A Review on Enterocin DD14, the Leaderless Two-Peptide Bacteriocin with Multiple Biological Functions and Unusual Transport Pathway
Abstract
:1. Introduction
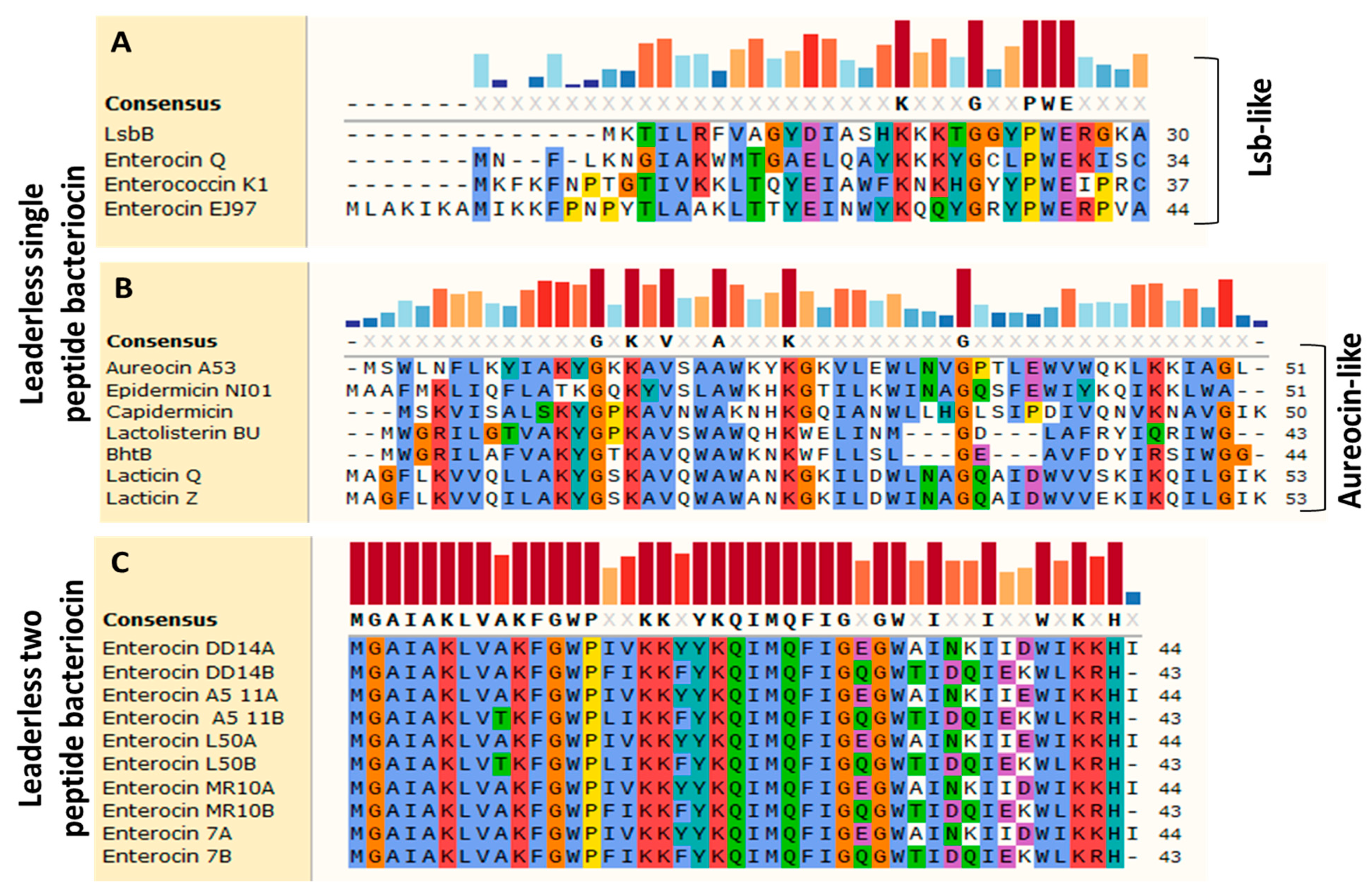
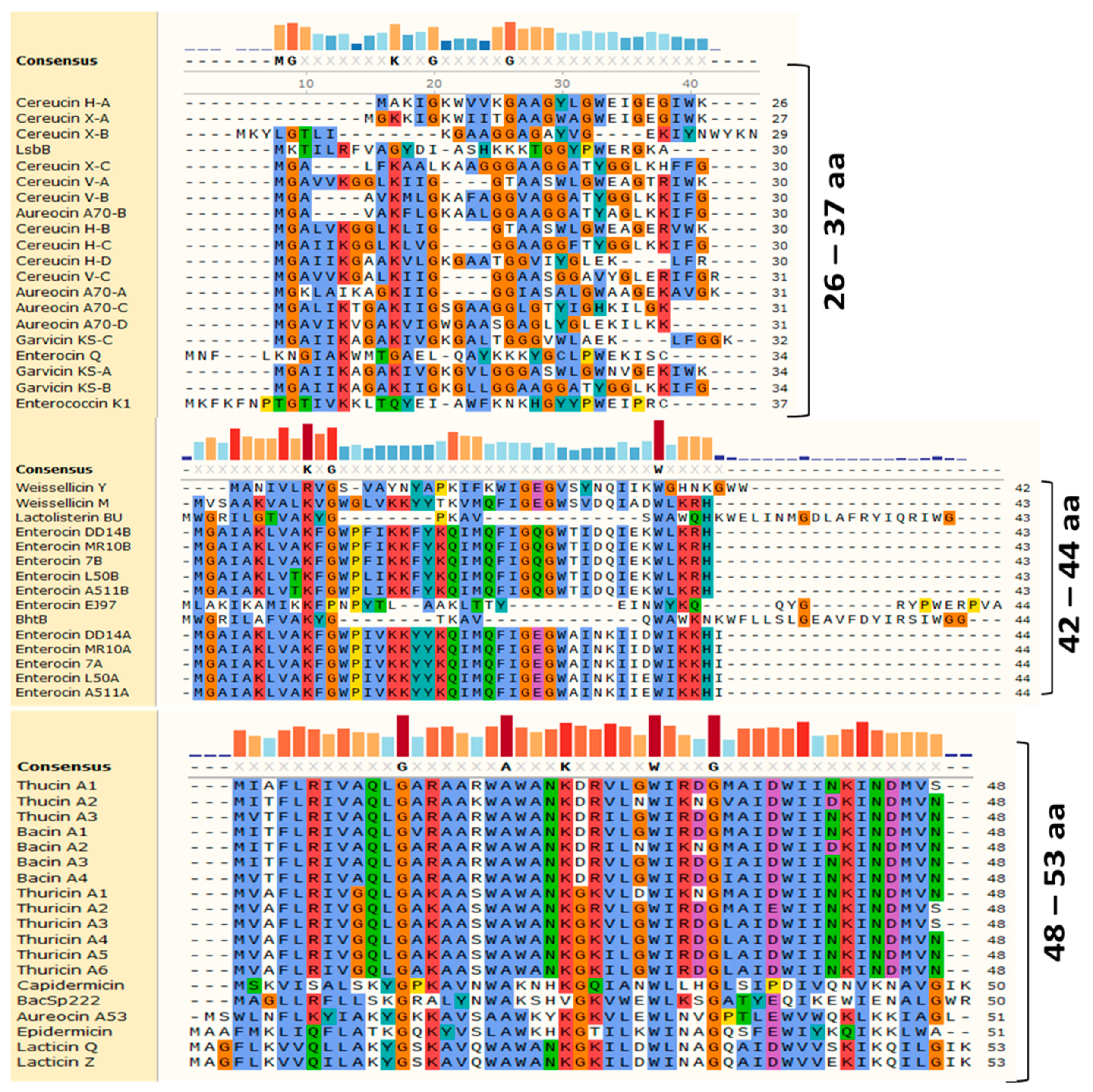
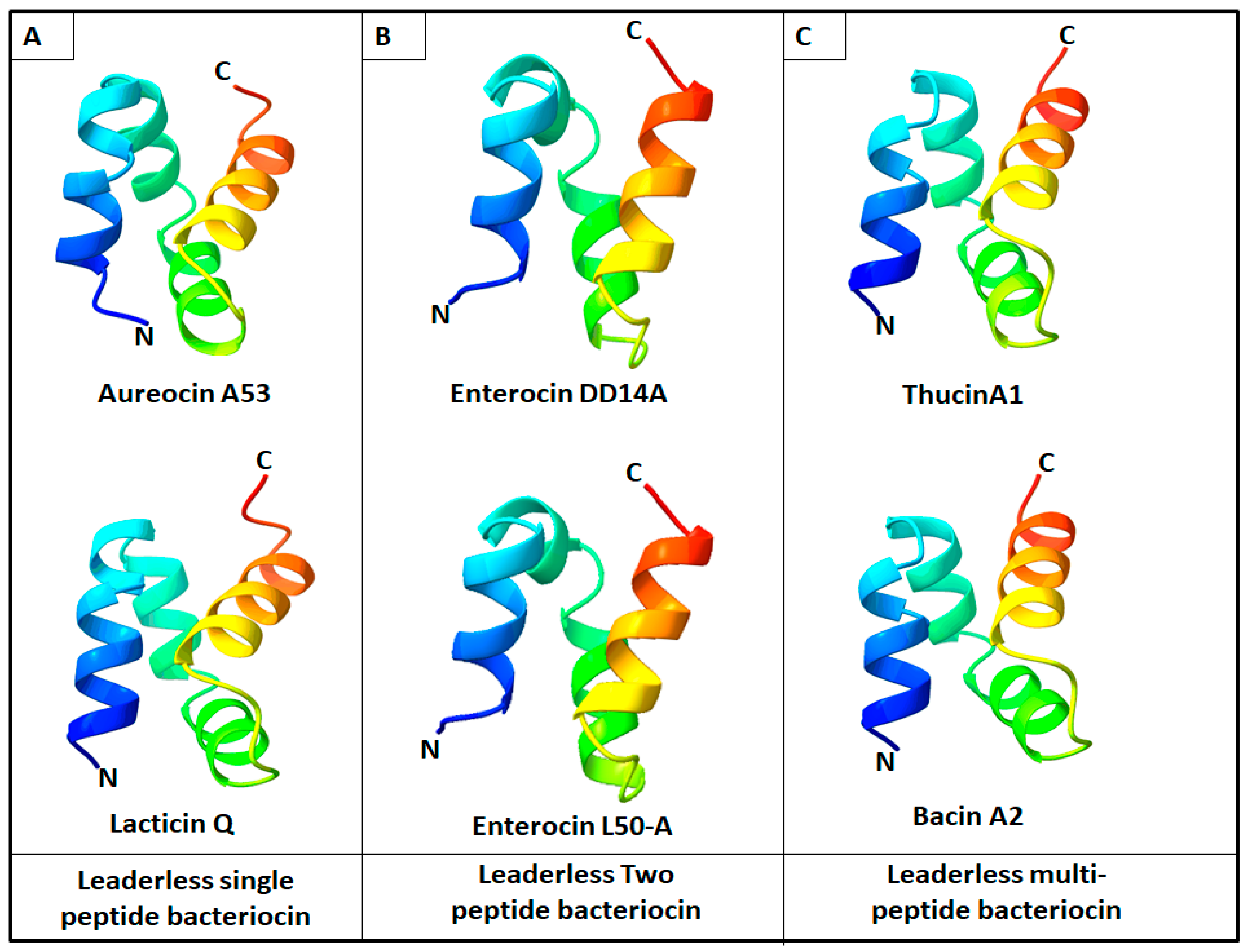
2. Diversity and Structures of Leaderless Bacteriocins
| Bacteriocin | Length (AA) | MW (Da) | pI | Net Charge | Hydrophobicity | Producer Bacterium | Reference | |
|---|---|---|---|---|---|---|---|---|
| Single peptide | Enterocin Q | 34 | 3979.73 | 9.78 | 4 | −0.359 | E. faecium L50 | [20] |
| Aureocin A53 | 51 | 6012.19 | 10.73 | 8 | −0.08 | Staphylococcus aureus A53 | [21] | |
| LsbB | 30 | 3437.99 | 10.75 | 6 | −0.683 | Lactococcus lactis subsp. lactis BGMN1-5 | [22] | |
| Enterocin EJ97 | 44 | 5350.29 | 10.39 | 6 | −0.589 | E. faecalis EJ97 | [23] | |
| BHT-B | 44 | 5193.06 | 10.57 | 4 | 0.241 | S. rattus strain BHT | [24] | |
| Lacticin Q | 53 | 5926.06 | 10.84 | 6 | 0.3 | L. lactis QU 5 | [25] | |
| Lacticin Z | 53 | 5968.1 | 10.63 | 5 | 0.28 | L. lactis QU 14 | [26] | |
| Weisselicin Y | 42 | 4923.68 | 10.36 | 5 | −0.09 | Weissella hellenica QU 13 | [27] | |
| Weissellicin M | 43 | 4966.85 | 10.39 | 5 | 0.037 | W. hellenica QU 13 | [27] | |
| Epidermicin NI01 | 51 | 6072.27 | 10.95 | 9 | −0.02 | S. epidermidis strain 224 | [28] | |
| BacSp222 | 50 | 5921.92 | 10.09 | 4 | −0.304 | S. pseudintermedius | [29] | |
| Enterocin K1 | 37 | 4592.37 | 10.24 | 6 | −0.7 | E. faecium EnGen0026 | [30] | |
| Lactolisterin BU | 43 | 5163.02 | 10.72 | 5 | −0.151 | L. lactis subsp. lactis. diacetylactis BGBU1-4 | [31] | |
| Capidermicin | 50 | 5464 | 10.22 | 5 | −0.064 | S. capitis CIT060 | [32] | |
| Toyoncin | 70 | 7817.1 | 9.94 | 4 | −0.02 | Bacillus toyonensis XIN-YC13 | ||
| Two peptides | Enterocin DD14 (MR10) (Ent7) | E. faecalis MRR 10-3; 14; 710C | ||||||
| DD14A (MR10A) (Ent7A) | 44 | 5204.32 | 10.68 | 7 | 0.202 | [33,34,35] | ||
| DD14B (MR10B) (Ent7B) | 43 | 5210.24 | 10.95 | 7 | −0.109 | |||
| Enterocin L50 | E. faecium L50 | [36] | ||||||
| L50A | 44 | 5218.35 | 10.68 | 7 | 0.202 | |||
| L50B | 43 | 5206.25 | 10.95 | 7 | −0.144 | |||
| Enterocin A5-11 | E. durans | [37] | ||||||
| A5-11A | 44 | 5190.31 | 10.57 | 7 | 0.202 | |||
| A5-11B | 43 | 5178.22 | 10.83 | 7 | −0.144 | |||
| Three peptides | Garvicin KS | L. garvieae KS1546 | [30] | |||||
| KS-A | 34 | 3481.14 | 10.98 | 4 | 0.403 | |||
| KS-B | 34 | 3188.84 | 11.02 | 5 | 0.691 | |||
| KS-C | 32 | 3127.76 | 10.98 | 4 | 0.588 | |||
| Cereucin X | B. cereus BAG2O-1 | [30] | ||||||
| X-A | 27 | 2972.48 | 10.33 | 2 | 0.004 | |||
| X-B | 29 | 3165.63 | 9.92 | 3 | −0.186 | |||
| X-C | 30 | 2826.25 | 10.64 | 4 | 0.52 | |||
| Cereucin V | B. cereus VD148 | [38] | ||||||
| V-A | 30 | 3142.69 | 11.08 | 3 | 0.373 | |||
| V-B | 30 | 2857.41 | 10.87 | 4 | 0.65 | |||
| V-C | 31 | 3004.52 | 10.88 | 3 | 0.69 | |||
| Thucin | B. thuringiensis P86 | [15] | ||||||
| ThuA1 | 48 | 5552.02 | 10.8 | 3 | 0.302 | |||
| ThuA2 | 48 | 5578.07 | 10 | 3 | 0.202 | |||
| ThuA3 | 48 | 5609.06 | 10.8 | 3 | 0.194 | |||
| Four peptides | Aureocin A70 | S. aureus A70 | [39] | |||||
| A70A | 31 | 2952.53 | 10.98 | 4 | 0.529 | |||
| A70B | 30 | 2825.34 | 10.87 | 4 | 0.707 | |||
| A70C | 31 | 2982.56 | 10.87 | 5 | 0.552 | |||
| A70D | 31 | 3114.76 | 10.7 | 4 | 0.632 | |||
| Cereucin H | B. cereus HuA2-4 | [38] | ||||||
| H-A | 26 | 2876.39 | 10.03 | 2 | 0.154 | |||
| H-B | 30 | 3170.7 | 10.5 | 2 | 0.233 | |||
| H-C | 30 | 2867.42 | 10.87 | 4 | 0.667 | |||
| H-D | 30 | 3018.63 | 10.86 | 4 | 0.62 | |||
| Bacin | Bacillus sp. TL12 | [14] | ||||||
| BacA1 | 48 | 5637.08 | 10.78 | 3 | 0.24375 | |||
| BacA2 | 48 | 5652.06 | 10.26 | 3 | 0.1479 | |||
| BacA3 | 48 | 5591.09 | 10.78 | 3 | 0.2479 | |||
| BacA4 | 48 | 5577.08 | 10.78 | 3 | 0.2416 | |||
| Six peptides | Thuricin | B. thuringiensis LX43 | [40] | |||||
| Thn A1 | 48 | 5396.98 | 9.82 | 2 | 0.308 | |||
| Thn A2 | 48 | 5382.96 | 10.27 | 2 | 0.404 | |||
| Thn A3 | 48 | 5322.99 | 9.99 | 2 | 0.45625 | |||
| Thn A4 | 48 | 5350 | 9.99 | 2 | 0.4 | |||
| Thn A5 | 48 | 5364.02 | 9.99 | 2 | 0.40625 | |||
| Thn A6 | 48 | 5364.02 | 9.99 | 2 | 0.40625 |
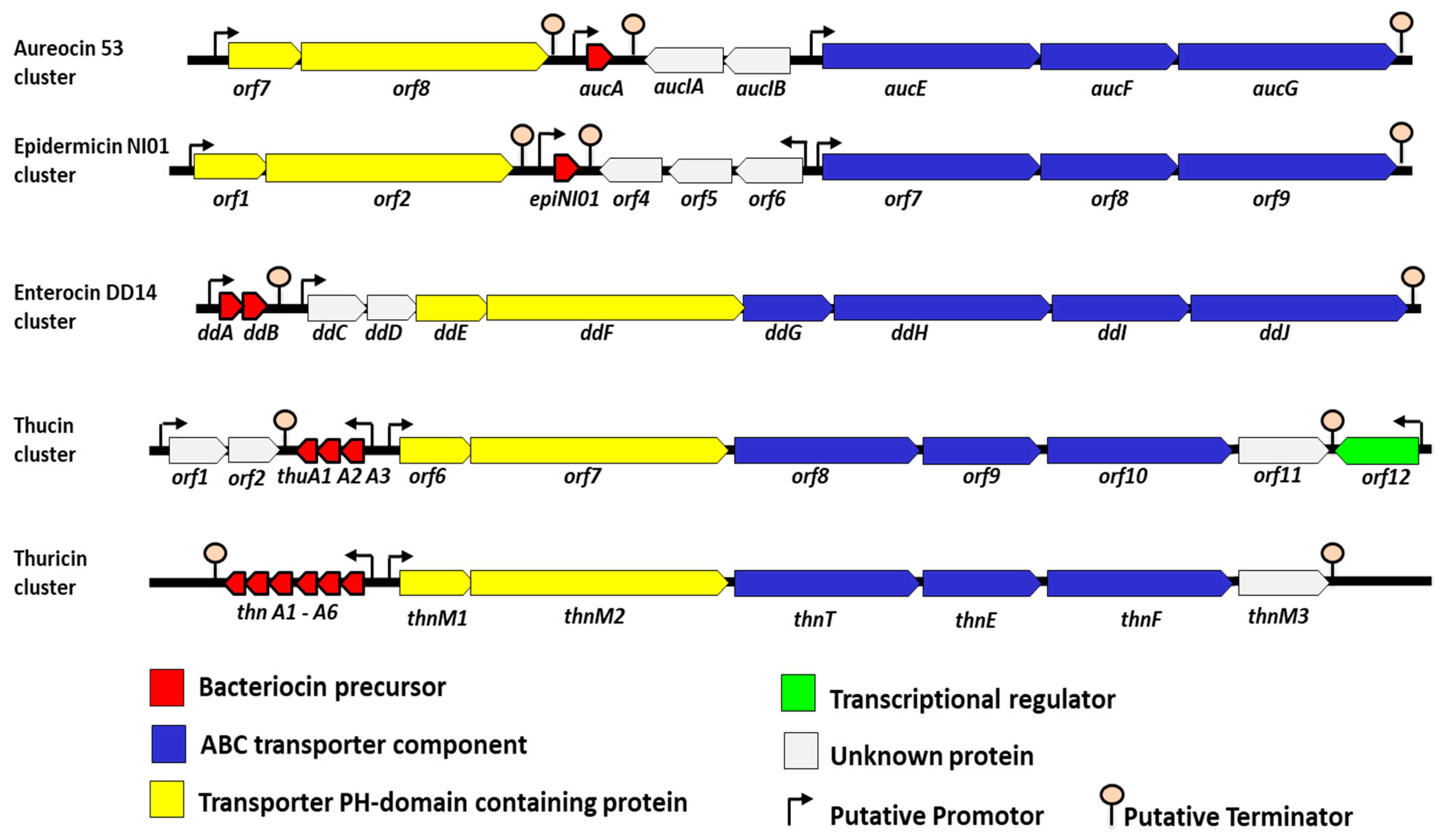
3. The Synthesis and Genetic Organization of the Leaderless Bacteriocins
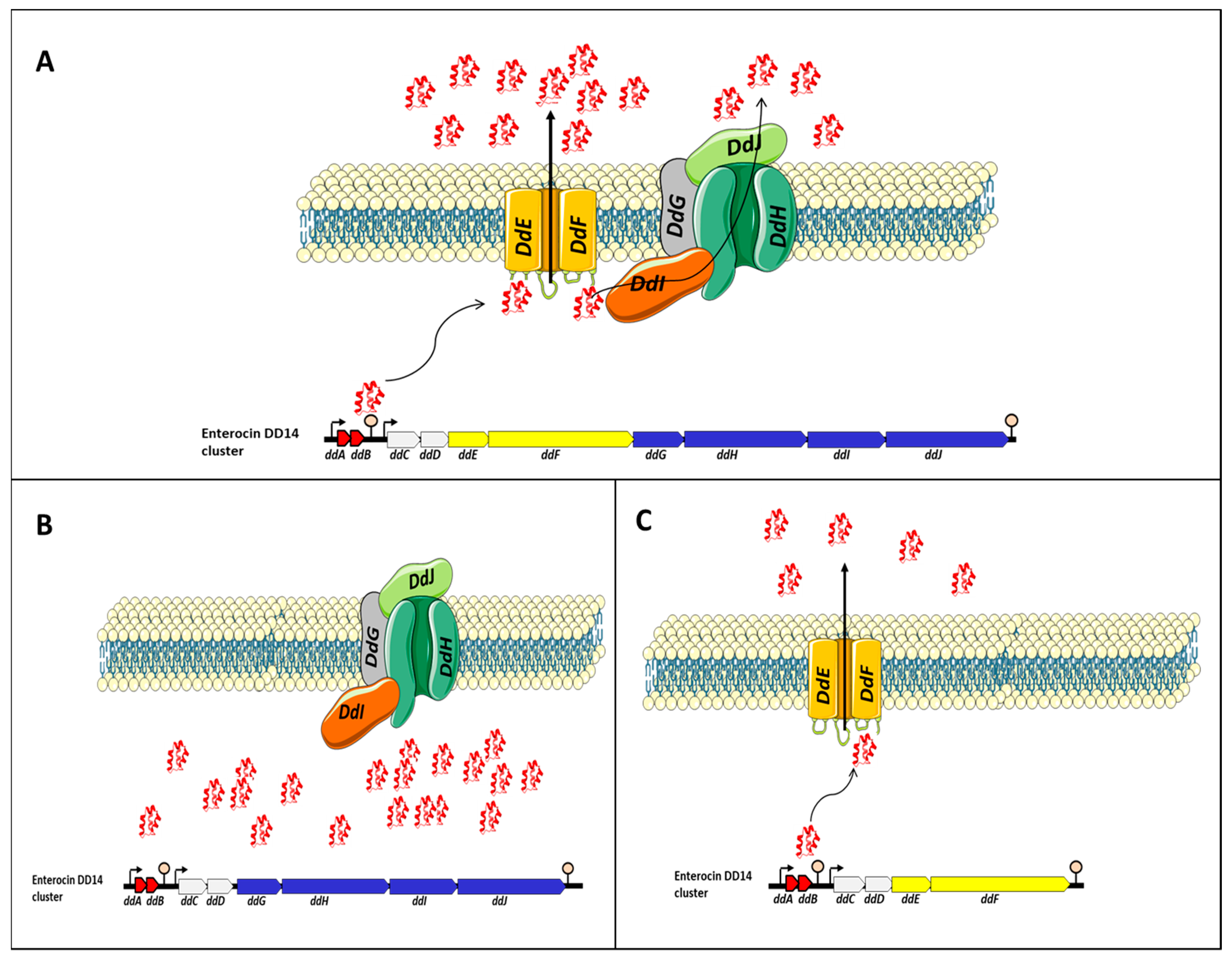
4. The Genetic and Synthesis Backgrounds of Enterocin DD14
5. Purification of Enterocin DD14
6. Antibacterial Spectrum of Enterocin DD14 and Potentiation of Antibiotic Activity
7. The Antiviral Spectrum of Enterocin DD14
8. Cytotoxicity of Enterocin DD14
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Montalbán-López, M.; Sánchez-Hidalgo, M.; Valdivia, E.; Martínez-Bueno, M.; Maqueda, M. Are Bacteriocins Underexploited? Novel Applications for Old Antimicrobials. Curr. Pharm. Biotechnol. 2011, 12, 1205–1220. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-C.; Lin, C.-H.; Sung, C.T.; Fang, J.-Y. Antibacterial Activities of Bacteriocins: Application in Foods and Pharmaceuticals. Front. Microbiol. 2014, 5, 241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teso-Pérez, C.; Martínez-Bueno, M.; Peralta-Sánchez, J.M.; Valdivia, E.; Maqueda, M.; Fárez-Vidal, M.E.; Martín-Platero, A.M. Enterocin Cross-Resistance Mediated by ABC Transport Systems. Microorganisms 2021, 9, 1411. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A Viable Alternative to Antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef]
- Sugrue, I.; O’Connor, P.M.; Hill, C.; Stanton, C.; Ross, R.P. Actinomyces Produces Defensin-Like Bacteriocins (Actifensins) with a Highly Degenerate Structure and Broad Antimicrobial Activity. J. Bacteriol. 2020, 202, e00529-19. [Google Scholar] [CrossRef] [Green Version]
- Verma, A.K.; Banerjee, R.; Dwivedi, H.P.; Juneja, V.K.; Carl, A.B. BACTERIOCINS|Potential in Food Preservation. In Encyclopedia of Food Microbiology; Academic Press: Cambridge, MA, USA; Elsevier Science: Amsterdam, The Netherlands, 2014; pp. 180–206. ISBN 978-0-12-384733-1. [Google Scholar]
- Johnson, M.E.M.; Jung, D.Y.-G.; Jin, D.Y.-Y.; Jayabalan, D.R.; Yang, D.S.H.; Suh, P.J.W. Bacteriocins as Food Preservatives: Challenges and Emerging Horizons. Crit. Rev. Food Sci. Nutr. 2018, 58, 2743–2767. [Google Scholar] [CrossRef]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Benítez-Chao, D.F.; León-Buitimea, A.; Lerma-Escalera, J.A.; Morones-Ramírez, J.R. Bacteriocins: An Overview of Antimicrobial, Toxicity, and Biosafety Assessment by in Vivo Models. Front. Microbiol. 2021, 12, 630695. [Google Scholar] [CrossRef]
- Al Atya, A.K.; Drider-Hadiouche, K.; Ravallec, R.; Silvain, A.; Vachee, A.; Drider, D. Probiotic Potential of Enterococcus faecalis Strains Isolated from Meconium. Front. Microbiol. 2015, 6, 227. [Google Scholar] [CrossRef]
- Klaenhammer, T.R. Genetics of Bacteriocins Produced by Lactic Acid Bacteria. FEMS Microbiol. Rev. 1993, 12, 39–85. [Google Scholar] [CrossRef]
- van der Meer, J.R.; Rollema, H.S.; Siezen, R.J.; Beerthuyzen, M.M.; Kuipers, O.P.; de Vos, W.M. Influence of Amino Acid Substitutions in the Nisin Leader Peptide on Biosynthesis and Secretion of Nisin by Lactococcus lactis. J. Biol. Chem. 1994, 269, 3555–3562. [Google Scholar] [CrossRef] [PubMed]
- Oman, T.J.; van der Donk, W.A. Follow the Leader: The Use of Leader Peptides to Guide Natural Product Biosynthesis. Nat. Chem. Biol. 2010, 6, 9–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Deng, S.; Liu, H.; Tang, L.; Wang, M.; Xin, B.; Li, F. Four Novel Leaderless Bacteriocins, Bacin A1, A2, A3, and A4 Exhibit Potent Antimicrobial and Antibiofilm Activities against Methicillin-Resistant Staphylococcus aureus. Microbiol. Spectr. 2022, 10, e00945-22. [Google Scholar] [CrossRef]
- Zhang, X.; Xin, N.; Zhu, Z.; Li, X.; Dai, D.; Pan, C.; Peng, D.; Sun, M. Three Novel Leaderless Bacteriocins Have Antimicrobial Activity against Gram-Positive Bacteria to Serve as Promising Food Biopreservative. Microb. Cell Fact. 2022, 21, 194. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xu, H.; Liu, S.; Song, B.; Liu, H.; Li, F.; Deng, S.; Wang, G.; Zeng, H.; Zeng, X.; et al. Toyoncin, a Novel Leaderless Bacteriocin That Is Produced by Bacillus toyonensis XIN-YC13 and Specifically Targets B. cereus and Listeria monocytogenes. Appl. Environ. Microbiol. 2021, 87, e0018521. [Google Scholar] [CrossRef] [PubMed]
- Lohans, C.T.; Towle, K.M.; Miskolzie, M.; McKay, R.T.; van Belkum, M.J.; McMullen, L.M.; Vederas, J.C. Solution Structures of the Linear Leaderless Bacteriocins Enterocin 7A and 7B Resemble Carnocyclin A, a Circular Antimicrobial Peptide. Biochemistry 2013, 52, 3987–3994. [Google Scholar] [CrossRef] [PubMed]
- Acedo, J.Z.; van Belkum, M.J.; Lohans, C.T.; Towle, K.M.; Miskolzie, M.; Vederas, J.C. Nuclear Magnetic Resonance Solution Structures of Lacticin Q and Aureocin A53 Reveal a Structural Motif Conserved among Leaderless Bacteriocins with Broad-Spectrum Activity. Biochemistry 2016, 55, 733–742. [Google Scholar] [CrossRef]
- Fimland, G.; Eijsink, V.G.H.; Nissen-Meyer, J. Mutational Analysis of the Role of Tryptophan Residues in an Antimicrobial Peptide. Biochemistry 2002, 41, 9508–9515. [Google Scholar] [CrossRef]
- Cintas, L.M.; Casaus, P.; Herranz, C.; Hâvarstein, L.S.; Holo, H.; Hernández, P.E.; Nes, I.F. Biochemical and Genetic Evidence That Enterococcus faecium L50 Produces Enterocins L50A and L50B, the Sec-Dependent Enterocin P, and a Novel Bacteriocin Secreted without an N-Terminal Extension Termed Enterocin Q. J. Bacteriol. 2000, 182, 6806–6814. [Google Scholar] [CrossRef] [Green Version]
- Netz, D.J.A.; Bastos, M.D.C.D.F.; Sahl, H.-G. Mode of Action of the Antimicrobial Peptide Aureocin A53 from Staphylococcus aureus. Appl. Environ. Microbiol. 2002, 68, 5274–5280. [Google Scholar] [CrossRef] [Green Version]
- Gajic, O.; Buist, G.; Kojic, M.; Topisirovic, L.; Kuipers, O.P.; Kok, J. Novel Mechanism of Bacteriocin Secretion and Immunity Carried out by Lactococcal Multidrug Resistance Proteins. J. Biol. Chem. 2003, 278, 34291–34298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-Hidalgo, M.; Maqueda, M.; Gálvez, A.; Abriouel, H.; Valdivia, E.; Martínez-Bueno, M. The Genes Coding for Enterocin EJ97 Production by Enterococcus faecalis EJ97 Are Located on a Conjugative Plasmid. Appl. Environ. Microbiol. 2003, 69, 1633–1641. [Google Scholar] [CrossRef] [Green Version]
- Hyink, O.; Balakrishnan, M.; Tagg, J.R. Streptococcus rattus Strain BHT Produces Both a Class I Two-Component Lantibiotic and a Class II Bacteriocin. FEMS Microbiol. Lett. 2005, 252, 235–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujita, K.; Ichimasa, S.; Zendo, T.; Koga, S.; Yoneyama, F.; Nakayama, J.; Sonomoto, K. Structural Analysis and Characterization of Lacticin Q, a Novel Bacteriocin Belonging to a New Family of Unmodified Bacteriocins of Gram-Positive Bacteria. Appl. Environ. Microbiol. 2007, 73, 2871–2877. [Google Scholar] [CrossRef] [Green Version]
- Iwatani, S.; Zendo, T.; Yoneyama, F.; Nakayama, J.; Sonomoto, K. Characterization and Structure Analysis of a Novel Bacteriocin, Lacticin Z, Produced by Lactococcus lactis QU 14. Biosci. Biotechnol. Biochem. 2007, 71, 1984–1992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masuda, Y.; Zendo, T.; Sawa, N.; Perez, R.H.; Nakayama, J.; Sonomoto, K. Characterization and Identification of Weissellicin Y and Weissellicin M, Novel Bacteriocins Produced by Weissella hellenica QU 13. J. Appl. Microbiol 2012, 112, 99–108. [Google Scholar] [CrossRef]
- Sandiford, S.; Upton, M. Identification, Characterization, and Recombinant Expression of Epidermicin NI01, a Novel Unmodified Bacteriocin Produced by Staphylococcus epidermidis That Displays Potent Activity against Staphylococci. Antimicrob. Agents Chemother. 2012, 56, 1539–1547. [Google Scholar] [CrossRef] [Green Version]
- Wladyka, B.; Piejko, M.; Bzowska, M.; Pieta, P.; Krzysik, M.; Mazurek, Ł.; Guevara-Lora, I.; Bukowski, M.; Sabat, A.J.; Friedrich, A.W.; et al. A Peptide Factor Secreted by Staphylococcus pseudintermedius Exhibits Properties of Both Bacteriocins and Virulence Factors. Sci. Rep. 2015, 5, 14569. [Google Scholar] [CrossRef] [Green Version]
- Ovchinnikov, K.V.; Kristiansen, P.E.; Straume, D.; Jensen, M.S.; Aleksandrzak-Piekarczyk, T.; Nes, I.F.; Diep, D.B. The Leaderless Bacteriocin Enterocin K1 Is Highly Potent against Enterococcus faecium: A Study on Structure, Target Spectrum and Receptor. Front. Microbiol. 2017, 8, 774. [Google Scholar] [CrossRef] [Green Version]
- Lozo, J.; Mirkovic, N.; O’Connor, P.M.; Malesevic, M.; Miljkovic, M.; Polovic, N.; Jovcic, B.; Cotter, P.D.; Kojic, M. Lactolisterin BU, a Novel Class II Broad-Spectrum Bacteriocin from Lactococcus lactis subsp. lactis Bv. Diacetylactis BGBU1-4. Appl. Environ. Microbiol. 2017, 83, e01519-17. [Google Scholar] [CrossRef] [Green Version]
- Lynch, D.; O’Connor, P.M.; Cotter, P.D.; Hill, C.; Field, D.; Begley, M. Identification and Characterisation of Capidermicin, a Novel Bacteriocin Produced by Staphylococcus capitis. PLoS ONE 2019, 14, e0223541. [Google Scholar] [CrossRef] [PubMed]
- Caly, D.L.; Chevalier, M.; Flahaut, C.; Cudennec, B.; Al Atya, A.K.; Chataigné, G.; D’Inca, R.; Auclair, E.; Drider, D. The Safe Enterocin DD14 Is a Leaderless Two-Peptide Bacteriocin with Anti-Clostridium Perfringens Activity. Int. J. Antimicrob. Agents 2017, 49, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Martín-Platero, A.M.; Valdivia, E.; Ruíz-Rodríguez, M.; Soler, J.J.; Martín-Vivaldi, M.; Maqueda, M.; Martínez-Bueno, M. Characterization of Antimicrobial Substances Produced by Enterococcus faecalis MRR 10-3, Isolated from the Uropygial Gland of the Hoopoe (Upupa epops). Appl. Environ. Microbiol. 2006, 72, 4245–4249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Vederas, J.C.; Whittal, R.M.; Zheng, J.; Stiles, M.E.; Carlson, D.; Franz, C.M.A.P.; McMullen, L.M.; van Belkum, M.J. Identification of an N-Terminal Formylated, Two-Peptide Bacteriocin from Enterococcus faecalis 710C. J. Agric. Food Chem. 2011, 59, 5602–5608. [Google Scholar] [CrossRef]
- Cintas, L.M.; Casaus, P.; Holo, H.; Hernandez, P.E.; Nes, I.F.; Håvarstein, L.S. Enterocins L50A and L50B, Two Novel Bacteriocins from Enterococcus faecium L50, Are Related to Staphylococcal Hemolysins. J. Bacteriol. 1998, 180, 1988–1994. [Google Scholar] [CrossRef] [Green Version]
- Batdorj, B.; Dalgalarrondo, M.; Choiset, Y.; Pedroche, J.; Métro, F.; Prévost, H.; Chobert, J.-M.; Haertlé, T. Purification and Characterization of Two Bacteriocins Produced by Lactic Acid Bacteria Isolated from Mongolian airag. J. Appl. Microbiol. 2006, 101, 837–848. [Google Scholar] [CrossRef]
- Ovchinnikov, K.V.; Chi, H.; Mehmeti, I.; Holo, H.; Nes, I.F.; Diep, D.B. Novel Group of Leaderless Multipeptide Bacteriocins from Gram-Positive Bacteria. Appl. Environ. Microbiol. 2016, 82, 5216–5224. [Google Scholar] [CrossRef] [Green Version]
- Netz, D.J.; Sahl, H.G.; Marcelino, R.; dos Santos Nascimento, J.; de Oliveira, S.S.; Soares, M.B.; do Carmo de Freire Bastos, M.; Marcolino, R. Molecular Characterisation of Aureocin A70, a Multi-Peptide Bacteriocin Isolated from Staphylococcus aureus. J. Mol. Biol. 2001, 311, 939–949. [Google Scholar] [CrossRef]
- Deng, S.; Liu, S.; Li, X.; Liu, H.; Li, F.; Liu, K.; Zeng, H.; Zeng, X.; Xin, B. Thuricins: Novel Leaderless Bacteriocins with Potent Antimicrobial Activity against Gram-Positive Foodborne Pathogens. J. Agric. Food Chem. 2022, 70, 9990–9999. [Google Scholar] [CrossRef]
- Liepinsh, E.; Andersson, M.; Ruysschaert, J.M.; Otting, G. Saposin Fold Revealed by the NMR Structure of NK-Lysin. Nat. Struct. Biol. 1997, 4, 793–795. [Google Scholar] [CrossRef]
- Bruhn, H. A Short Guided Tour through Functional and Structural Features of Saposin-like Proteins. Biochem. J. 2005, 389, 249–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, V.E.; Leyko, P.; Alattia, J.-R.; Chen, L.; Privé, G.G. Crystal Structures of Saposins A and C. Protein Sci. 2006, 15, 1849–1857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Towle, K.M.; Vederas, J.C. Structural Features of Many Circular and Leaderless Bacteriocins Are Similar to Those in Saposins and Saposin-like Peptides. MedChemComm 2017, 8, 276–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez, R.H.; Zendo, T.; Sonomoto, K. Circular and Leaderless Bacteriocins: Biosynthesis, Mode of Action, Applications, and Prospects. Front. Microbiol. 2018, 9, 2085. [Google Scholar] [CrossRef] [Green Version]
- Nascimento, J.D.S.; Coelho, M.L.V.; Ceotto, H.; Potter, A.; Fleming, L.R.; Salehian, Z.; Nes, I.F.; Bastos, M.D.C.D.F. Genes Involved in Immunity to and Secretion of Aureocin A53, an Atypical Class II Bacteriocin Produced by Staphylococcus aureus A53. J. Bacteriol. 2012, 194, 875–883. [Google Scholar] [CrossRef] [Green Version]
- Iwatani, S.; Yoneyama, F.; Miyashita, S.; Zendo, T.; Nakayama, J.; Sonomoto, K. Identification of the Genes Involved in the Secretion and Self-Immunity of Lacticin Q, an Unmodified Leaderless Bacteriocin from Lactococcus lactis QU 5. Microbiology 2012, 158, 2927–2935. [Google Scholar] [CrossRef] [Green Version]
- Iwatani, S.; Horikiri, Y.; Zendo, T.; Nakayama, J.; Sonomoto, K. Bifunctional Gene Cluster LnqBCDEF Mediates Bacteriocin Production and Immunity with Differential Genetic Requirements. Appl. Environ. Microbiol. 2013, 79, 2446–2449. [Google Scholar] [CrossRef] [Green Version]
- Iwatani, S.; Ishibashi, N.; Flores, F.P.; Zendo, T.; Nakayama, J.; Sonomoto, K. LnqR, a TetR-Family Transcriptional Regulator, Positively Regulates Lacticin Q Production in Lactococcus lactis QU 5. FEMS Microbiol. Lett. 2016, 363, fnw200. [Google Scholar] [CrossRef] [Green Version]
- Coelho, M.L.V.; Coutinho, B.G.; da Silva Santos, O.C.; Nes, I.F.; de Freire Bastos, M.D.C. Immunity to the Staphylococcus aureus Leaderless Four-Peptide Bacteriocin Aureocin A70 Is Conferred by AurI, an Integral Membrane Protein. Res. Microbiol. 2014, 165, 50–59. [Google Scholar] [CrossRef]
- Ladjouzi, R.; Lucau-Danila, A.; Benachour, A.; Drider, D. A Leaderless Two-Peptide Bacteriocin, Enterocin DD14, Is Involved in Its Own Self-Immunity: Evidence and Insights. Front. Bioeng. Biotechnol. 2020, 8, 644. [Google Scholar] [CrossRef]
- Pérez-Ramos, A.; Ladjouzi, R.; Benachour, A.; Drider, D. Evidence for the Involvement of Pleckstrin Homology Domain-Containing Proteins in the Transport of Enterocin DD14 (EntDD14); a Leaderless Two-Peptide Bacteriocin. Int. J. Mol. Sci. 2021, 22, 12877. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Ramos, A.; Ladjouzi, R.; Mihasan, M.; Teiar, R.; Benachour, A.; Drider, D. Advances in Characterizing the Transport Systems of and Resistance to EntDD14, A Leaderless Two-Peptide Bacteriocin with Potent Inhibitory Activity. Int. J. Mol. Sci. 2023, 24, 1517. [Google Scholar] [CrossRef] [PubMed]
- Criado, R.; Gutiérrez, J.; Martín, M.; Herranz, C.; Hernández, P.E.; Cintas, L.M. Immunochemical Characterization of Temperature-Regulated Production of Enterocin L50 (EntL50A and EntL50B), Enterocin P, and Enterocin Q by Enterococcus faecium L50. Appl. Environ. Microbiol. 2006, 72, 7634–7643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coelho, M.L.V.; Fleming, L.R.; Bastos, M.D.C.D.F. Insights into Aureocin A70 Regulation: Participation of Regulator AurR, Alternative Transcription Factor σ(B) and Phage Φ11 Regulator CI. Res. Microbiol. 2016, 167, 90–102. [Google Scholar] [CrossRef] [PubMed]
- Ladjouzi, R.; Lucau-Danila, A.; Drider, D. Metabolic Shift of an Isogenic Strain of Enterococcus Faecalis 14, Deficient in Its Own Bacteriocin Synthesis, as Revealed by a Transcriptomic Analysis. Int. J. Mol. Sci. 2020, 21, 4653. [Google Scholar] [CrossRef]
- Bierbaum, G.; Sahl, H.-G. Lantibiotics: Mode of Action, Biosynthesis and Bioengineering. Curr. Pharm. Biotechnol. 2009, 10, 2–18. [Google Scholar] [CrossRef]
- Nissen-Meyer, J.; Rogne, P.; Oppegard, C.; Haugen, H.; Kristiansen, P. Structure-Function Relationships of the Non-Lanthionine-Containing Peptide (Class II) Bacteriocins Produced by Gram-Positive Bacteria. Curr. Pharm. Biotechnol. 2009, 10, 19–37. [Google Scholar] [CrossRef]
- Diaz, M.; Valdivia, E.; Martínez-Bueno, M.; Fernández, M.; Soler-González, A.S.; Ramírez-Rodrigo, H.; Maqueda, M. Characterization of a New Operon, as-48EFGH, from the as-48 Gene Cluster Involved in Immunity to Enterocin AS-48. Appl. Environ. Microbiol. 2003, 69, 1229–1236. [Google Scholar] [CrossRef] [Green Version]
- Klein, C.; Entian, K.D. Genes Involved in Self-Protection against the Lantibiotic Subtilin Produced by Bacillus subtilis ATCC 6633. Appl. Environ. Microbiol. 1994, 60, 2793–2801. [Google Scholar] [CrossRef] [Green Version]
- Siegers, K.; Entian, K.D. Genes Involved in Immunity to the Lantibiotic Nisin Produced by Lactococcus lactis 6F3. Appl. Environ. Microbiol. 1995, 61, 1082–1089. [Google Scholar] [CrossRef] [Green Version]
- Yarmus, M.; Mett, A.; Shapira, R. Cloning and Expression of the Genes Involved in the Production of and Immunity against the Bacteriocin Lacticin RM. Biochim. Biophys. Acta 2000, 1490, 279–290. [Google Scholar] [CrossRef]
- Beis, K.; Rebuffat, S. Multifaceted ABC Transporters Associated to Microcin and Bacteriocin Export. Res. Microbiol. 2019, 170, 399–406. [Google Scholar] [CrossRef]
- Rincé, A.; Dufour, A.; Uguen, P.; Le Pennec, J.P.; Haras, D. Characterization of the Lacticin 481 Operon: The Lactococcus lactis Genes LctF, LctE, and LctG Encode a Putative ABC Transporter Involved in Bacteriocin Immunity. Appl. Environ. Microbiol. 1997, 63, 4252–4260. [Google Scholar] [CrossRef] [Green Version]
- Cintas, L.M.; Casaus, P.; Håvarstein, L.S.; Hernández, P.E.; Nes, I.F. Biochemical and Genetic Characterization of Enterocin P, a Novel Sec-Dependent Bacteriocin from Enterococcus faecium P13 with a Broad Antimicrobial Spectrum. Appl. Environ. Microbiol. 1997, 63, 4321–4330. [Google Scholar] [CrossRef] [Green Version]
- De Vuyst, L.; Leroy, F. Bacteriocins from Lactic Acid Bacteria: Production, Purification, and Food Applications. J. Mol. Microbiol. Biotechnol. 2007, 13, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Jamaluddin, N.; Stuckey, D.C.; Ariff, A.B.; Faizal Wong, F.W. Novel Approaches to Purifying Bacteriocin: A Review. Crit. Rev. Food Sci. Nutr. 2018, 58, 2453–2465. [Google Scholar] [CrossRef] [PubMed]
- Carlin Fagundes, P.; Nascimento de Sousa Santos, I.; Silva Francisco, M.; Mattos Albano, R.; de Freire Bastos, M.D.C. Genetic and Biochemical Characterization of Hyicin 3682, the First Bacteriocin Reported for Staphylococcus hyicus. Microbiol. Res. 2017, 198, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Dezwaan, D.C.; Mequio, M.J.; Littell, J.S.; Allen, J.P.; Rossbach, S.; Pybus, V. Purification and Characterization of Enterocin 62-6, a Two-Peptide Bacteriocin Produced by a Vaginal Strain of Enterococcus faecium: Potential Significance in Bacterial Vaginosis. Microb. Ecol. Health Dis. 2007, 19, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, V.; Jamal, Q.M.S.; Siddiqui, M.U.; Shukla, A.K.; Alzohairy, M.A.; Al Karaawi, M.A.; Kamal, M.A. Methods of Screening-Purification and Antimicrobial Potentialities of Bacteriocin in Health Care. Curr. Drug Metab. 2017, 18, 814–830. [Google Scholar] [CrossRef] [PubMed]
- Pingitore, E.V.; Salvucci, E.; Sesma, F.; Nader-Macias, M.E. Different Strategies for Purification of Antimicrobial Peptides from Lactic Acid Bacteria (LAB). Commun. Curr. Res. Educ. Top. Trends Appl. Microbiol. 2007, 1, 557–568. [Google Scholar]
- Cui, Y.; Luo, L.; Wang, X.; Lu, Y.; Yi, Y.; Shan, Y.; Liu, B.; Zhou, Y.; Lü, X. Mining, Heterologous Expression, Purification, Antibactericidal Mechanism, and Application of Bacteriocins: A Review. Compr. Rev. Food Sci. Food Saf. 2021, 20, 863–899. [Google Scholar] [CrossRef] [PubMed]
- Guyonnet, D.; Fremaux, C.; Cenatiempo, Y.; Berjeaud, J.M. Method for Rapid Purification of Class IIa Bacteriocins and Comparison of Their Activities. Appl. Environ. Microbiol. 2000, 66, 1744–1748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borzenkov, V.; Surovtsev, V.; Dyatlov, I. Obtaining Bacteriocins by Chromatographic Methods. Adv. Biosci. Biotechnol. 2014, 2014, 55054. [Google Scholar] [CrossRef] [Green Version]
- Yap, P.G.; Lai, Z.W.; Tan, J.S. Bacteriocins from Lactic Acid Bacteria: Purification Strategies and Applications in Food and Medical Industries: A Review. Beni-Suef Univ. J. Basic Appl. Sci. 2022, 11, 51. [Google Scholar] [CrossRef]
- Zgheib, H.; Belguesmia, Y.; Boukherroub, R.; Drider, D. Alginate Nanoparticles Enhance Anti-Clostridium Perfringens Activity of the Leaderless Two-Peptide Enterocin DD14 and Affect Expression of Some Virulence Factors. Probiotics Antimicrob. Proteins 2021, 13, 1213–1227. [Google Scholar] [CrossRef]
- Ugras, S.; Sezen, K.; Kati, H.; Demirbag, Z. Purification and Characterization of the Bacteriocin Thuricin Bn1 Produced by Bacillus thuringiensis subsp. kurstaki Bn1 Isolated from a Hazelnut Pest. J. Microbiol. Biotechnol. 2013, 23, 167–176. [Google Scholar] [CrossRef] [Green Version]
- Berić, T.; Stanković, S.; Draganić, V.; Kojić, M.; Lozo, J.; Fira, D. Novel Antilisterial Bacteriocin Licheniocin 50.2 from Bacillus licheniformis VPS50.2 Isolated from Soil Sample. J. Appl. Microbiol. 2014, 116, 502–510. [Google Scholar] [CrossRef]
- Forestier, A.; Belguesmia, Y.; Krier, F.; Drider, D.; Dhulster, P.; Firdaous, L. Recovery of Nisin from Culture Supernatants of Lactococcus lactis by Ultrafiltration: Flux Properties and Separation Efficiency. Food Bioprod. Process. 2022, 136, 196–210. [Google Scholar] [CrossRef]
- Izquierdo, E.; Wagner, C.; Marchioni, E.; Aoude-Werner, D.; Ennahar, S. Enterocin 96, a Novel Class II Bacteriocin Produced by Enterococcus faecalis WHE 96, Isolated from Munster Cheese. Appl. Environ. Microbiol. 2009, 75, 4273–4276. [Google Scholar] [CrossRef] [Green Version]
- Khan, H.; Flint, S.H.; Yu, P.-L. Development of a Chemically Defined Medium for the Production of Enterolysin A from Enterococcus faecalis B9510. J. Appl. Microbiol. 2013, 114, 1092–1102. [Google Scholar] [CrossRef]
- Al Atya, A.K.; Belguesmia, Y.; Chataigne, G.; Ravallec, R.; Vachée, A.; Szunerits, S.; Boukherroub, R.; Drider, D. Anti-MRSA Activities of Enterocins DD28 and DD93 and Evidences on Their Role in the Inhibition of Biofilm Formation. Front. Microbiol. 2016, 7, 817. [Google Scholar] [CrossRef] [Green Version]
- Belguesmia, Y.; Spano, G.; Drider, D. Potentiating Effects of Leaderless Enterocin DD14 in Combination with Methicillin on Clinical Methicillin-Resistant Staphylococcus aureus S1 Strain. Microbiol. Res. 2021, 252, 126864. [Google Scholar] [CrossRef] [PubMed]
- Bendjeddou, K.; Hamma-Faradji, S.; Meddour, A.A.; Belguesmia, Y.; Cudennec, B.; Bendali, F.; Daube, G.; Taminiau, B.; Drider, D. Gut Microbiota, Body Weight and Histopathological Examinations in Experimental Infection by Methicillin-Resistant Staphylococcus aureus: Antibiotic versus Bacteriocin. Benef. Microbes 2021, 12, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Zgheib, H.; Drider, D.; Belguesmia, Y. Broadening and Enhancing Bacteriocins Activities by Association with Bioactive Substances. Int. J. Environ. Res. Public Health 2020, 17, 7835. [Google Scholar] [CrossRef]
- Naghmouchi, K.; Baah, J.; Hober, D.; Jouy, E.; Rubrecht, C.; Sané, F.; Drider, D. Synergistic Effect between Colistin and Bacteriocins in Controlling Gram-Negative Pathogens and Their Potential to Reduce Antibiotic Toxicity in Mammalian Epithelial Cells. Antimicrob. Agents Chemother. 2013, 57, 2719–2725. [Google Scholar] [CrossRef] [Green Version]
- Teiar, R.; Pérez-Ramos, A.; Zgheib, H.; Cudennec, B.; Belguesmia, Y.; Drider, D. Anti-Adhesion and Anti-Inflammatory Potential of the Leaderless Class IIb Bacteriocin Enterocin DD14. Probiotics Antimicrob. Proteins 2022, 14, 613–619. [Google Scholar] [CrossRef]
- Messaoudi, S.; Kergourlay, G.; Dalgalarrondo, M.; Choiset, Y.; Ferchichi, M.; Prévost, H.; Pilet, M.F.; Chobert, J.M.; Manai, M.; Dousset, X. Purification and Characterization of a New Bacteriocin Active against Campylobacter Produced by Lactobacillus salivarius SMXD51. Food Microbiol. 2012, 32, 129–134. [Google Scholar] [CrossRef]
- Line, J.E.; Svetoch, E.A.; Eruslanov, B.V.; Perelygin, V.V.; Mitsevich, E.V.; Mitsevich, I.P.; Levchuk, V.P.; Svetoch, O.E.; Seal, B.S.; Siragusa, G.R.; et al. Isolation and Purification of Enterocin E-760 with Broad Antimicrobial Activity against Gram-Positive and Gram-Negative Bacteria. Antimicrob. Agents Chemother. 2008, 52, 1094–1100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Atya, A.K.; Abriouel, H.; Kempf, I.; Jouy, E.; Auclair, E.; Vachée, A.; Drider, D. Effects of Colistin and Bacteriocins Combinations on the In Vitro Growth of Escherichia coli Strains from Swine Origin. Probiotics Antimicrob. Proteins 2016, 8, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, M.A. Determination of 50% Endpoint Titer Using a Simple Formula. World J. Virol. 2016, 5, 85. [Google Scholar] [CrossRef]
- Wachsman, M.B.; Castilla, V.; de Ruiz Holgado, A.P.; de Torres, R.A.; Sesma, F.; Coto, C.E. Enterocin CRL35 Inhibits Late Stages of HSV-1 and HSV-2 Replication in Vitro. Antivir. Res. 2003, 58, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Cavicchioli, V.Q.; Carvalho, O.V.; Paiva, J.C.; Todorov, S.D.; Silva Júnior, A.; Nero, L.A. Inhibition of Herpes Simplex Virus 1 (HSV-1) and Poliovirus (PV-1) by Bacteriocins from Lactococcus lactis subsp. lactis and Enterococcus Durans Strains Isolated from Goat Milk. Int. J. Antimicrob. Agents 2018, 51, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Erol, I.; Kotil, S.E.; Ortakci, F.; Durdagi, S. Exploring the Binding Capacity of Lactic Acid Bacteria Derived Bacteriocins against RBD of SARS-CoV-2 Omicron Variant by Molecular Simulations. J. Biomol. Struct. Dyn. 2023, 1–11. [Google Scholar] [CrossRef]
- Omer, A.A.M.; Hinkula, J.; Tran, P.-T.-H.; Melik, W.; Zattarin, E.; Aili, D.; Selegård, R.; Bengtsson, T.; Khalaf, H. Plantaricin NC8 Aβ Rapidly and Efficiently Inhibits Flaviviruses and SARS-CoV-2 by Disrupting Their Envelopes. PLoS ONE 2022, 17, e0278419. [Google Scholar] [CrossRef]
- Bhattacharya, R.; Gupta, A.M.; Mitra, S.; Mandal, S.; Biswas, S.R. A Natural Food Preservative Peptide Nisin Can Interact with the SARS-CoV-2 Spike Protein Receptor Human ACE2. Virology 2021, 552, 107–111. [Google Scholar] [CrossRef]
- Kourmentza, K.; Gromada, X.; Michael, N.; Degraeve, C.; Vanier, G.; Ravallec, R.; Coutte, F.; Karatzas, K.A.; Jauregi, P. Antimicrobial Activity of Lipopeptide Biosurfactants against Foodborne Pathogen and Food Spoilage Microorganisms and Their Cytotoxicity. Front. Microbiol. 2021, 11, 561060. [Google Scholar] [CrossRef] [PubMed]
- Abengózar, M.Á.; Cebrián, R.; Saugar, J.M.; Gárate, T.; Valdivia, E.; Martínez-Bueno, M.; Maqueda, M.; Rivas, L. Enterocin AS-48 as Evidence for the Use of Bacteriocins as New Leishmanicidal Agents. Antimicrob. Agents Chemother. 2017, 61, e02288-16. [Google Scholar] [CrossRef] [Green Version]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ladjouzi, R.; Dussert, E.; Teiar, R.; Belguesmia, Y.; Drider, D. A Review on Enterocin DD14, the Leaderless Two-Peptide Bacteriocin with Multiple Biological Functions and Unusual Transport Pathway. Antibiotics 2023, 12, 1188. https://doi.org/10.3390/antibiotics12071188
Ladjouzi R, Dussert E, Teiar R, Belguesmia Y, Drider D. A Review on Enterocin DD14, the Leaderless Two-Peptide Bacteriocin with Multiple Biological Functions and Unusual Transport Pathway. Antibiotics. 2023; 12(7):1188. https://doi.org/10.3390/antibiotics12071188
Chicago/Turabian StyleLadjouzi, Rabia, Elodie Dussert, Radja Teiar, Yanath Belguesmia, and Djamel Drider. 2023. "A Review on Enterocin DD14, the Leaderless Two-Peptide Bacteriocin with Multiple Biological Functions and Unusual Transport Pathway" Antibiotics 12, no. 7: 1188. https://doi.org/10.3390/antibiotics12071188
APA StyleLadjouzi, R., Dussert, E., Teiar, R., Belguesmia, Y., & Drider, D. (2023). A Review on Enterocin DD14, the Leaderless Two-Peptide Bacteriocin with Multiple Biological Functions and Unusual Transport Pathway. Antibiotics, 12(7), 1188. https://doi.org/10.3390/antibiotics12071188







