Antimicrobial Resistance of Staphylococcus aureus Isolated between 2017 and 2022 from Infections at a Tertiary Care Hospital in Romania
Abstract
1. Introduction
2. Results
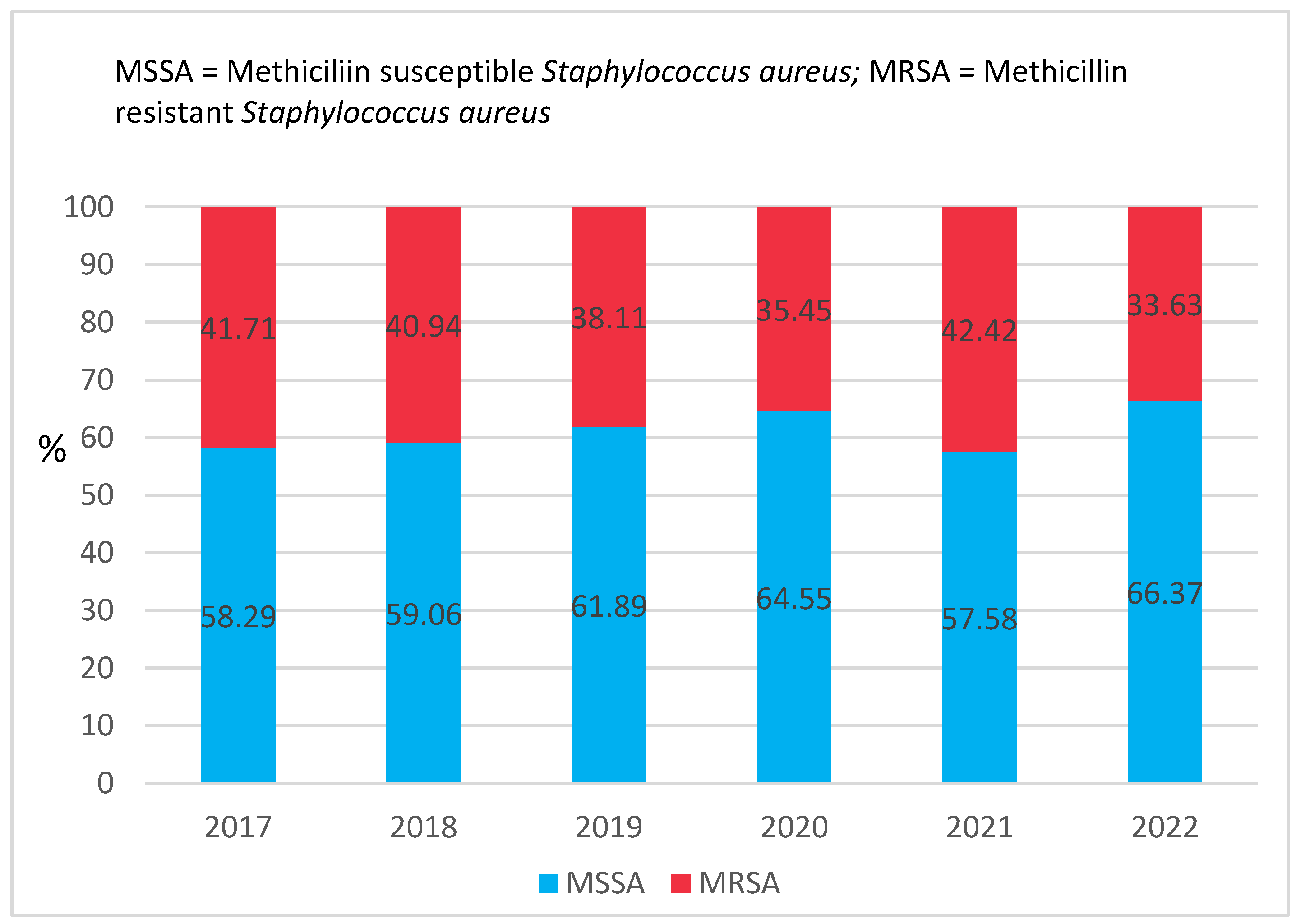
2.1. The Source of Staphylococcus Aureus Strains
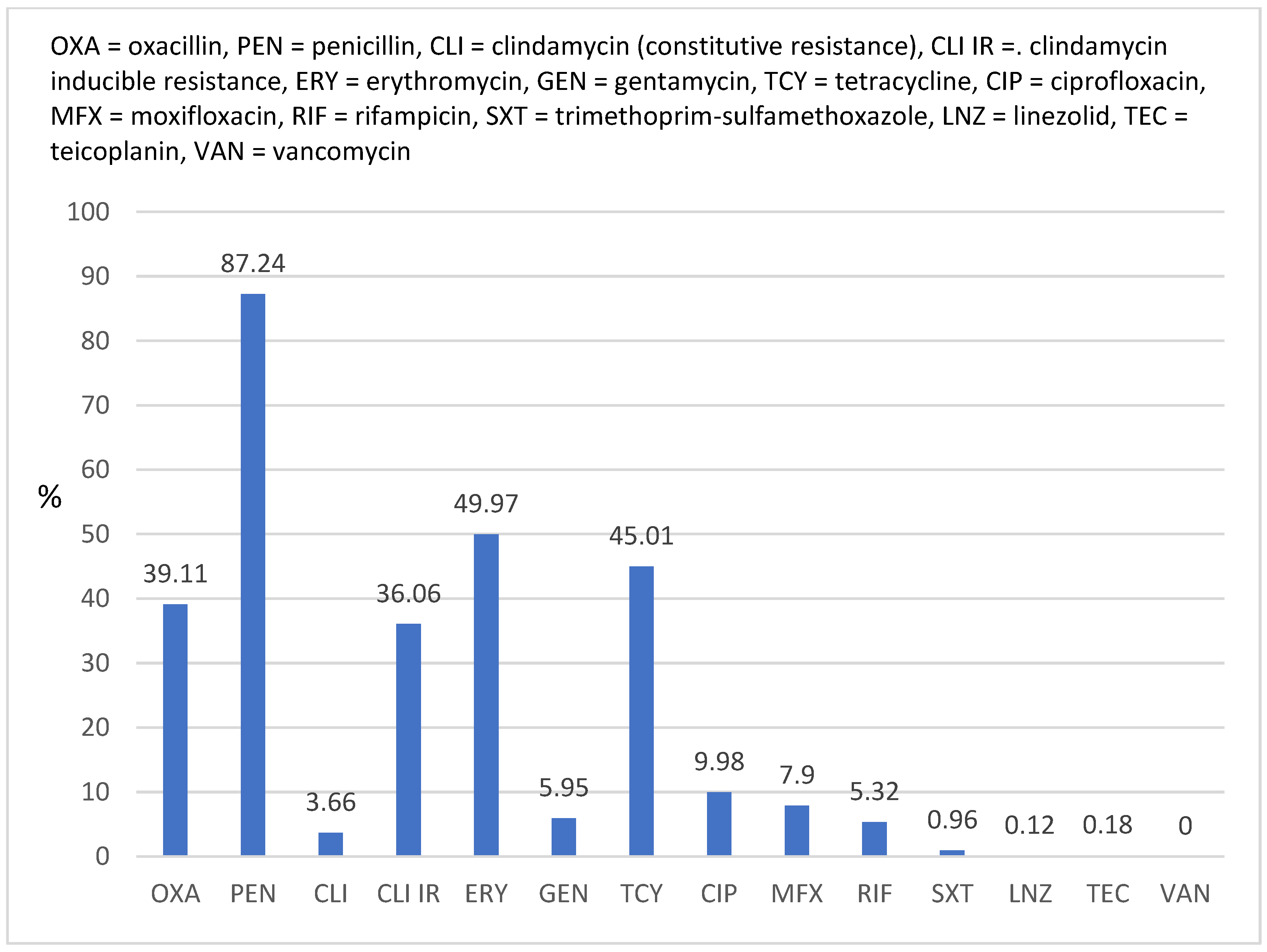
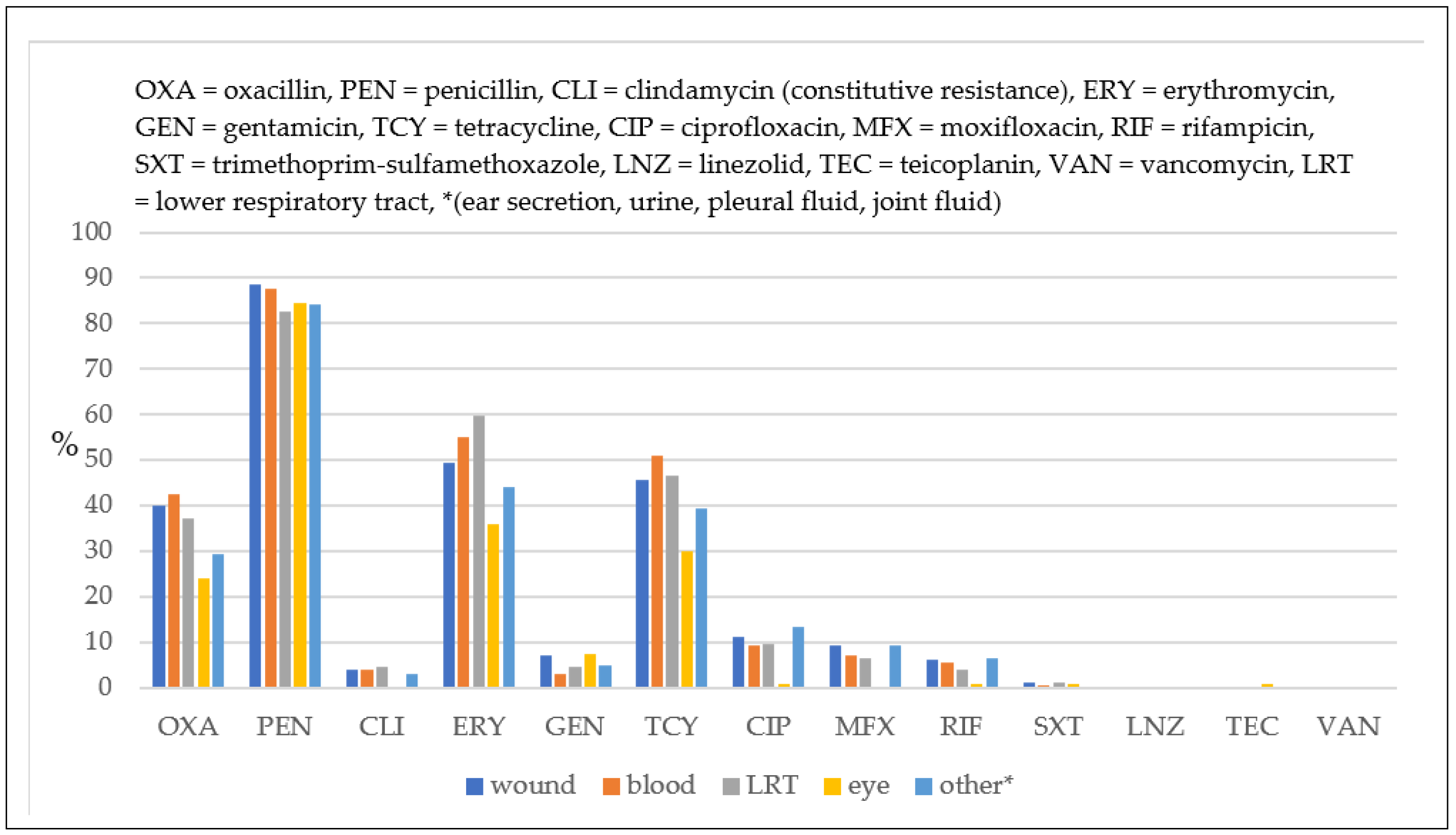
2.2. Antimicrobial Susceptibility
3. Discussion
4. Materials and Methods
4.1. Study Design and Study Setting
4.2. Bacterial Culture
4.3. Staphylococcus aureus Identification
4.4. Antimicrobial Susceptibility Testing
4.5. Quality Control
4.6. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviation
| MRSA | Methicillin Resistant Staphylococcus aureus |
| MSSA | Methicillin Susceptible Staphylococcus aureus |
| MIC | Minimum Inhibitory Concentration |
| AST | Antimicrobial Susceptibility Testing |
| PBP | Penicillin Binding Protein |
| EUCAST | European Committee on Antimicrobial Susceptibility Testing |
References
- Chambers, H.F.; DeLeo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef]
- Tsouklidis, N.; Kumar, R.; Heindl, S.E.; Soni, R.; Khan, S. Understanding the Fight Against Resistance: Hospital-Acquired Methicillin-Resistant Staphylococcus aureus vs. Community-Acquired Methicillin-Resistant Staphylococcus aureus. Cureus 2020, 12, e8867. [Google Scholar] [CrossRef]
- Onyeka, F.I.; Nwobodo, D.; Umenne, I.C.; Atada, E.E.; Ojukwu, C.A.; Aniekwe, M.A.; Philomena, J.J.; Ikem, J.C. Antibiotic Resistance Pattern of Staphylococcus aureus Isolated from Nostrils of Healthy Undergraduates of Madonna University Elele Campus, Rivers State, Nigeria. Microbes Infect. Dis. 2020, 2, 280–285. [Google Scholar] [CrossRef]
- Kluytmans, J.; van Belkum, A.; Verbrugh, H. Nasal carriage of Staphylococcus aureus: Epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 1997, 10, 505–520. [Google Scholar] [CrossRef]
- Saba, C.K.S.; Amenyona, J.K.; Kpordze, S.W. Prevalence and pattern of antibiotic resistance of Staphylococcus aureus isolated from door handles and other points of contact in public hospitals in Ghana. Antimicrob. Resist. Infect. Control. 2017, 6, 44. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, E.; Adlerberth, I.; Wold, A. Antibiotic resistance in Staphylococcus aureus colonising the intestines of Swedish infants. Clin. Microbiol. Infect. 2004, 10, 890–894. [Google Scholar] [CrossRef]
- Gurung, R.R.; Maharjan, P.; Chhetri, G.G. Antibiotic resistance pattern of Staphylococcus aureus with reference to MRSA isolates from pediatric patients. Futur. Sci. OA 2020, 6, FSO464. [Google Scholar] [CrossRef]
- CDC. COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022; Center for Disease Control and Prevention (CDC): Atlanta, GA, USA, 2022; pp. 1–42. [CrossRef]
- Akya, A.; Lorestani, R.C.; Shahveisi-Zadeh, J.; Bozorgomid, A. Antimicrobial Resistance of Staphylococcus aureus Isolated from Hospital Wastewater in Kermanshah, Iran. Risk Manag. Health Policy 2020, 13, 1035–1042. [Google Scholar] [CrossRef] [PubMed]
- Willis, J.A.; Cheburkanov, V.; Chen, S.; Soares, J.M.; Kassab, G.; Blanco, K.C.; Bagnato, V.S.; de Figueiredo, P.; Yakovlev, V.V. Breaking down antibiotic resistance in methicillin-resistant Staphylococcus aureus: Combining antimicrobial photodynamic and antibiotic treatments. Proc. Natl. Acad. Sci. USA 2022, 119, e2208378119. [Google Scholar] [CrossRef]
- Schito, G.C. The importance of the development of antibiotic resistance in Staphylococcus aureus. Clin. Microbiol. Infect. 2006, 12, 3–8. [Google Scholar] [CrossRef]
- Arenz, S.; Wilson, D. Blast from the Past: Reassessing Forgotten Translation Inhibitors, Antibiotic Selectivity, and Resistance Mechanisms to Aid Drug Development. Mol. Cell 2016, 61, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Deyno, S.; Toma, A.; Worku, M.; Bekele, M. Antimicrobial resistance profile of Staphylococcus aureus isolates isolated from ear discharges of patients at University of Hawassa comprehensive specialized hospital. BMC Pharmacol. Toxicol. 2017, 18, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Santosaningsih, D.; Santoso, S.; Budayanti, N.S.; Suata, K.; Lestari, E.S.; Wahjono, H.; Djamal, A.; Kuntaman, K.; van Belkum, A.; Laurens, M.; et al. Characterisation of clinical Staphylococcus aureus isolates harbouring mecA or Panton-Valentine leukocidin genes from four tertiary care hospitals in Indonesia. Trop. Med. Int. Health 2016, 21, 610–618. [Google Scholar] [CrossRef]
- Paterson, G.K.; Harrison, E.M.; Holmes, M.A. The emergence of mecC methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2013, 22, 42–47. [Google Scholar] [CrossRef]
- Mlynarczyk-Bonikowska, B.; Kowalewski, C.; Krolak-Ulinska, A.; Marusza, W. Molecular Mechanisms of Drug Resistance in Staphylococcus aureus. Int. J. Mol. Sci. 2022, 23, 8088. [Google Scholar] [CrossRef]
- Zeng, D.; Debabov, D.; Hartsell, T.L.; Cano, R.J.; Adams, S.; Schuyler, J.A.; McMillan, R.; Pace, J.L. Approved Glycopeptide Antibacterial Drugs: Mechanism of Action and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a026989. [Google Scholar] [CrossRef] [PubMed]
- Long, K.S.; Vester, B. Resistance to Linezolid Caused by Modifications at Its Binding Site on the Ribosome. Antimicrob. Agents Chemother. 2012, 56, 603–612. [Google Scholar] [CrossRef]
- Zhou, D.; Steitz, T.A.; Polikanov, Y.S.; Gagnon, M.G. Ribosome-Targeting Antibiotics: Modes of Action, Mechanisms of Resistance, and Implications for Drug Design. Annu. Rev. Biochem. 2018, 87, 451–478. [Google Scholar] [CrossRef]
- Bassetti, M.; Magnasco, L.; Vena, A.; Portunato, F.; Giacobbe, D.R. Methicillin-resistant Staphylococcus aureus lung infection in coronavirus disease 2019: How common? Curr. Opin. Infect. Dis. 2022, 35, 149–162. [Google Scholar] [CrossRef]
- Alshaikh, F.S.; Godman, B.; Sindi, O.N.; Seaton, R.A.; Kurdi, A. Prevalence of bacterial coinfection and patterns of antibiotics prescribing in patients with COVID-19: A systematic review and meta-analysis. PLoS ONE 2022, 17, e0272375. [Google Scholar] [CrossRef]
- Arientová, S.; Jícha, Z.; Beran, O.; Holub, M. Decreased quality of care for Staphylococcus aureus bacteremia during the COVID-19 pandemic. BMC Infect. Dis. 2022, 22, 1–5. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, R.F. The interface between COVID-19 and bacterial healthcare-associated infections. Clin. Microbiol. Infect. 2021, 27, 1772–1776. [Google Scholar] [CrossRef]
- Adalbert, J.R.; Varshney, K.; Tobin, R.; Pajaro, R. Clinical outcomes in patients co-infected with COVID-19 and Staphylococcus aureus: A scoping review. BMC Infect. Dis. 2021, 21, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Habib, G.; Mahmood, K.; Gul, H.; Tariq, M.; Ain, Q.U.; Hayat, A.; Rehman, M.U. Pathophysiology of Methicillin-Resistant Staphylococcus aureus Superinfection in COVID-19 Patients. Pathophysiology 2022, 29, 405–413. [Google Scholar] [CrossRef]
- CDC. Antibiotic Resistance Threats in the United States; Center for Disease Control and Prevention (CDC): Atlanta, GA, USA, 2019; pp. 1–140. [CrossRef]
- European Centre for Disease Prevention and Control. Surveillance Atlas of Infectious Diseases. Available online: http://atlas.ecdc.europa.eu/public/index.aspx (accessed on 4 April 2023).
- Dorneanu, O.; Miftode, E.; Vremeră, T.; Năstase, E.; Filip, O.; Luca, V. Prevalence and characteristics of Staphylococcus aureus isolated from infections in Northeast Romania. J. Prev. Med. 2006, 14, 66–70. [Google Scholar]
- Szekely, E.; Lőrinczi, L.; Bilca, D.; Fodor, E.; Soki, J.; Sabau, M. Incidence, antibiotic resistance and clonal relations of MRSA strains isolated from a Romanian university hospital. Acta Microbiol. et Immunol. Hung. 2008, 55, 1–13. [Google Scholar] [CrossRef]
- Ionescu, R.; Mediavilla, J.R.; Chen, L.; Grigorescu, D.O.; Idomir, M.; Kreiswirth, B.N.; Roberts, R.B. Molecular Characterization and Antibiotic Susceptibility of Staphylococcus aureus from a Multidisciplinary Hospital in Romania. Microb. Drug Resist. 2010, 16, 263–272. [Google Scholar] [CrossRef]
- Nica, M.; Biolan, T.; Dascalu, A.; Mozes, E.; Toderan, A.; Calistru, P.; Ceauşu, E. Bacterial strains isolated from systemic infections and reported for evaluation and antibiotic resistance surveillance by the “Dr. Victor Babes” Clinical Hospital for Infectious and Tropical Diseases, Bucharest. Bacteriol. Virusol. Parazitol. Epidemiol. 2010, 55, 161–168. [Google Scholar]
- Dorobăţ, O.M.; Bădicuţ, I.; Tălăpan, D.; Tenea, C.; Rafila, A. Antibiotic resistance of Gram-positive cocci isolated in 2008. Bacteriol. Virusol. Parazitol. Epidemiol. 2011, 55, 83–92. [Google Scholar]
- Nastase, E.; Dorneanu, O.; Vremera, T.; Logigan, C.; Miftode, E.; Dorobăţ, C.M. MecA and pvl genes detection in Staphylococcus aureus strains isolated from lower respiratory tract infections. Rev. Med. Chir. Soc. Med. Nat. Iasi. 2010, 114, 1162–1168. [Google Scholar]
- De Santis, V.; Corona, A.; Vitale, D.; Nencini, C.; Potalivo, A.; Prete, A.; Zani, G.; Malfatto, A.; Tritapepe, L.; Taddei, S.; et al. Bacterial infections in critically ill patients with SARS-2-COVID-19 infection: Results of a prospective observational multicenter study. Infection 2021, 50, 139–148. [Google Scholar] [CrossRef]
- Gardete, S.; Tomasz, A. Mechanisms of vancomycin resistance in Staphylococcus aureus. J. Clin. Investig. 2014, 124, 2836–2840. [Google Scholar] [CrossRef]
- Foster, T.J. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 2017, 41, 430–449. [Google Scholar] [CrossRef]
- Bakthavatchalam, Y.D.; Babu, P.; Munusamy, E.; Dwarakanathan, H.T.; Rupali, P.; Zervos, M.; Victor, P.J.; Veeraraghavan, B. Genomic insights on heterogeneous resistance to vancomycin and teicoplanin in Methicillin-resistant Staphylococcus aureus: A first report from South India. PLoS ONE 2019, 14, e0227009. [Google Scholar] [CrossRef] [PubMed]
- Cepeda, J.; Hayman, S.; Whitehouse, T.; Kibbler, C.C.; Livermore, D.; Singer, M.; Wilson, A.P.R. Teicoplanin resistance in methicillin-resistant Staphylococcus aureus in an intensive care unit. J. Antimicrob. Chemother. 2003, 52, 533–534. [Google Scholar] [CrossRef]
- McCallum, N.; Karauzum, H.; Getzmann, R.; Bischoff, M.; Majcherczyk, P.; Berger-Bächi, B.; Landmann, R. In Vivo Survival of Teicoplanin-Resistant Staphylococcus aureus and Fitness Cost of Teicoplanin Resistance. Antimicrob. Agents Chemother. 2006, 50, 2352–2360. [Google Scholar] [CrossRef] [PubMed]
- Vaudaux, P.; Francois, P.; Berger-Bächi, B.; Lew, D.P. In vivo emergence of subpopulations expressing teicoplanin or vancomycin resistance phenotypes in a glycopeptide-susceptible, methicillin-resistant strain of Staphylococcus aureus. J. Antimicrob. Chemother. 2001, 47, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Szymanek-Majchrzak, K.; Mlynarczyk, A.; Mlynarczyk, G. Characteristics of glycopeptide-resistant Staphylococcus aureus strains isolated from inpatients of three teaching hospitals in Warsaw, Poland. Antimicrob. Resist. Infect. Control. 2018, 7, 105. [Google Scholar] [CrossRef] [PubMed]
- Tsakris, A.; Papadimitriou, E.; Douboyas, J.; Stylianopoulou, F.; Manolis, E. Emergence of Vancomycin-Intermediate Staphylococcus aureus and S. sciuri, Greece. Emerg. Infect. Dis. 2002, 8, 536–537. [Google Scholar] [CrossRef]
- Cataño-Correa, J.C.; Cardona-Arias, J.A.; Mancilla, J.P.P.; García, M.T. Bacterial superinfection in adults with COVID-19 hospitalized in two clinics in Medellín-Colombia, 2020. PLoS ONE 2021, 16, e0254671. [Google Scholar] [CrossRef]
- Jian, Y.; Lv, H.; Liu, J.; Huang, Q.; Liu, Y.; Liu, Q.; Li, M. Dynamic Changes of Staphylococcus aureus Susceptibility to Vancomycin, Teicoplanin, and Linezolid in a Central Teaching Hospital in Shanghai, China, 2008–2018. Front. Microbiol. 2020, 11, 908. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.-Y.; Choy, B.N.K.; Zhou, M.-M.; Zhao, Z.-Y. Antibiotic Resistance Pattern of Staphylococcus aureus Isolated from Pediatrics with Ocular Infections: A 6-Year Hospital-Based Study in China. Front. Pediatr. 2021, 9, 728634. [Google Scholar] [CrossRef] [PubMed]
- The European Committee on Antimicrobial Susceptibility Testing. Available online: https://www.eucast.org/ast_of_bacteria (accessed on 23 November 2022).
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. EUCAST, versions 7.1–12.0, 2017–2022. Available online: https://www.eucast.org/ast_of_bacteria/previous_versions_of_documents/ (accessed on 21 February 2023).
- The European Committee on Antimicrobial Susceptibility Testing. EUCAST guidelines for detection of resistance mechanisms and specific resistance of clinical and/or epidemiological importance. EUCAST, version 2.0, July 2017. Available online: https://www.eucast.org/resistance_mechanisms/ (accessed on 9 May 2022).
- EUCAST. The European Committee on Antimicrobial Susceptibility Testing. Routine and Extended Internal Quality Control for MIC Determination and Disk Diffusion as Recommended by EUCAST, version 6.0–10.0, 2017–2022. Available online: http://www.eucast.org (accessed on 25 November 2022).


| Year | Wound Secretion (N) | Blood (N) | LRT a (N) | Eye Secretion (N) | Other b (N) | Total N (%) |
|---|---|---|---|---|---|---|
| 2017 | 245 | 68 | 21 | 45 | 31 | 410 (24.52) |
| 2018 | 237 | 75 | 23 | 33 | 13 | 381 (22.79) |
| 2019 | 276 | 87 | 22 | 31 | 30 | 446 (26.67) |
| 2020 | 54 | 27 | 15 | 6 | 8 | 110 (6.58) |
| 2021 | 25 | 17 | 43 | 4 | 10 | 99 (5.92) |
| 2022 | 129 | 39 | 33 | 15 | 10 | 226 (13.52) |
| Total N (%) | 966 (57.78) | 313 (18.72) | 157 (9.39) | 134 (8.01) | 102 (6.1) | 1672 (100) |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GC | VAN 0.25 | VAN 0.5 | VAN 1 | VAN 2 | VAN 4 | TEC 0.25 | TEC 0.5 | TEC 1 | TEC 2 | TEC 4 | TEC 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tălăpan, D.; Sandu, A.-M.; Rafila, A. Antimicrobial Resistance of Staphylococcus aureus Isolated between 2017 and 2022 from Infections at a Tertiary Care Hospital in Romania. Antibiotics 2023, 12, 974. https://doi.org/10.3390/antibiotics12060974
Tălăpan D, Sandu A-M, Rafila A. Antimicrobial Resistance of Staphylococcus aureus Isolated between 2017 and 2022 from Infections at a Tertiary Care Hospital in Romania. Antibiotics. 2023; 12(6):974. https://doi.org/10.3390/antibiotics12060974
Chicago/Turabian StyleTălăpan, Daniela, Andreea-Mihaela Sandu, and Alexandru Rafila. 2023. "Antimicrobial Resistance of Staphylococcus aureus Isolated between 2017 and 2022 from Infections at a Tertiary Care Hospital in Romania" Antibiotics 12, no. 6: 974. https://doi.org/10.3390/antibiotics12060974
APA StyleTălăpan, D., Sandu, A.-M., & Rafila, A. (2023). Antimicrobial Resistance of Staphylococcus aureus Isolated between 2017 and 2022 from Infections at a Tertiary Care Hospital in Romania. Antibiotics, 12(6), 974. https://doi.org/10.3390/antibiotics12060974






