The Emergence of Antimicrobial Resistance and Virulence Characteristics in Enterococcus Species Isolated from Bovine Milk
Abstract
:1. Introduction
2. Results and Discussion
2.1. Prevalence of Enterococcus spp. in Bovine Milk
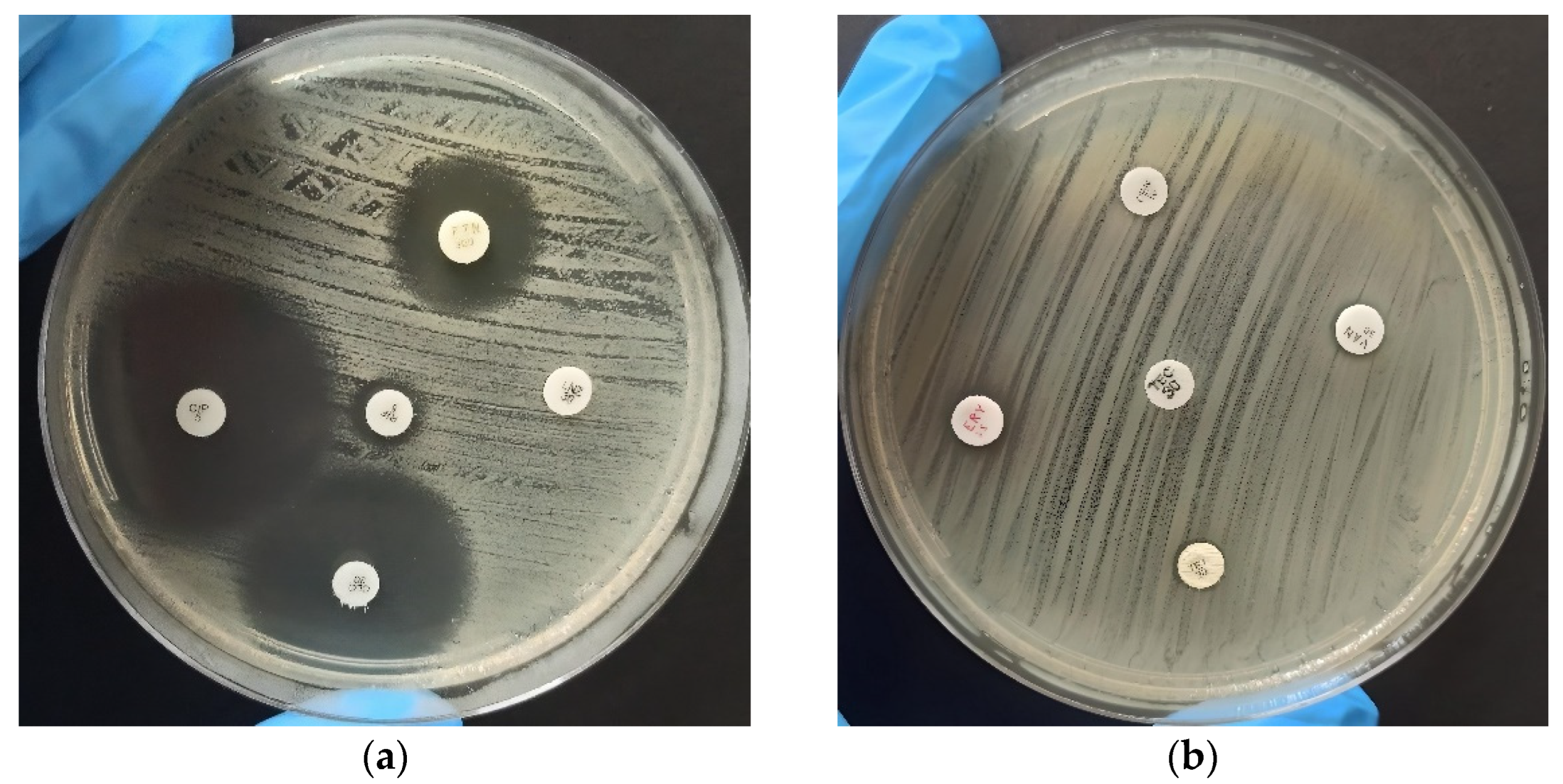
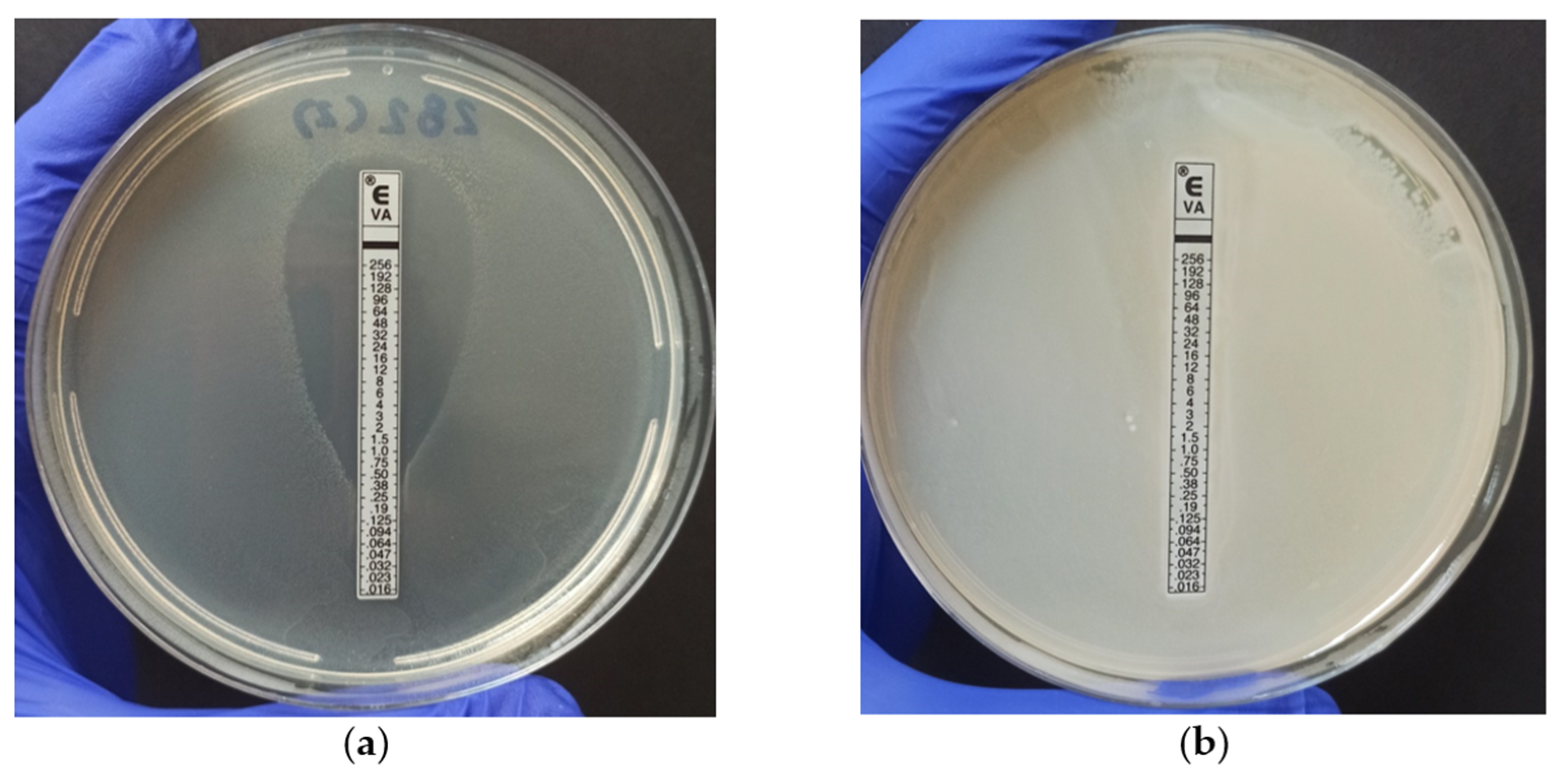
2.2. Antimicrobial Resistance
2.3. Detection of Virulence Genes
3. Materials and Methods
3.1. Identification of Enterococcal Isolates
3.2. Identification of E. faecium and E. faecalis by PCR
3.3. Identification of Enterococcus spp. by MALDI-TOF MS
3.4. Antimicrobial Resistance
3.5. Virulence Genes
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reshi, A.A.; Husain, I.; Bhat, S.A.; Rehman, M.U.; Razak, R.; Bilal, S.; Mir, M.R. Bovine Mastitis as an Evolving Disease and Its Impact on the Dairy Industry. Int. J. Curr. Res. Rev. 2015, 7, 48. [Google Scholar]
- Ruegg, P.L. A 100-Year Review: Mastitis detection, management, and prevention. J. Dairy Sci. 2017, 100, 10381–10397. [Google Scholar] [CrossRef] [Green Version]
- Gonçalves, J.L.; Kamphuis, C.; Martins, C.M.M.R.; Barreiro, J.R.; Tomazi, T.; Gameiro, A.H.; Hogeveen, H.; dos Santos, M.V. Bovine subclinical mastitis reduces milk yield and economic return. Livest. Sci. 2018, 210, 25–32. [Google Scholar] [CrossRef]
- Dyson, R.; Charman, N.; Hodge, A.; Rowe, S.; Taylor, L. A survey of mastitis pathogens including antimicrobial susceptibility in southeastern Australian dairy herds. J. Dairy Sci. 2022, 105, 1504–1518. [Google Scholar] [CrossRef] [PubMed]
- Sharun, K.; Dhama, K.; Tiwari, R.; Gugjoo, M.B.; Yatoo, M.I.; Patel, S.K.; Pathak, M.; Karthik, K.; Khurana, S.K.; Singh, R.; et al. Advances in therapeutic and managemental approaches of bovine mastitis: A comprehensive review. Veter.-Q. 2021, 41, 107–136. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, S.; Shang, X.; Wang, X.; Yan, Z.; Li, H.; Li, J. Short communication: Antimicrobial resistance and virulence genes of Enterococcus faecalis isolated from subclinical bovine mastitis cases in China. J. Dairy Sci. 2019, 102, 140–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, Z.; Zhang, W.; Song, Y.; Liu, W.; Xu, H.; Xi, X.; Menghe, B.; Zhang, H.; Sun, Z. Comparative genomic analysis of the genus Enterococcus. Microbiol. Res. 2017, 196, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Torres, C.; Alonso, C.A.; Ruiz-Ripa, L.; León-Sampedro, R.; Del Campo, R.; Coque, T.M. Antimicrobial Resistance in Enterococcus spp. of animal origin. Microbiol. Spectr. 2018, 6, 185–227. [Google Scholar] [CrossRef] [PubMed]
- Eggesbø, M.; Moen, B.; Peddada, S.; Baird, D.; Rugtveit, J.; Midtvedt, T.; Bushel, P.R.; Sekelja, M.; Rudi, K. Development of gut microbiota in infants not exposed to medical interventions. Apmis 2011, 119, 17–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Oliveira, R.P.; Aragão, B.B.; de Melo, R.P.B.; da Silva, D.M.S.; de Carvalho, R.G.; Juliano, M.A.; Farias, M.P.O.; de Lira, N.S.C.; Mota, R.A. Bovine mastitis in northeastern Brazil: Occurrence of emergent bacteria and their phenotypic and genotypic profile of antimicrobial resistance. Comp. Immunol. Microbiol. Infect. Dis. 2022, 85, 101802. [Google Scholar] [CrossRef]
- Różańska, H.; Lewtak-Piłat, A.; Kubajka, M.; Weiner, M. Occurrence of enterococci in mastitic cow’s milk and their antimicrobial resistance. J. Veter.-Res. 2019, 63, 93–97. [Google Scholar] [CrossRef] [Green Version]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Lebreton, F.; Willems, R.J.L.; Gilmore, M.S. Enterococcus Diversity, Origins in Nature, and Gut Colonization; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Frieri, M.; Kumar, K.; Boutin, A. Antibiotic resistance. J. Infect. Public Health 2017, 10, 369–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selleck, E.M.; Van Tyne, D.; Gilmore, M.S. Pathogenicity of enterococci. Microbiol. Spectr. 2019, 7, 7. [Google Scholar] [CrossRef]
- Raza, T.; Ullah, S.R.; Mehmood, K.; Andleeb, S. Vancomycin resistant Enterococci: A brief review. J. Pak. Med. Assoc. 2018, 68, 768–772. [Google Scholar] [PubMed]
- Kim, H.-J.; Youn, H.-Y.; Kang, H.-J.; Moon, J.-S.; Jang, Y.-S.; Song, K.-Y.; Seo, K.-H. Prevalence and Virulence Characteristics of Enterococcus faecalis and Enterococcus faecium in Bovine Mastitis Milk Compared to Bovine Normal Raw Milk in South Korea. Animals 2022, 12, 1407. [Google Scholar] [CrossRef]
- Kiruthiga, A.; Padmavathy, K.; Shabana, P.; Naveenkumar, V.; Gnanadesikan, S.; Malaiyan, J. Improved detection of esp, hyl, asa1, gelE, cylA virulence genes among clinical isolates of Enterococci. BMC Res. Notes 2020, 13, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Aung, M.S.; Urushibara, N.; Kawaguchiya, M.; Ohashi, N.; Hirose, M.; Kudo, K.; Tsukamoto, N.; Ito, M.; Kobayashi, N. Antimicrobial Resistance, Virulence Factors, and Genotypes of Enterococcus faecalis and Enterococcus faecium Clinical Isolates in Northern Japan: Identification of optrA in ST480 E. faecalis. Antibiotics 2023, 12, 108. [Google Scholar] [CrossRef]
- El-Zamkan, M.A.; Mohamed, H.M.A. Antimicrobial resistance, virulence genes and biofilm formation in Enterococcus species isolated from milk of sheep and goat with subclinical mastitis. PLoS ONE 2021, 16, e0259584. [Google Scholar] [CrossRef]
- Stogios, P.J.; Savchenko, A. Molecular mechanisms of vancomycin resistance. Protein Sci. 2020, 29, 654–669. [Google Scholar] [CrossRef]
- Yoon, S.; Lee, Y.J. Molecular Characteristics of Enterococcus faecalis and Enterococcus faecium from Bulk Tank Milk in Korea. Animals 2021, 11, 661. [Google Scholar] [CrossRef]
- Margalho, L.P.; van Schalkwijk, S.; Bachmann, H.; Sant’ana, A.S. Enterococcus spp. in Brazilian artisanal cheeses: Occurrence and assessment of phenotypic and safety properties of a large set of strains through the use of high throughput tools combined with multivariate statistics. Food Control 2020, 118, 107425. [Google Scholar] [CrossRef]
- Výrostková, J.; Regecová, I.; Dudriková, E.; Marcinčák, S.; Vargová, M.; Kováčová, M.; Maľová, J. Antimicrobial Resistance of Enterococcus sp. Isolated from Sheep and Goat Cheeses. Foods 2021, 10, 1844. [Google Scholar] [CrossRef] [PubMed]
- Addis, M.F.; Tanca, A.; Uzzau, S.; Oikonomou, G.; Bicalho, R.C.; Moroni, P. The bovine milk microbiota: Insights and perspectives from -omics studies. Mol. Biosyst. 2016, 12, 2359–2372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaczorowski, Ł.; Powierska-Czarny, J.; Wolko, Ł.; Piotrowska-Cyplik, A.; Cyplik, P.; Czarny, J. The Influence of Bacteria Causing Subclinical Mastitis on the Structure of the Cow’s Milk Microbiome. Molecules 2022, 27, 1829. [Google Scholar] [CrossRef] [PubMed]
- CLSI M100-S24; Performance Standards for Antimicrobial Testing Susceptibility. 32nd ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022; Volume 32 M100, ISBN 978-1-68440-135-2.
- Fernandes, S.C.; Dhanashree, B. Drug resistance & virulence determinants in clinical isolates of Enterococcus species. Indian J. Med. Res. 2013, 137, 981–985. [Google Scholar]
- Jahansepas, A.; Aghazadeh, M.; Rezaee, M.A.; Hasani, A.; Sharifi, Y.; Aghazadeh, T.; Mardaneh, J. Occurrence of Enterococcus faecalis and Enterococcus faecium in Various Clinical Infections: Detection of Their Drug Resistance and Virulence Determinants. Microb. Drug Resist. 2018, 24, 76–82. [Google Scholar] [CrossRef]
- Nasaj, M.; Mousavi, S.M.; Hosseini, S.M.; Arabestani, M.R. Prevalence of Virulence Factors and Vancomycin-resistant Genes among Enterococcus faecalis and E. faecium Isolated from Clinical Specimens. Iran. J. Public Health 2016, 45, 806–813. [Google Scholar]
- Aslam, M.; Diarra, M.S.; Service, C.; Rempel, H. Characterization of Antimicrobial Resistance in Enterococcus Spp. Recovered from a Commercial Beef Processing Plant. Foodborne Pathog. Dis. 2010, 7, 235–241. [Google Scholar] [CrossRef]
- Enne, V.I.; Delsol, A.A.; Roe, J.M.; Bennett, P.M. Rifampicin resistance and its fitness cost in Enterococcus faecium. J. Antimicrob. Chemother. 2004, 53, 203–207. [Google Scholar] [CrossRef] [Green Version]
- Deshpande, L.M.; Fritsche, T.R.; Moet, G.J.; Biedenbach, D.J.; Jones, R.N. Antimicrobial resistance and molecular epidemiology of vancomycin-resistant enterococci from North America and Europe: A report from the SENTRY antimicrobial surveillance program. Diagn. Microbiol. Infect. Dis. 2007, 58, 163–170. [Google Scholar] [CrossRef]
- Sharifi, Y.; Hasani, A.; Ghotaslou, R.; Naghili, B.; Aghazadeh, M.; Milani, M.; Bazmany, A. Virulence and Antimicrobial Resistance in Enterococci Isolated from Urinary Tract Infections. Adv. Pharm. Bull. 2013, 3, 197–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontana, R.; Aldegheri, M.; Ligozzi, M.; Lopez, H.; Sucari, A.; Satta, G. Overproduction of a low-affinity penicillin-binding protein and high-level ampicillin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 1994, 38, 1980–1983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zapun, A.; Contreras-Martel, C.; Vernet, T. Penicillin-binding proteins and β-lactam resistance. FEMS Microbiol. Rev. 2008, 32, 361–385. [Google Scholar] [CrossRef] [Green Version]
- Arias, C.; Contreras, G.; Murray, B. Management of multidrug-resistant enterococcal infections. Clin. Microbiol. Infect. 2010, 16, 555–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Hou, S.; Zhang, Q.; Ma, Y.; Zhang, Y.; Kan, W.; Zhao, X. Prevalence of virulence and resistance to antibiotics in pathogenic enterococci isolated from mastitic cows. J. Vet.-Med. Sci. 2016, 78, 1663–1668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, W.R.; Munita, J.M.; Arias, A.C. Mechanisms of antibiotic resistance in enterococci. Expert Rev. Anti-Infect. Ther. 2014, 12, 1221–1236. [Google Scholar] [CrossRef] [PubMed]
- WHO. Global Action Plan on Antimicrobial Resistance 2017; WHO: Geneva, Switzerland, 2015.
- Cortés, C.; Delafuente, R.; Contreras, A.; Sánchez, A.; Corrales, J.; Ruizsantaquiteria, J.; Orden, J. Occurrence and preliminary study of antimicrobial resistance of enterococci isolated from dairy goats in Spain. Int. J. Food Microbiol. 2006, 110, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Mannu, L.; Paba, A.; Daga, E.; Comunian, R.; Zanetti, S.; Duprè, I.; Sechi, L. Comparison of the incidence of virulence determinants and antibiotic resistance between Enterococcus faecium strains of dairy, animal and clinical origin. Int. J. Food Microbiol. 2003, 88, 291–304. [Google Scholar] [CrossRef]
- Ahmed, M.O.; Baptiste, K.E. Vancomycin-Resistant Enterococci: A Review of Antimicrobial Resistance Mechanisms and Perspectives of Human and Animal Health. Microb. Drug Resist. 2018, 24, 590–606. [Google Scholar] [CrossRef] [Green Version]
- Xanthopoulou, K.; Peter, S.; Tobys, D.; Behnke, M.; Dinkelacker, A.G.; Eisenbeis, S.; Falgenhauer, J.; Falgenhauer, L.; Fritzenwanker, M.; Gölz, H.; et al. Vancomycin-resistant Enterococcus faecium colonizing patients on hospital admission in Germany: Prevalence and molecular epidemiology. J. Antimicrob. Chemother. 2020, 75, 2743–2751. [Google Scholar] [CrossRef] [PubMed]
- ECDC. Surveillance of Antimicrobial Resistance in Europe 2018; European Centre for Disease Prevention and Control: Stockholm, Sweden, 2019. [CrossRef]
- Martín-Platero, A.M.; Valdivia, E.; Maqueda, M.; Martínez-Bueno, M. Characterization and safety evaluation of enterococci isolated from Spanish goats’ milk cheeses. Int. J. Food Microbiol. 2009, 132, 24–32. [Google Scholar] [CrossRef]
- Song, H.; Bae, Y.; Jeon, E.; Kwon, Y.; Joh, S. Multiplex PCR analysis of virulence genes and their influence on antibiotic resistance in Enterococcus spp. isolated from broiler chicken. J. Vet.-Sci. 2019, 20, e26. [Google Scholar] [CrossRef]
- Perin, L.M.; Miranda, R.O.; Todorov, S.D.; Franco, B.D.G.d.M.; Nero, L.A. Virulence, antibiotic resistance and biogenic amines of bacteriocinogenic lactococci and enterococci isolated from goat milk. Int. J. Food Microbiol. 2014, 185, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.R.; Fedorka-Cray, P.J.; Barrett, J.B. Use of a Genus- and Species-Specific Multiplex PCR for Identification of Enterococci. J. Clin. Microbiol. 2004, 42, 3558–3565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barcelos, M.M.; Martins, L.; Grenfell, R.C.; Juliano, L.; Anderson, K.L.; dos Santos, M.V.; Gonçalves, J.L. Comparison of standard and on-plate extraction protocols for identification of mastitis-causing bacteria by MALDI-TOF MS. Braz. J. Microbiol. 2019, 50, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Peltroche-Llacsahuanga, H.; Top, J.; Weber-Heynemann, J.; Lütticken, R.; Haase, G. Comparison of Two Chromogenic Media for Selective Isolation of Vancomycin-Resistant Enterococci from Stool Specimens. J. Clin. Microbiol. 2009, 47, 4113–4116. [Google Scholar] [CrossRef] [Green Version]
- Bauer, A.W.; Kirby, W.M.M.; Sherris, J.C.; Turck, M. Antibiotic Susceptibility Testing by a Standardized Single Disk Method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef]
- Vankerckhoven, V.; Van Autgaerden, T.; Vael, C.; Lammens, C.; Chapelle, S.; Rossi, R.; Jabes, D.; Goossens, H. Development of a Multiplex PCR for the Detection of asa1, gelE, cylA, esp, and hyl Genes in Enterococci and Survey for Virulence Determinants among European Hospital Isolates of Enterococcus faecium. J. Clin. Microbiol. 2004, 42, 4473–4479. [Google Scholar] [CrossRef] [Green Version]
- Eaton, T.J.; Gasson, M.J. Molecular Screening of Enterococcus Virulence Determinants and Potential for Genetic Exchange between Food and Medical Isolates. Appl. Environ. Microbiol. 2001, 67, 1628–1635. [Google Scholar] [CrossRef] [Green Version]
- Creti, R.; Imperi, M.; Bertuccini, L.; Fabretti, F.; Orefici, G.; Di Rosa, R.; Baldassarri, L. Survey for virulence determinants among Enterococcus faecalis isolated from different sources. J. Med. Microbiol. 2004, 53, 13–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Species | Isolation (n = 100) | Frequency (%) | Sample Type | |||
|---|---|---|---|---|---|---|
| Clinical Mastitis (n = 4) | Subclinical Mastitis (n = 34) | No Mastitis (Healthy) (n = 59) | Not Classified (n = 3) | |||
| E. casseliflavus | 1 | 1 | 0 | 0 | 1 | 0 |
| E. durans | 22 | 22 | 1 | 2 | 18 | 1 |
| E. faecalis | 22 | 22 | 1 | 11 | 9 | 1 |
| E. faecium | 26 | 26 | 2 | 8 | 15 | 1 |
| E. gallinarum | 11 | 11 | 0 | 5 | 6 | 0 |
| E. hirae | 13 | 13 | 0 | 4 | 9 | 0 |
| E. saccharolyticus | 5 | 5 | 0 | 4 | 1 | 0 |
| Total | 100 | 100 | 4 | 34 | 59 | 3 |
| Distribution of Strains that Displayed Susceptibility, Intermediate Resistance, or Resistance (%) | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antimicrobial 1 | E. faecalis (n = 22) | E. faecium (n = 26) | E. durans (n = 22) | E. hirae (n = 13) | E. casseliflavus (n = 1) | E. gallinarum (n = 11) | E. saccharolyticus (n = 5) | ||||||||||||||
| S | I | R | S | I | R | S | I | R | S | I | R | S | I | R | S | I | R | S | I | R | |
| PEN | 90.9 | 0 | 9.1 | 65.4 | 0 | 34.6 | 100 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 0 |
| VAN | 95.5 | 0 | 4.5 | 92.3 | 0 | 7.7 | 100 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 0 |
| TEI | 95.5 | 0 | 4.5 | 92.3 | 0 | 7.7 | 100 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 0 |
| ERY | 63.6 | 13.6 | 22.7 | 46.2 | 3.8 | 50 | 100 | 0 | 0 | 69.2 | 30.8 | 0 | 100 | 0 | 0 | 100 | 0 | 0 | 80 | 20 | 0 |
| TET | 68.2 | 4.5 | 27.3 | 50 | 3.8 | 46.2 | 90.9 | 9.1 | 0 | 92.3 | 7.7 | 0 | 100 | 0 | 0 | 9.1 | 72.7 | 18.2 | 80 | 20 | 0 |
| CIP | 77.3 | 18.2 | 4.5 | 84.6 | 15.4 | 0 | 95.5 | 0 | 4.5 | 100 | 0 | 0 | 100 | 0 | 0 | 36.4 | 63.6 | 0 | 80 | 0 | 20 |
| NIT | 100 | 0 | 0 | 92.3 | 0 | 7.7 | 59.1 | 0 | 40.9 | 100 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 0 |
| RIF | 72.7 | 13.6 | 13.6 | 30.8 | 26.9 | 42.3 | 36.4 | 4.5 | 59.1 | 100 | 0 | 0 | 100 | 0 | 0 | 90.9 | 9.1 | 0 | 100 | 0 | 0 |
| CHL | 100 | 0 | 0 | 96.2 | 3.8 | 0 | 95.5 | 4.5 | 0 | 100 | 0 | 0 | 100 | 0 | 0 | 90.9 | 9.1 | 0 | 100 | 0 | 0 |
| LIN | 95.5 | 0 | 4.5 | 88.5 | 0 | 11.5 | 81.8 | 13.6 | 4.5 | 100 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 0 |
| Species | Virulence Genes (%) | ||||||
|---|---|---|---|---|---|---|---|
| cylA | esp | efaA | ace | asa1 | gelE | hyl | |
| E. caseliflavus (n = 1) | - | - | - | - | - | - | - |
| E. durans (n = 22) | - | - | - | - | - | - | - |
| E. faecalis (n = 22) | - | - | - | - | 8 (36%) | 10 (45.5%) | - |
| E. faecium (n = 26) | - | - | - | - | - | - | - |
| E. gallinarum (n = 11) | - | - | - | - | 1 (9.1%) | - | - |
| E. hirae (n = 13) | - | - | - | - | - | - | - |
| E. saccharolyticus (n = 5) | - | - | - | - | - | - | - |
| Gene | Primers Sequence (5′ to 3′) 1 | Product Size (bp) | Reference |
|---|---|---|---|
| gelE | gelE-F TATGACAATGCTTTTTGGGAT | 213 | [53] |
| gelE-R AGATGCACCCGAAATAATATA | |||
| hyl | hyl-F ACAGAAGAGCTGCAGGAAATG | 276 | [53] |
| hylR GACTGACGTCCAAGTTTCCAA | |||
| asa1 | asa1-F GCACGCTATTACGAACTATGA | 375 | [53] |
| asa1-R AAGAAAGAACATCACCACGA | |||
| esp | esp-F AGATTTCATCTTTGATTCTTGG | 510 | [53] |
| esp-R AATTGATTCTTTAGCATCTGG | |||
| cylA | cylA-F ACTCGGGGATTGATAGGC | 688 | [53] |
| cylA-R GCTGCTAAAGCTGCGCTT | |||
| efaA | efaA-F GACAGACCCTCACGAATA | 705 | [54] |
| efaA-R AGTTCATCATGCTGTAGTA | |||
| ace | ace-F GGAATGACCGAGAACGATGGC | 616 | [55] |
| ace-R GCTTGATGTTGGCCTGCTTCCG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paschoalini, B.R.; Nuñez, K.V.M.; Maffei, J.T.; Langoni, H.; Guimarães, F.F.; Gebara, C.; Freitas, N.E.; dos Santos, M.V.; Fidelis, C.E.; Kappes, R.; et al. The Emergence of Antimicrobial Resistance and Virulence Characteristics in Enterococcus Species Isolated from Bovine Milk. Antibiotics 2023, 12, 1243. https://doi.org/10.3390/antibiotics12081243
Paschoalini BR, Nuñez KVM, Maffei JT, Langoni H, Guimarães FF, Gebara C, Freitas NE, dos Santos MV, Fidelis CE, Kappes R, et al. The Emergence of Antimicrobial Resistance and Virulence Characteristics in Enterococcus Species Isolated from Bovine Milk. Antibiotics. 2023; 12(8):1243. https://doi.org/10.3390/antibiotics12081243
Chicago/Turabian StylePaschoalini, Beatriz Rizzo, Karen Vanessa Munive Nuñez, Juliana Takahashi Maffei, Hélio Langoni, Felipe Freitas Guimarães, Clarice Gebara, Natylane Eufransino Freitas, Marcos Veiga dos Santos, Carlos Eduardo Fidelis, Roberto Kappes, and et al. 2023. "The Emergence of Antimicrobial Resistance and Virulence Characteristics in Enterococcus Species Isolated from Bovine Milk" Antibiotics 12, no. 8: 1243. https://doi.org/10.3390/antibiotics12081243
APA StylePaschoalini, B. R., Nuñez, K. V. M., Maffei, J. T., Langoni, H., Guimarães, F. F., Gebara, C., Freitas, N. E., dos Santos, M. V., Fidelis, C. E., Kappes, R., Gonçalves, M. C., & Silva, N. C. C. (2023). The Emergence of Antimicrobial Resistance and Virulence Characteristics in Enterococcus Species Isolated from Bovine Milk. Antibiotics, 12(8), 1243. https://doi.org/10.3390/antibiotics12081243









