Differential Antimicrobial Effect of Three-Sized Biogenic Silver Nanoparticles as Broad-Spectrum Antibacterial Agents against Plant Pathogens
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of the Cyanobacterial Strain and Pathogenic Bacteria
2.2. DNA Extraction
2.3. Polymerase Chain Reaction (PCR)
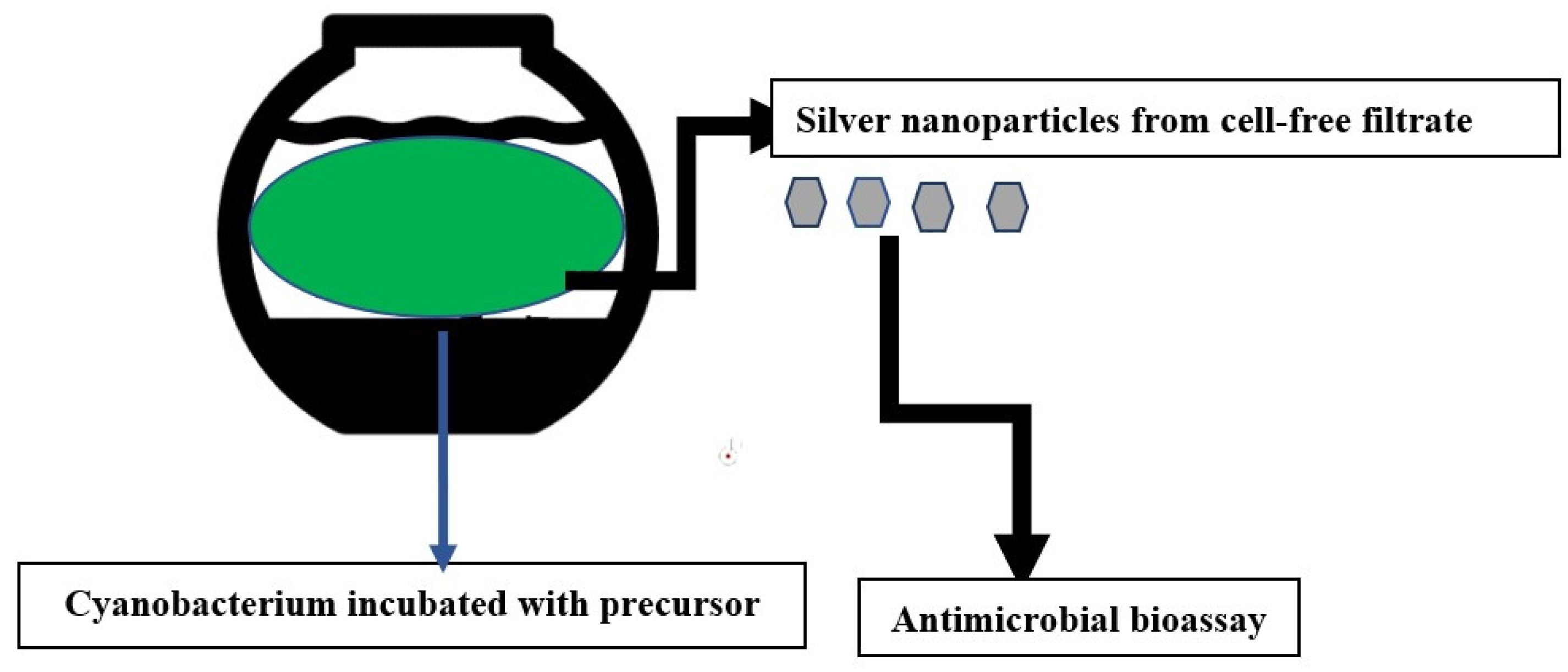
2.4. Biosynthesis of Silver Nanoparticles by Cyanobacterial Cultures
2.5. Characterization of Silver Nanoparticles
2.6. Scanning Electron Microscopy
2.7. Antimicrobial Bioassay
2.7.1. Antibacterial Action and Measurement of Minimum Inhibitory Concentration (MIC)
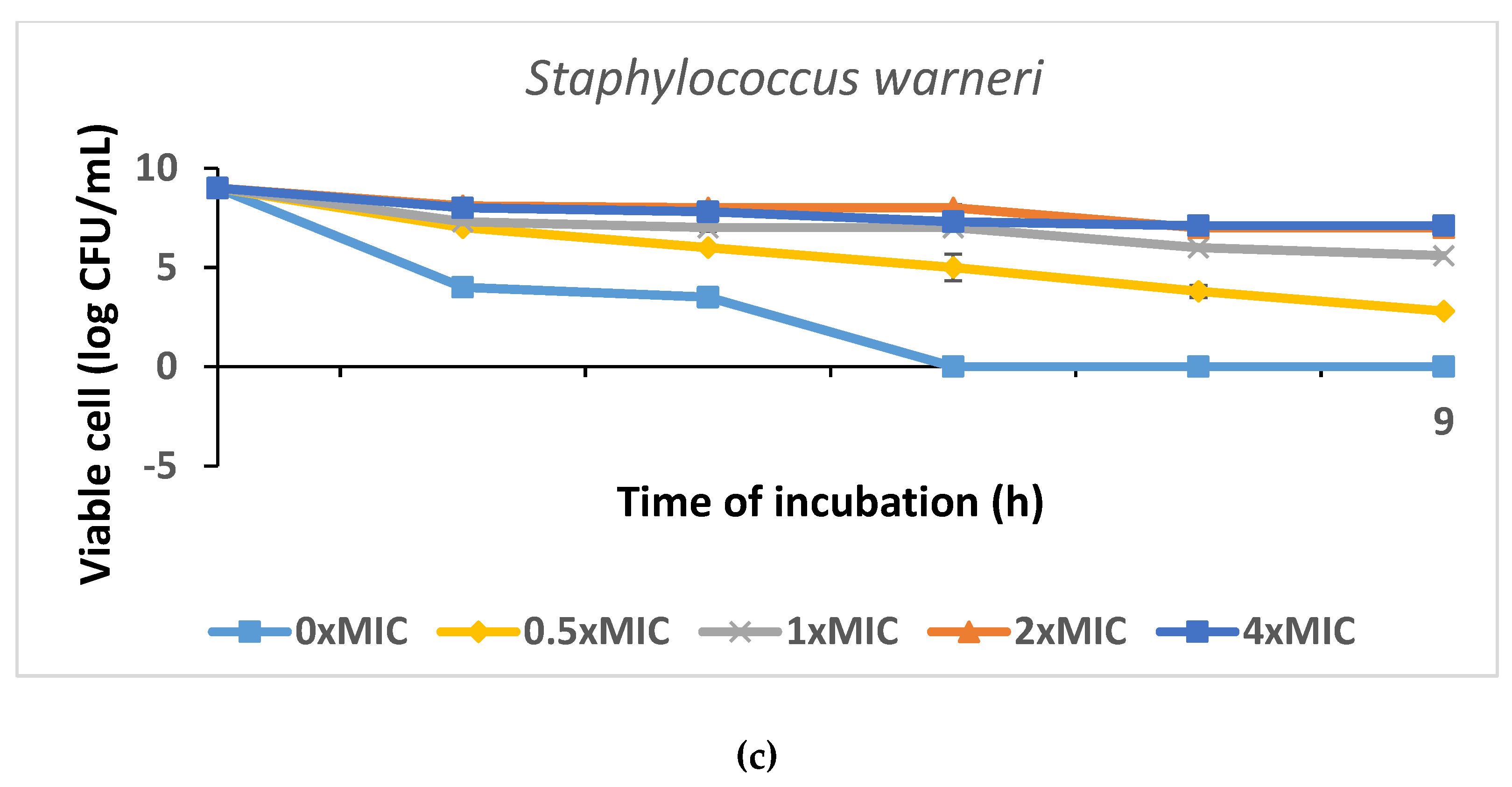
2.7.2. Time-Kill Test of AgNPs
3. Results
3.1. Molecular Characterization and Phylogenetic Inference of Pathogenic Bacteria
3.2. UV-Visible Spectroscopy
3.3. FTIR Spectroscopy
3.4. Scanning Electron Microscopy
3.5. The Antimicrobial Bioassay of Biogenic Nanosilver Samples against Plant Pathogenic Bacteria
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Abou El-Nour, K.M.; Eftaiha, A.A.; Al-Warthan, A.; Ammar, R.A. Synthesis and applications of silver nanoparticles. Arab. J. Chem. 2010, 3, 135–140. [Google Scholar] [CrossRef]
- Younis, N.S.; El Semary, N.A.; Mohamed, M.E. Silver nanoparticles green synthesis via cyanobacterium Phormidium sp.: Characterization, wound healing, antioxidant, antibacterial, and anti-Inflammatory activities. Eur. Rev. Med. Pharam. Sci. 2021, 25, 3083–3096. [Google Scholar]
- Mohamed, M.E.; El Semary, N.A.; Younis, N.S. Silver nanoparticle production by the cyanobacterium Cyanothece sp.: De Novo manipulation of nano-biosynthesis by phytohormones. Life 2022, 12, 139. [Google Scholar] [CrossRef] [PubMed]
- Lengke, M.F.; Fleet, M.E.; Southam, G. Biosynthesis of silver nanoparticles by filamentous cyanobacteria from a silver(I) nitrate complex. Langmuir 2007, 23, 2694–2699. [Google Scholar] [CrossRef]
- Patel, V.; Berthold, D.; Puranik, P.; Gantar, M. Screening of cyanobacteria and microalgae for their ability to synthesize silver nanoparticles with antibacterial activity. Biotechnol. Rep. 2015, 5, 112–119. [Google Scholar] [CrossRef]
- Parial, D.; Patra, H.K.; Roychoudhury, P.; Dasgupta, A.K.; Pal, R. Gold nanorod production by cyanobacteria—A green chemistry approach. J. Appl. Phycol. 2012, 24, 55–60. [Google Scholar] [CrossRef]
- Hamida, R.S.; Ali, M.A.; Redhwan, A.; Bin-Meferij, M.M. Cyanobacteria—A promising platform in green nanotechnology: A Review on nanoparticles fabrication and their prospective applications. Int. J. Nanomed. 2020, 15, 6033–6066. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.; Wen, W.J.; Xiong, X.; Hu, Y. Silver nanowire-carbon fiber cloth nanocomposites synthesized by UV curing adhesive for electrochemical point-of-use water disinfection. Chemosphere 2016, 154, 537–545. [Google Scholar] [CrossRef]
- Pal, S.; Yu, K.T.; Song, J.M. Does the antibacterial activity of silver nanoparticles depend on the shape of the nanoparticle? A study of the gram-negative bacterium Escherichia coli. Appl. Environ. Microbiol. 2007, 73, 1712–1720. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.; Lim, D.; Choi, J. Assessment of Size-Dependent Antimicrobial and Cytotoxic Properties of Silver Nanoparticles. Adv. Mater. Sci. Eng. 2014, 2014, 763807. [Google Scholar] [CrossRef]
- Oves, M.; Rauf, M.A.; Qari, H.A. Therapeutic Applications of Biogenic Silver Nanomaterial Synthesized from the Paper Flower of Bougainvillea glabra (Miami, Pink). Nanomaterials 2023, 13, 615. [Google Scholar] [CrossRef]
- More, P.R.; Pandit, S.; Filippis, A.; Franci, G.; Mijakovic, I.; Galdiero, M. Silver Nanoparticles: Bactericidal and Mechanistic Approach against Drug Resistant Pathogens. Microorganisms 2023, 11, 369. [Google Scholar] [CrossRef] [PubMed]
- Sampath, G.; Chen, Y.-Y.; Rameshkumar, N.; Krishnan, M.; Nagarajan, K.; Shyu, D.J.H. Biologically Synthesized Silver Nanoparticles and Their Diverse Applications. Nanomaterials 2022, 12, 3126. [Google Scholar] [CrossRef]
- Trzcińska-Wencel, J.; Wypij, M.; Rai, M.; Golińska, P. Biogenic nanosilver bearing antimicrobial and antibiofilm activities and its potential for application in agriculture and industry. Front. Microbiol. 2023, 14, 1125685. [Google Scholar] [CrossRef]
- Hamida, R.S.; Abdelmeguid, N.E.; Ali, M.A.; Bin-Meferij, M.M.; Khalil, M.I. Synthesis of Silver Nanoparticles Using a Novel Cyanobacteria Desertifilum sp. extract: Their Antibacterial and Cytotoxicity Effects. Int. J. Nanomed. 2020, 15, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979, 111, 1–61. [Google Scholar] [CrossRef]
- El Semary, N.; Bakir, E. Multidrug-resistant bacterial pathogens and public health: The antimicrobial effect of cyanobacterial-biosynthesized silver nanoparticles. Antibiotics 2022, 11, 1003. [Google Scholar] [CrossRef] [PubMed]
- Patra, J.K.; Baek, K.-H. Antibacterial Activity and Synergistic Antibacterial Potential of Biosynthesized Silver Nanoparticles against Foodborne Pathogenic Bacteria along with its Anticandidal and Antioxidant Effects. Front. Microbiol. 2017, 8, 167. [Google Scholar] [CrossRef] [PubMed]
- Kubo, I.; Fujita, K.; Kubo, A.; Nihei, K.; Ogura, T. Antibacterial activity of coriander volatile compounds against Salmonella choleraesuis. J. Agric. Food Chem. 2004, 52, 3329–3332. [Google Scholar] [CrossRef]
- Dong, Y.; Zhu, H.; Shen, Y.; Zhang, W.; Zhang, L. Antibacterial activity of silver nanoparticles of different particle size against Vibrio natriegens. PLoS ONE 2019, 14, e0222322. [Google Scholar] [CrossRef]
- Zhang, X.-F.; Liu, Z.-G.; Shen, W.; Gurunathan, S. Silver Nanoparticles: Synthesis, Characterization, Properties, Applications, and Therapeutic Approaches. Int. J. Mol. Sci. 2016, 17, 1534. [Google Scholar] [CrossRef]
- Mota, R.; Flores, C.; Tamagnini, P. Cyanobacterial Extracellular Polymeric Substances (EPS). In Polysaccharides of Microbial Origin; Oliveira, J.M., Radhouani, H., Reis, R.L., Eds.; Springer: Cham, Switzerland, 2021. [Google Scholar] [CrossRef]
- Gharibshahi, L.; Saion, E.; Gharibshahi, E.; Shaari, A.H.; Matori, K.A. Structural and Optical Properties of Ag Nanoparticles Synthesized by Thermal Treatment Method. Materials 2017, 10, 402. [Google Scholar] [CrossRef] [PubMed]
- Ferdous, Z.; Nemmar, A. Health impact of silver nanoparticles: A Review of the biodistribution and toxicity following various routes of exposure. Int. J. Mol. Sci. 2020, 21, 2375. [Google Scholar] [CrossRef] [PubMed]
- Durán, N.; Silveira, C.P.; Durán, M.; Martinez, D.S.T. Silver nanoparticle protein corona and toxicity: A mini-review. J. Nanobiotechnol. 2015, 13, 55. [Google Scholar] [CrossRef] [PubMed]
- Ali, D.M.; Sasikala, M.; Gunasekaran, M.; Thajuddin, N. Biosynthesis and characterization of silver nanoparticles using marine cyanobacterium, Oscillatoria willei NTDM01. Dig. J. Nanomater. Biostruct. 2011, 6, 385–390. [Google Scholar]
- Qing, Y.; Cheng, L.; Li, R.; Liu, G.; Zhang, Y.; Tang, X.; Wang, J.; Liu, H.; Qin, Y. Potential antibacterial mechanism of silver nanoparticles and the optimization of orthopedic implants by advanced modification technologies. Int. J. Nanomed. 2018, 13, 3311. [Google Scholar] [CrossRef]
- Lee, S.H.; Jun, B.-H. Silver nanoparticles: Synthesis and application for nanomedicine. Int. J. Mol. Sci. 2019, 20, 865. [Google Scholar] [CrossRef]
- Das, R.; Gang, S.; Nath, S.S. Preparation and antibacterial activity of silver nanoparticles. J. Biomater. Nanobiotechnol. 2011, 2, 472. [Google Scholar] [CrossRef]
- Shang, L.; Nienhaus, K.; Nienhaus, G.U. Engineered nanoparticles interacting with cells: Size matters. J. Nanobiotechnol. 2014, 12, 5. [Google Scholar] [CrossRef]
- Long, Y.-M.; Hu, L.-G.; Yan, X.-T.; Zhao, X.-C.; Zhou, Q.-F.; Cai, Y.; Jiang, G.-B. Surface ligand controls silver ion release of nanosilver and its antibacterial activity against Escherichia coli. Int. J. Nanomed. 2017, 12, 3193. [Google Scholar] [CrossRef]
- Baysal, G.; Demirci, C.; Özpinar, H. Proporties and synthesis of biosilver nanofilms for antimicrobial food packaging. Polymers 2023, 15, 689. [Google Scholar] [CrossRef] [PubMed]




| Bacteria | Inhibition Zone of Smallest AgNPs (cm) | Inhibition Zone of Medium AgNPs (cm) | Inhibition Zone of Largest AgNPs (cm) |
|---|---|---|---|
| Erwinia pyrifoliae | 1.0 ± 0.2 | 0.8 ± 0.1 | 0.6 ± 0.1 |
| Staphylococcus warneri | 1.7 ± 0.1 | 1.5 ± 0.3 | 1.0 ± 0.1 |
| Xanthomonas citri | 1.0 ± 0.1 | 0.9 ± 0.3 | 0.9 ± 0.1 |
| Incubation Time (Hours) | MIC (% w/v) | ||||
|---|---|---|---|---|---|
| 0 | 0.5 | 1 | 2 | 4 | |
| 0 | a 8 a ± 0.9 | a 8 a ± 0.87 | a 8 a ± 0.77 | a 8.1 a ± 0.91 | a 8 a ± 0.78 |
| 0.5 | b 7.5 a ± 0.6 | b 7.1 b ± 0.68 | b 6.3 c ± 0.35 | b 5.2 d ± 0.35 | b 3.1 e ± 0.32 |
| 1 | b 7.5 a ± 0.4 | b 7 b ± 0.56 | c 5.7 c ± 0.39 | c 4.6 d ± 0.69 | c 2.5 e ± 0.21 |
| 2 | b 7.2 a ± 0.4 | b 7 b ± 0.43 | c 5.7 c ± 0.21 | d 3.3 d ± 0.28 | d 0 e ± 0.0 |
| 4 | d 7 a ± 0.22 | c 6 b ± 0.2 | d 4.7 c ± 0.11 | e 2.8 d ± 0.12 | d 0 e ± 0.0 |
| 6 | d 7 a ± 0.35 | b 6 c ± 0.4 | e 4 c ± 0.22 | f 1.7 d ± 0.13 | d 0 e ± 0.0 |
| Incubation Time (Hours) | MIC (% w/v) | ||||
|---|---|---|---|---|---|
| 0 | 0.5 | 1 | 2 | 4 | |
| 0 | a 8 a ± 0.8 | a 8 a ± 0.91 | a 8 a ± 0.7 | a 8.1 a ± 0.81 | a 8 a ± 0.8 |
| 0.5 | b 7.4 a ± 0.5 | b 7 b ± 0.68 | b 6 c ± 0.5 | b 5 d ± 0.3 | b 3 e ± 0.1 |
| 1 | b 7.4 a ± 0.3 | b 6.7 b ± 0.56 | c 5.5 c ± 0.29 | c 4.4 d ± 0.9 | c 2.1 e ± 0.14 |
| 2 | c 7 a ± 0.2 | b 6.7 b ± 0.43 | c 5.5 c ± 0.21 | d 3.1 d ± 0.2 | d 0 e ± 0.0 |
| 4 | d 6 a ± 0.12 | c 6 b ± 0.2 | d 4.2 c ± 0.1 | e 2.2 d ± 0.1 | d 0 e ± 0.0 |
| 6 | d 6 a ± 0.44 | b 6 c ± 0.4 | e 4 c ± 0.12 | f 1.5 d ± 0.1 | d 0 e ± 0.0 |
| Incubation Time (Hours) | MIC (% w/v) | ||||
|---|---|---|---|---|---|
| 0 | 0.5 | 1 | 2 | 4 | |
| 0 | a 8 a ± 0.9 | a 8 a ± 0.8 | a 8 a ± 0.1 | a 8.1 a ± 0.1 | a 8 a ± 0.71 |
| 0.5 | b 7.1 a ± 0.4 | b 7 b ± 0.2 | b 6.1 c ± 0.4 | b 5 d ± 0.5 | b 4.1 e ± 0.4 |
| 1 | b 7.1 a ± 0.3 | b 6.6 b ± 0.4 | c 5.1 c ± 0.29 | c 4.5 d ± 0.9 | c 3.5 e ± 0.3 |
| 2 | c 7 a ± 0.3 | b 6.6 b ± 0.3 | c 5.1 c ± 0.22 | d 3.2 d ± 0.8 | d 0 e ± 0.0 |
| 4 | d 6.5 a ± 0.1 | c 5.5 b ± 0.1 | d 4.3 c ± 0.1 | e 2.4 d ± 0.3 | d 0 e ± 0.0 |
| 6 | d 6.5 a ± 0.5 | b 5.5 c ± 0.3 | e 4 c ± 0.3 | f 1.1 d ± 0.2 | d 0 e ± 0.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aldayel, M.F.; El Semary, N.; Adams, D.G. Differential Antimicrobial Effect of Three-Sized Biogenic Silver Nanoparticles as Broad-Spectrum Antibacterial Agents against Plant Pathogens. Antibiotics 2023, 12, 1114. https://doi.org/10.3390/antibiotics12071114
Aldayel MF, El Semary N, Adams DG. Differential Antimicrobial Effect of Three-Sized Biogenic Silver Nanoparticles as Broad-Spectrum Antibacterial Agents against Plant Pathogens. Antibiotics. 2023; 12(7):1114. https://doi.org/10.3390/antibiotics12071114
Chicago/Turabian StyleAldayel, Munirah F., Nermin El Semary, and David G. Adams. 2023. "Differential Antimicrobial Effect of Three-Sized Biogenic Silver Nanoparticles as Broad-Spectrum Antibacterial Agents against Plant Pathogens" Antibiotics 12, no. 7: 1114. https://doi.org/10.3390/antibiotics12071114
APA StyleAldayel, M. F., El Semary, N., & Adams, D. G. (2023). Differential Antimicrobial Effect of Three-Sized Biogenic Silver Nanoparticles as Broad-Spectrum Antibacterial Agents against Plant Pathogens. Antibiotics, 12(7), 1114. https://doi.org/10.3390/antibiotics12071114








