.; Zhang, J.; Thacker, P.A.; Qiao, S. Effects of the antimicrobial peptide cecropin AD on performance and intestinal health in weaned piglets challenged with Escherichia coli. Peptides 2012, 35, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhan, Y.; Ma, W.; Zhu, Y.; Wang, Z. Effects of Antimicrobial peptides on egg production, egg quality and caecal microbiota of hens during the late laying period. Anim. Sci. J. 2020, 91, e13387. [Google Scholar] [CrossRef] [PubMed]
- Keymanesh, K.; Soltani, S.; Sardari, S. Application of antimicrobial peptides in agriculture and food industry. World J. Microbiol. Biotechnol. 2009, 25, 933–944. [Google Scholar] [CrossRef]
- León Madrazo, A.; Segura Campos, M.R. Review of antimicrobial peptides as promoters of food safety: Limitations and possibilities within the food industry. J. Food Saf. 2020, 40, e12854. [Google Scholar] [CrossRef]
- Hwanhlem, N.; Ivanova, T.; Haertlé, T.; Jaffrès, E.; Dousset, X. Inhibition of food-spoilage and foodborne pathogenic bacteria by a nisin Z-producing Lactococcus lactis subsp. lactis KT2W2L. LWT Food Sci. Technol. 2017, 82, 170–175. [Google Scholar] [CrossRef]
- Hwang, D.; Lee, S.H.; Goo, T.-W.; Yun, E.-Y. Potential of Antimicrobial Peptide-Overexpressed Tenebrio molitor larvae extract as a natural preservative for korean traditional sauces. Insects 2022, 13, 381. [Google Scholar] [CrossRef]
- Hou, L.; Shi, Y.; Zhai, P.; Le, G. Inhibition of foodborne pathogens by Hf-1, a novel antibacterial peptide from the larvae of the housefly (Musca domestica) in medium and orange juice. Food Control 2007, 18, 1350–1357. [Google Scholar] [CrossRef]
- Shen, P.; Ding, K.; Wang, L.; Tian, J.; Huang, X.; Zhang, M.; Dang, X. In vitro and in vivo antimicrobial activity of antimicrobial peptide Jelleine-I against foodborne pathogen Listeria monocytogenes. Int. J. Food Microbiol. 2023, 387, 110050. [Google Scholar] [CrossRef]
- Dolezal, T.; Krejcova, G.; Bajgar, A.; Nedbalova, P.; Strasser, P. Molecular regulations of metabolism during immune response in insects. Insect Biochem. Mol. Biol. 2019, 109, 31–42. [Google Scholar] [CrossRef]
- Clark, R.I.; Tan, S.W.; Péan, C.B.; Roostalu, U.; Vivancos, V.; Bronda, K.; Pilátová, M.; Fu, J.; Walker, D.W.; Berdeaux, R. MEF2 is an in vivo immune-metabolic switch. Cell 2013, 155, 435–447. [Google Scholar] [CrossRef]
- Grau, T.; Vilcinskas, A.; Joop, G. Sustainable farming of the mealworm Tenebrio molitor for the production of food and feed. Z. Für Nat. C 2017, 72, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Melis, R.; Braca, A.; Sanna, R.; Spada, S.; Mulas, G.; Fadda, M.L.; Sassu, M.M.; Serra, G.; Anedda, R. Metabolic response of yellow mealworm larvae to two alternative rearing substrates. Metabolomics 2019, 15, 113. [Google Scholar] [CrossRef] [PubMed]
- Ruschioni, S.; Loreto, N.; Foligni, R.; Mannozzi, C.; Raffaelli, N.; Zamporlini, F.; Pasquini, M.; Roncolini, A.; Cardinali, F.; Osimani, A. Addition of olive pomace to feeding substrate affects growth performance and nutritional value of mealworm (Tenebrio molitor L.) larvae. Foods 2020, 9, 317. [Google Scholar] [CrossRef] [PubMed]
- Spranghers, T.; Ottoboni, M.; Klootwijk, C.; Ovyn, A.; Deboosere, S.; De Meulenaer, B.; Michiels, J.; Eeckhout, M.; De Clercq, P.; De Smet, S. Nutritional composition of black soldier fly (Hermetia illucens) prepupae reared on different organic waste substrates. J. Sci. Food Agric. 2017, 97, 2594–2600. [Google Scholar] [CrossRef]
- Abisgold, J.; Simpson, S. The physiology of compensation by locusts for changes in dietary protein. J. Exp. Biol. 1987, 129, 329–346. [Google Scholar] [CrossRef]
- Krams, I.; Kecko, S.; Kangassalo, K.; Moore, F.R.; Jankevics, E.; Inashkina, I.; Krama, T.; Lietuvietis, V.; Meija, L.; Rantala, M.J. Effects of food quality on trade-offs among growth, immunity and survival in the greater wax moth Galleria Mellonella. Insect Sci. 2015, 22, 431–439. [Google Scholar] [CrossRef]
- Lee, K.P.; Cory, J.S.; Wilson, K.; Raubenheimer, D.; Simpson, S.J. Flexible diet choice offsets protein costs of pathogen resistance in a caterpillar. Proc. R. Soc. B-Biol. Sci. 2006, 273, 823–829. [Google Scholar] [CrossRef]
- Povey, S.; Cotter, S.C.; Simpson, S.J.; Lee, K.P.; Wilson, K. Can the protein costs of bacterial resistance be offset by altered feeding behaviour? J. Anim. Ecol. 2009, 78, 437–446. [Google Scholar] [CrossRef]
- Catalán, T.P.; Barceló, M.; Niemeyer, H.M.; Kalergis, A.M.; Bozinovic, F. Pathogen-and diet-dependent foraging, nutritional and immune ecology in mealworms. Evol. Ecol. Res. 2011, 13, 711–723. [Google Scholar]
- Ponton, F.; Wilson, K.; Cotter, S.C.; Raubenheimer, D.; Simpson, S.J. Nutritional immunology: A multi-dimensional approach. PLoS Pathog. 2011, 7, e1002223. [Google Scholar] [CrossRef]
- Cotter, S.C.; Reavey, C.E.; Tummala, Y.; Randall, J.L.; Holdbrook, R.; Ponton, F.; Simpson, S.J.; Smith, J.A.; Wilson, K. Diet modulates the relationship between immune gene expression and functional immune responses. Insect Biochem. Mol. Biol. 2019, 109, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Unckless, R.L.; Rottschaefer, S.M.; Lazzaro, B.P. The complex contributions of genetics and nutrition to immunity in Drosophila melanogaster. PLoS Genet. 2015, 11, e1005030. [Google Scholar] [CrossRef] [PubMed]
- Srygley, R.; Jaronski, S. Protein deficiency lowers resistance of Mormon crickets to the pathogenic fungus Beauveria bassiana. J. Insect Physiol. 2018, 105, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Hassan, B.; Siddiqui, J.A.; Xu, Y. Vertically transmitted gut bacteria and nutrition influence the immunity and fitness of Bactrocera dorsalis larvae. Front Microbiol. 2020, 11, 596352. [Google Scholar] [CrossRef]
- Danihlík, J.; Škrabišová, M.; Lenobel, R.; Šebela, M.; Omar, E.; Petřivalský, M.; Crailsheim, K.; Brodschneider, R. Does the pollen diet influence the production and expression of antimicrobial peptides in individual honey bees? Insects 2018, 9, 79. [Google Scholar] [CrossRef]
- Choi, W.H.; Choi, H.J.; Goo, T.W.; Quan, F.S. Novel antibacterial peptides induced by probiotics in Hermetia illucens (Diptera: Stratiomyidae) larvae. Entomol. Res. 2018, 48, 237–247. [Google Scholar] [CrossRef]
- Bruno, D.; Montali, A.; Mastore, M.; Brivio, M.F.; Mohamed, A.; Tian, L.; Grimaldi, A.; Casartelli, M.; Tettamanti, G. Insights into the immune response of the black soldier fly larvae to bacteria. Front. Immunol. 2021, 12, 4866. [Google Scholar] [CrossRef]
- Jing, X.; Vogel, H.; Grebenok, R.J.; Zhu-Salzman, K.; Behmer, S.T. Dietary sterols/steroids and the generalist caterpillar Helicoverpa zea: Physiology, biochemistry and midgut gene expression. Insect Biochem. Mol. Biol. 2012, 42, 835–845. [Google Scholar] [CrossRef]
- Engel, P.; Moran, N.A. The gut microbiota of insects–diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef]
- Douglas, A.E. Multiorganismal insects: Diversity and function of resident microorganisms. Annu. Rev. Entomol. 2015, 60, 17–34. [Google Scholar] [CrossRef]
- Moret, Y.; Schmid-Hempel, P. Survival for immunity: The price of immune system activation for bumblebee workers. Science 2000, 290, 1166–1168. [Google Scholar] [CrossRef] [PubMed]
- Ponton, F.; Wilson, K.; Holmes, A.J.; Cotter, S.C.; Raubenheimer, D.; Simpson, S.J. Integrating nutrition and immunology: A new frontier. J. Insect Physiol. 2013, 59, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Di Angelo, J.R.; Bland, M.L.; Bambina, S.; Cherry, S.; Birnbaum, M.J. The immune response attenuates growth and nutrient storage in Drosophila by reducing insulin signaling. PNAS 2009, 106, 20853–20858. [Google Scholar] [CrossRef] [PubMed]
- Becker, T.; Loch, G.; Beyer, M.; Zinke, I.; Aschenbrenner, A.C.; Carrera, P.; Inhester, T.; Schultze, J.L.; Hoch, M. FOXO-dependent regulation of innate immune homeostasis. Nature 2010, 463, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Gedeon, T.; Bokes, P. Delayed protein synthesis reduces the correlation between mRNA and protein fluctuations. Biophys. J. 2012, 103, 377–385. [Google Scholar] [CrossRef]
- Jovanovic, M.; Rooney, M.S.; Mertins, P.; Przybylski, D.; Chevrier, N.; Satija, R.; Rodriguez, E.H.; Fields, A.P.; Schwartz, S.; Raychowdhury, R. Dynamic profiling of the protein life cycle in response to pathogens. Science 2015, 347, 1259038. [Google Scholar] [CrossRef]
- Hinnebusch, A.G.; Natarajan, K. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell 2002, 1, 22–32. [Google Scholar] [CrossRef]
- Brackley, C.A.; Romano, M.C.; Thiel, M. The dynamics of supply and demand in mRNA translation. PLoS Comput. Biol. 2011, 7, e1002203. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dho, M.; Candian, V.; Tedeschi, R. Insect Antimicrobial Peptides: Advancements, Enhancements and New Challenges. Antibiotics 2023, 12, 952. https://doi.org/10.3390/antibiotics12060952
Dho M, Candian V, Tedeschi R. Insect Antimicrobial Peptides: Advancements, Enhancements and New Challenges. Antibiotics. 2023; 12(6):952. https://doi.org/10.3390/antibiotics12060952
Chicago/Turabian StyleDho, Matteo, Valentina Candian, and Rosemarie Tedeschi. 2023. "Insect Antimicrobial Peptides: Advancements, Enhancements and New Challenges" Antibiotics 12, no. 6: 952. https://doi.org/10.3390/antibiotics12060952
APA StyleDho, M., Candian, V., & Tedeschi, R. (2023). Insect Antimicrobial Peptides: Advancements, Enhancements and New Challenges. Antibiotics, 12(6), 952. https://doi.org/10.3390/antibiotics12060952







