Nanosilver: An Old Antibacterial Agent with Great Promise in the Fight against Antibiotic Resistance
Abstract
:1. Brief History of Silver (Ag) and Its Old Antimicrobial Applications
2. Modern Antimicrobial Applications of Nanosilver
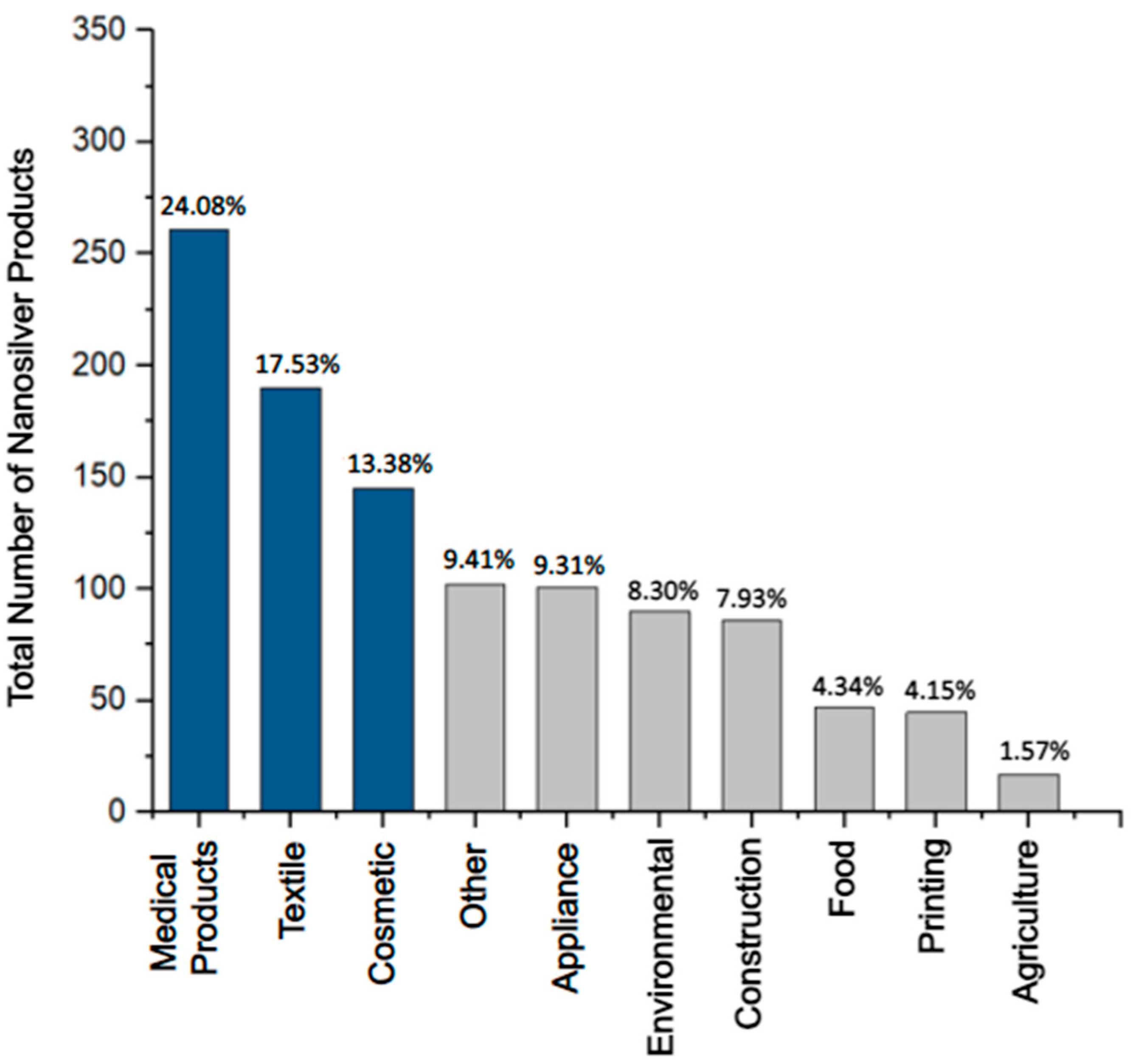
| Product Type | Search Result | Vendor | [Ag] and Ag Form | Purpose | U.S. FDA Approval |
|---|---|---|---|---|---|
| Silver-based wound dressings | “silver wound dressing” n = 2214 |
|
|
| YES |
| Ankle socks with silver | “silver textile” n = 1155 |
|
|
| NO |
| Platinum silver nanocolloid cream | “silver cosmetic” n = 2292 |
|
|
| NO |
3. Gram-Negative Bacteria (GNB) Versus Gram-Positive Bacteria (GPB) Models
4. Silver Nanoparticle (AgNP) Models
5. Antibacterial Mechanisms of AgNPs
5.1. Cell Membrane Damage
5.2. Cell DNA Damage
5.3. Collateral Cell Damage
6. Antibiotics against Bacteria
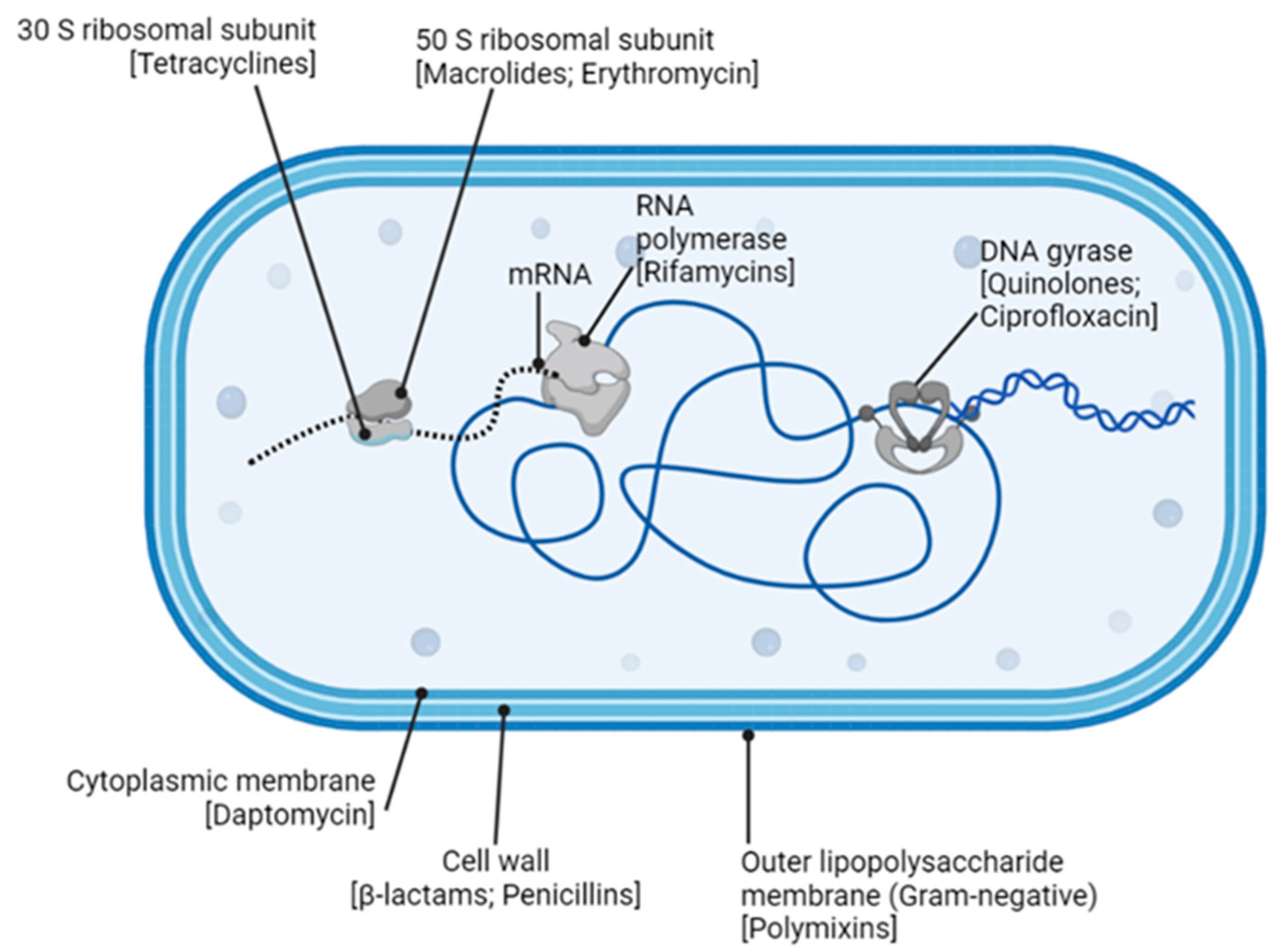
7. Mechanisms of Antibiotic Resistance
8. Synergistic Effects of AgNP-Antibiotic Conjugates on Antibiotic-Resistant Bacteria
| Fabrication Method and Reducing Agent | AgNP Size and Coating | Synergy, Bacterial Model, and Method of Evaluation |
|---|---|---|
| Biological Bacillus sp. [260] | 14–42 nm primary and aromatic amines | ZoI; all combinations of fusidic acid, gentamycin, ciprofloxacin, erythromycin, penicillin, chloramphenicol, levofloxacin, nalidixic acid, and ampicillin against S. epidermidis, S. aureus, V. cholerae, S. aureus, Salmonella Typhi, and Salmonella Paratyphi |
| Chemical [261] | 10–30 nm nanosilver colloid | MIC; allicin against MRSA |
| Biological Trichoderma viride Aspergillus flavus [262,263] | 5–40 nm | ZoI; ampicillin, kanamycin, erythromycin, and chloramphenicol against E. coli, S. Typhi, S. aureus, Micrococcus luteus, P. aeruginosa, E. faecalis, A. baumanii, K. pneumoniae, and Bacillus spp. |
| Biological Phytophthora Infestans [264] | 5–30 nm | ZoI, MIC, mupirocin, neomycin, vancomycin against S aureus; cefazolin, mupirocin, gentamycin, vancomycin against P. aeruginosa; and cefazolin, mupirocin, gentamycin, neomycin, tetracycline against E. coli. |
| Chemical Maltose [265] | 28 nm and 8 nm | MIC; amoxycillin, colistin, and gentamycin against A. pleuropneumoniae, and P. multocida. Penicillin G against A. pleuropneumoniae |
| Biological Mukia Maderaspatana [266] | N. A. | ZoI; biofilm microplate; cefriaxone with B. subtilis, K. pneumoniae, S. aureus, S. Typhi, and Pseudomonas fluorescens |
| Chemical ascorbic acid [267] | 20 nm | MIC; amoxicillin against E. coli |
| Biological E. hermannii, C. sedlakii, and P. putida [268] | 4–12 nm | ZoI; gentamicin against P. aeruginosa and vancomycin against S. aureus and MRSA |
| Chemical solid silver [269] | N. A. | MIC, FIC, biofilm, and hydroxyl radical assay; E. faecium, S. mutans, and E. Coli with ampicillin; E. faecium and P. aeruginosa with chloramphenicol; S. aureus, S. mutans, E. coli, and P. aeruginosa with kanamycin |
| Biological K. pneumoniae [270] | 50 nm | ZoI; penicillin G, amoxicillin, erythromycin, clindamycin, and vancomycin against S. aureus and E. coli |
| Biological Dioscorea bulbifera [271] | 8–20 nm | ZoI; chloramphenicol and vancomycin against P. aeruginosa, streptomycin with E. coli |
| Chemical sodium citrate and garlic [272] | - citrate-coated | ZoI; S. Typhi, E. coli, P. aeruginosa, M. luteus, S. aureus with amoxclav and S. Typhi with ampicillin |
| Chemical [273] | 3.0 nm | FI; E. faecium, ampicillin and chloramphenicol; S. mutans, ampicillin and kanamycin; E. coli, ampicillin and kanamycin; P. aeruginosa, chloramphenicol and kanamycin |
| Chemical NaBH4/citrate [274] | 5.0−12.0 nm citrate-coated | A. baumannii with polymyxin B and rifampicin |
| Chemical NaBH4/maltose [265] | 8.0 nm gelatin-coated | FIC; A. pleuropneumoniae, penicillin G; E. coli, colistin; S. aureus, gentamicin |
| Chemical Gallic acid [275] | 8.6 nm Gallic-acid-coated | FIC; E. faecium, A. baumannii, K. pneumoniae, Morganella morganii, and P. aeruginosa, ampicillin and amikacin; S. aureus, E. coli, and Enterobacter cloacae, amikacin (FIC) |
| Chemical NaBH4/citrate/hydrazine [276] | 10 nm PVP- and citrate-coated | ZoI; S. aureus, cephalexin |
| Chemical NaBH4/citrate [277] | 16 nm PVP-coated | ZoI; E. coli, streptomycin, ampicillin, and tetracycline; S. aureus, streptomycin, ampicillin, and tetracycline |
| Chemical NaBH4/citrate [278] | 19.3 nm SDS-coated | ZoI; E. coli, streptomycin, ampicillin, and tetracycline; S. aureus, streptomycin, ampicillin, and tetracycline |
| Chemical ascorbic acid [279] | 20.0 nm | MIC; E. coli, amoxicillin |
| Chemical NaBH4 [280] | 20.0 nm PVP-coated | ZoI; all combinations of vancomycin and amikacin and S. aureus and E. coli |
| Chemical Tween 80 [281] | 20.0–40.0 nm Tween 80-coated | FIC; S. epidermidis and gentamicin |
| Chemical citrate [246] | 23.0 nm citrate-coated | MIC and inhibition (plate counting); S. Typhimurium, tetracycline, neomycin, and penicillin G |
| Chemical ethylene glycol [251] | 25.0 nm PVP-coated | FIC; E. coli and S. aureus, gentamicin |
| Chemical Maltose [282] | 26.0 nm gelatin | MIC; E. coli, ampicillin, ampicillin/sulbactam, aztreonam, cefazolin, cefoxitin, cefuroxime, cotrimoxazole, colistin, gentamicin, ofloxacin, oxolinic acid, and tetracycline; P. aeruginosa, amikacin, aztreonam, cefepime, cefoperazone, ceftazidime, ciprofloxacin, colistin, gentamicin, meropenem, ofloxacin, piperacillin, and piperacillin/tazobactam; S. aureus, ampicillin/sulbactam, chloramphenicol, ciprofloxacin, clindamycin, cotrimoxazole, erythromycin, gentamicin, oxacillin, penicillin, teicoplanin, tetracycline, and vancomycin |
| Chemical NaBH4/maltose [265] | 28.0 nm gelatin | Amoxycillin, penicillin G, gentamicin, and colistin |
| Chemical Maltose [283] | 28.0 nm Maltose | FIC; E. coli and K. pneumoniae with cefotaxime, ceftazidime, meropenem, ciprofloxacin, and gentamicin; synergism in all resistant strains except to K. pneumonia carbapenemase |
| Chemical Citrate [253] | 29.8 nm citrate-coated | Ampicillin, penicillin, enoxacin, kanamycin, neomycin, and tetracycline, and S. Typhimurium |
| Chemical Citrate [284] | 29.8 nm citrate-coated | Inhibition (plate counting); S. Typhimurium, enoxacin, kanamycin, neomycin, and tetracycline |
| Chemical NaBH4/citrate [271] | 38.3 nm citrate-coated | ZoI; E. coli, streptomycin, ampicillin, and tetracycline; S. aureus, streptomycin, ampicillin, and tetracycline |
| Chemical Citrate [251] | 70.0 nm Citrate-coated | ZoI; vancomycin and S. aureus and E. coli |
| Biological Streptomyces cali- diresistants IF17 strain [284] | 5.0–20.0 nm biomolecules from actinobacterial strains | FIC: E. coli, tetracycline; S. aureus, ampicillin, kanamycin, and tetracycline; B. subtilis, ampicillin, kanamycin, and tetracycline |
| Biological Klebsiella pneumoniae extract [271] | 5.0–32.0 nm proteins from biomass | ZoI; E. coli, amoxicillin, erythromycin, penicillin, and vancomycin; S. aureus, amoxicillin, erythromycin, penicillin, and vancomycin |
| Biological Streptomyces calidiresistants IF11 [284] | 5.0–50.0 nm biomolecules from reducing strains | FIC; B. subtilis, kanamycin |
| Biological Actinomycetes strains [277] | 17.0 nm proteins from biomass | MIC and ZoI; E. coli, K. pneumoniae, and P. aeruginosa, ampicillin |
| Biological Klebsiella pneumoniae [285] | 20.0 nm - | ZoI; E. faecalis, chloramphenicol and gentamicin |
| Biological silver-resistant estuarine P. aeruginosa strain [286] | 35.0–60.0 nm biomolecules from reducing strains | ZoI; all combinations of ampicillin and ciprofloxacin with resistant S. aureus strain VN3 and ciprofloxacin-resistant V. cholera strain VN1 |
| Biological Trichoderma viride [287] | 5.0–40.0 nm proteins from biomass | ZoI; E. coli, ampicillin, chloramphenicol, erythromycin, and kanamycin; M. luteus, ampicillin, chloramphenicol, and kanamycin; S. Typhi, ampicillin, chloramphenicol, erythromycin, and kanamycin; S. aureus, ampicillin, chloramphenicol, erythromycin, and kanamycin |
| Biological Acinetobacter calcoaceticus [288] | 8.0–12.0 nm biomolecules from reducing strains | ZoI or MIC; A. baumannii, amikacin, amoxicillin, ampicillin, chloramphenicol, ciprofloxacin, doxycycline, gentamicin, tetracycline, trimethoprim, and vancomycin; Klebsiella (previously known as Enterobacter) aerogenes, amikacin, amoxicillin, ampicillin, ceftriaxone, chloramphenicol, ciprofloxacin, doxycycline, gentamicin, kanamycin, penicillin, tetracycline, trimethoprim, and vancomycin; E. coli, amikacin, amoxicillin, ampicillin, ceftazidime, ceftriaxone, chloramphenicol, ciprofloxacin, doxycycline, gentamicin, kanamycin, penicillin, tetracycline, trimethoprim, and vancomycin; P. aeruginosa, amikacin, amoxicillin, ampicillin, ceftazidime, ceftriaxone, chloramphenicol, ciprofloxacin, doxycycline, gentamicin, kanamycin, penicillin, tetracycline, trimethoprim, and vancomycin; S. Typhimurium, amikacin, ampicillin, ceftazidime, ceftriaxone, chloramphenicol, ciprofloxacin, doxycycline, gentamicin, kanamycin, penicillin, tetracycline, trimethoprim, and vancomycin; Shigella sonnei, amikacin, amoxicillin, ampicillin, ceftazidime, ceftriaxone, ciprofloxacin, chloramphenicol, ciprofloxacin, doxycycline, gentamicin, kanamycin, tetracycline, trimethoprim, and vancomycin; S. aureus, amikacin, amoxicillin, ampicillin, ceftazidime, ceftriaxone, chloramphenicol, ciprofloxacin, doxycycline, gentamicin, kanamycin, penicillin, tetracycline, trimethoprim, and vancomycin; S. mutans, amikacin, amoxicillin, ampicillin, ceftazidime, ceftriaxone, chloramphenicol, ciprofloxacin, doxycycline, kanamycin, penicillin, tetracycline, trimethoprim, and vancomycin |
| Biological Cryphonectria sp. [289] | 30–70 nm | ZoI; S. aureus, S. Typhi, and E. coli, streptomycin |
| Biological Emericella nidulans [290] | 66.7 nm biomolecules from biomass | FIC; E. coli, amikacin and streptomycin |
| Biological Aspergillus flavus [290] | 81.1 nm biomolecules from biomass | FIC; E. coli, amikacin and streptomycin; S. aureus, kanamycin, oxytetracycline, and streptomycin |
| Biological Dioscorea bulbfera [272] | 2.0 nm biomolecules from biomass | ZoI; all combinations of treptomycin, rifampicin, chloramphenicol, novobiocin, and ampicillin in E. coli, P. aeruginosa, and S. aureus |
| Biological Dioscorea bulbifera [291] | 5.0–30.0 nm proteins from biomass | ZoI; A. baumannii, amoxicillin, ampicillin, cefotaxime, erythromycin, gentamycin, kanamycin, nalidixic acid, nitrofurantoin, penicillin, piperacillin, rifampicin, and rimethoprim; B. subtilis, ampicillin, cefotaxime, chloramphenicol, nalidixic acid, nitrofurantoin, penicillin, piperacillin, streptomycin, trimethoprim, and vancomycin; E. cloacae, amikacin, amoxicillin, erythromycin, nalidixic acid, and penicillin; E. coli, amikacin, erythromycin, kanamycin, nalidixic acid, polymyxin, streptomycin, and trimethoprim; Haemophilus influenzae, cefotaxime, ceftriaxone, nitrofurantoin, and trimethoprim; K. pneumoniae, amoxicillin, ampicillin, chloramphenicol, erythromycin, feropenem, nitrofurantoin, penicillin, rifampicin, trimethoprim, and vancomycin; Neisseria mucosa, amikacin, ampicillin, erythromycin, feropenem, gentamycin, nitrofurantoin, penicillin, polymyxin, tetracycline, trimethoprim, and vancomycin; Proteus mirabilis, erythromycin, nalidixic acid, and vancomycin; P. aeruginosa, amikacin, amoxicillin, ampicillin, chloramphenicol, doxycycline, erythromycin, feropenem, gentamycin, kanamycin, nalidixic acid, nitrofurantoin, penicillin, streptomycin, trimethoprim, and vancomycin; S. Typhi, amikacin, amoxicillin, ampicillin, cefotaxime, ceftriaxone, chloramphenicol, erythromycin, gentamycin, kanamycin, nalidixic acid, nitrofurantoin, penicillin, piperacillin, polymyxin, streptomycin, trimethoprim, and vancomycin; Serratia odorifera, ceftazidme, erythromycin, nalidixic acid, nitrofurantoin, trimethoprim, and vancomycin; S. aureus, amikacin, amoxicillin, ampicillin, ceftazidme, erythromycin, kanamycin, nalidixic acid, polymyxin, streptomycin, and trimethoprim; Vibrio parahemolyticus, ampicillin, cefotaxime, ceftriaxone, kanamycin, nalidixic acid, nitrofurantoin, polymyxin, and trimethoprim |
| Biological Argyreia nervosa [292] | 5.0–40.0 nm biomolecules from biomass | ZoI; S. aureus, amoxicillin/clavulamic acid, ciprofloxacin, erythromycin, gentamicin, streptomycin, tetracycline, and vancomycin; E. coli, amoxicillin/clavulamic acid, erythromycin, streptomycin, tetracycline, and vancomycin |
| Biological Gum kondagogu [293] | 5.8 nm biomolecules from biomass | FIC; S. aureus, gentamicin and streptomicin; S. aureus, streptomicin; E. coli, streptomicin; P. aeruginosa, streptomicin |
| Biological Rosa damascenes [294] | 7.4–18.3 nm | ZoI; cefotaxime with E. coli and MRSA |
| Biological Ulva fasciata [295] | 15.0 nm | ZoI; E. coli, cefotaxime, cefuroxime, fosfomycin, chloramphenicol, azithromycin, and gentamicin; Salmonella enterica, azithromycin, gentamicin, oxacillin, cefotaxime, neomycin, ampicillin/sulbactam, cefuroxime, fosfomycin, chloramphenicol, and oxytetracycline; S. aureus, azithromycin, oxacillin, cefotaxime, neomycin, ampicillin/sulbactam, cefuroxime, fosfomycin, chloramphenicol, and oxytetracycline |
| Biological Eurotium cristatum [296] | 15.0–20.0 nm biomolecules from biomass | ZoI; all combinations of vancomycin, oleandomycin, ceftazidime, rifampicin, penicillin G, neomycin, cephazolin, novobiocin, carbenicillin, lincomycin, tetracycline, and erythromycin, and Candida albicans, P. aeruginosa, and E. coli |
| Biological Urtica dioica Linn. [297] | 20.0–30.0 nm biomolecules from biomass | ZoI; B. cereus, streptomycin, amikacin, kanamycin, vancomycin, tetracycline, ampicillin, cefepime, amoxicillin, and cefotaxime; S. epidermidis, streptomycin, amikacin, kanamycin, tetracycline, ampicillin, cefepime, and amoxicillin; S. aureus, streptomycin, amikacin, kanamycin, vancomycin, tetracycline, cefepime, amoxicillin, and cefotaxime; B. subtilis, streptomycin, amikacin, kanamycin, vancomycin, tetracycline, ampicillin, cefepime, amoxicillin, and cefotaxime; E. coli, streptomycin, amikacin, vancomycin, tetracycline, ampicillin, cefepime, amoxicillin, and cefotaxime; S. Typhimurium, streptomycin, amikacin, kanamycin, vancomycin, tetracycline, ampicillin, cefepime, amoxicillin, and cefotaxime; K. pneumoniae, streptomycin, amikacin, kanamycin, vancomycin, tetracycline, ampicillin, cefepime, amoxicillin, and cefotaxime; Serratia marcescens, streptomycin, kanamycin, tetracycline, ampicillin, amoxicillin, and cefotaxime |
| Biological Zea may [298] | 45.3 nm biomolecules from biomass | ZoI; B. cereus, E. coli, Listeria. monocytogenes, Salmonella Typhimurium, and S. aureus, kanamycin and rifampicin |
| Commercial N. A. [299] | 10.0–15.0 nm N. A. | Optical density; ampicillin, kanamycin, gentamycin, and clindamycin with A. baumannii |
| Commercial N. A. [300] | 15.2 nm starch | FIC; Burkholderia pseudomallei with meropenem and gentamicin sulfate |
| Commercial N. A. [301] | 35.0 nm PVP | FIC; E. coli, Salmonella Typhimurium, and S. aureus, kanamycin |
9. Toxicity of AgNPs
9.1. In Vitro Studies
9.2. In Vivo Studies
9.3. Human Studies
10. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alexander, J.W. History of the medical use of silver. Surg. Infect. 2009, 10, 289. [Google Scholar] [CrossRef] [Green Version]
- Lane, N. The unseen world: Reflections on Leeuwenhoek (1677) ‘Concerning Little Animals’. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140344. [Google Scholar] [CrossRef] [PubMed]
- Lobanovska, M.; Pilla, G. Penicillin’s Discovery and Antibiotic Resistance: Lessons for the Future? Yale J. Biol. Med. 2017, 90, 135–145. [Google Scholar]
- Barillo, D.J.; Marx, D.E. Silver in medicine: A brief history BC 335 to present. Burns 2014, 40, S3–S8. [Google Scholar] [CrossRef] [PubMed]
- Mijnendonckx, K.; Leys, N.; Mahillon, J.; Silver, S.; Van Houdt, R. Antimicrobial silver: Uses, toxicity and potential for resistance. Biometals 2013, 26, 609–621. [Google Scholar] [CrossRef] [PubMed]
- Barras, F.; Aussel, L.; Ezraty, B. Silver and Antibiotic, New Facts to an Old Story. Antibiotics 2018, 7, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, R.J. An historical overview of the use of silver in wound management. Br. J. Nurs. 2001, 10, 16079. [Google Scholar] [CrossRef]
- Medici, S.; Peana, M.; Nurchi, V.M.; Zoroddu, M.A. Medical Uses of Silver: History, Myths, and Scientific Evidence. J. Med. Chem. 2019, 62, 5923–5943. [Google Scholar] [CrossRef]
- Turner, R.J. Is Silver the Ultimate Antimicrobial Bullet? Antibiotics 2018, 7, 112. [Google Scholar] [CrossRef] [Green Version]
- Wallner, C.; Moormann, E.; Lulof, P.; Drysch, M.; Lehnhardt, M.; Behr, B. Burn Care in the Greek and Roman Antiquity. Medicina 2020, 56, 657. [Google Scholar] [CrossRef] [PubMed]
- Davies, R.L.; Etris, S.F. The development and functions of silver in water purification and disease control. Catal. Today 1997, 36, 107–114. [Google Scholar] [CrossRef]
- Sim, W.; Barnard, R.T.; Blaskovich, M.A.T.; Ziora, Z.M. Antimicrobial Silver in Medicinal and Consumer Applications: A Patent Review of the Past Decade (2007–2017). Antibiotics 2018, 7, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lara, H.H.; Garza-Treviño, E.N.; Ixtepan-Turrent, L.; Singh, D.K. Silver nanoparticles are broad-spectrum bactericidal and virucidal compounds. J. Nanobiotechnol. 2011, 9, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dingledine, R.; Hassel, B. A New Approach for Epilepsy. Cerebrum 2016, 2016, cer-07-16. [Google Scholar] [PubMed]
- Borsuk, D.E.; Gallant, M.; Richard, D.; Williams, H.B. Silver-coated nylon dressings for pediatric burn victims. Can. J. Plast. Surg. 2007, 15, 29–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernigou, P. Ambroise Paré’s life (1510–1590): Part I. Int. Orthop. 2013, 37, 543–547. [Google Scholar] [CrossRef] [Green Version]
- Jerger, S.E.; Parekh, U. Argyria. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Park, S.W.; Shin, H.T.; Lee, K.T.; Lee, D.Y. Medical concern for colloidal silver supplementation: Argyria of the nail and face. Ann. Dermatol. 2013, 25, 111–112. [Google Scholar] [CrossRef] [Green Version]
- Yano, M.; Furukawa, Y. Two-dimensional constrained chaos and industrial revolution cycles. Proc. Natl. Acad. Sci. USA 2023, 120, e2117497120. [Google Scholar] [CrossRef]
- Landecker, H. Antimicrobials before antibiotics: War, peace, and disinfectants. Palgrave Commun. 2019, 5, 45. [Google Scholar] [CrossRef] [Green Version]
- Fiani, B.; Covarrubias, C.; Jarrah, R.; Kondilis, A.; Doan, T.M. Bellevue Hospital, the Oldest Public Health center in the United States of America. World Neurosurg. 2022, 167, 57–61. [Google Scholar] [CrossRef]
- CDC.gov. Available online: https://www.cdc.gov/smallpox/history/history.html#:~:text=Edward%20Jenner%20(1749%E2%80%931823 (accessed on 25 June 2023).
- Riedel, S. Edward Jenner and the history of smallpox and vaccination. In Baylor University Medical Center Proceedings; Taylor & Francis: New York, NY, USA, 2005; Volume 18, pp. 21–25. [Google Scholar]
- Sakai, T.; Morimoto, Y. The History of Infectious Diseases and Medicine. Pathogens 2022, 11, 1147. [Google Scholar] [CrossRef]
- National Research Council (US) Committee to Update Science, Medicine, and Animals. A Theory of Germs. In Science, Medicine, and Animals, 2nd ed.; National Academy Press: Washington, DC, USA, 2004; pp. 7–8. [Google Scholar]
- Casanova, J.-L.; Abel, L. The Genetic Theory of Infectious Diseases: A Brief History and Selected Illustrations. Annu. Rev. Genom. Hum. Genet. 2016, 14, 215–243. [Google Scholar] [CrossRef] [Green Version]
- Christensen, S.B. Drugs That Changed Society: History and Current Status of the Early Antibiotics: Salvarsan, Sulfonamides, and β-Lactams. Molecules 2021, 26, 6057. [Google Scholar] [CrossRef]
- Williams, K.J. The introduction of ‘chemotherapy’using arsphenamine–The first magic bullet. J. R. Soc. Med. 2009, 102, 343–348. [Google Scholar] [CrossRef] [Green Version]
- ACS.org. Available online: https://www.acs.org/education/whatischemistry/landmarks/flemingpenicillin.html (accessed on 25 June 2023).
- Simon, J.W. Povidone-iodine prophylaxis of ophthalmia neonatorum. Br. J. Ophthalmol. 2003, 87, 1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Politano, A.D.; Campbell, K.T.; Rosenberger, L.H.; Sawyer, R.G. Use of Silver in the Prevention and Treatment of Infections: Silver Review. Surg. Infect. 2013, 14, 8–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- NIH. Available online: https://www.nccih.nih.gov/health/colloidal-silver-what-you-need-to-know (accessed on 25 June 2023).
- Nowack, B.; Krug, H.F.; Height, M. 120 Years of Nanosilver History: Implications for Policy Makers. Environ. Sci. Technol. 2011, 45, 1177–1183. [Google Scholar] [CrossRef] [PubMed]
- Vorobyev, S.; Vishnyakova, E.; Likhatski, M.; Romanchenko, A.; Nemtsev, I.; Mikhlin, Y. Reactivity and Chemical Sintering of Carey Lea Silver Nanoparticles. Nanomaterials 2019, 9, 1525. [Google Scholar] [CrossRef] [Green Version]
- Dolai, J.; Mandal, K.; Jana, N.R. Nanoparticle Size Effects in Biomedical Applications. ACS Appl. Nano Mater. 2021, 4, 6471–6496. [Google Scholar] [CrossRef]
- Sandhu, A. Who invented nano? Nat. Nanotechnol. 2006, 1, 87. [Google Scholar] [CrossRef]
- Wikimedia Commons. File: Moneda Venezolana de 5 Bolivares de 1919.jpg. Available online: https://commons.wikimedia.org/wiki/File:Moneda_Venezolana_de_5_Bolivares_de_1919.jpg (accessed on 25 June 2023).
- Macroscale. Available online: https://www.sciencedirect.com/topics/engineering/macroscale (accessed on 25 June 2023).
- Microscale. Available online: https://www.sciencedirect.com/topics/engineering/microscale (accessed on 25 June 2023).
- Royal Society of Chemistry. Carbon. Available online: https://www.rsc.org/periodic-table/element/6/carbon (accessed on 25 June 2023).
- Wikimedia Commons. File: Canine Roundworm 1.JPG. Available online: https://commons.wikimedia.org/wiki/File:Canine_roundworm_1.JPG (accessed on 25 June 2023).
- Wikimedia Commons. File: Legionella Pneumophila 01.jpg. Available online: https://commons.wikimedia.org/wiki/File:Legionella_pneumophila_01.jpg (accessed on 25 June 2023).
- Wikimedia Commons. File: Monkeypox Virion—CDC.jpg. Available online: https://commons.wikimedia.org/wiki/File:Monkeypox_Virion_-_CDC.jpg (accessed on 25 June 2023).
- Wikimedia Commons. File: Viroid1.png. Available online: https://commons.wikimedia.org/wiki/File:Viroid1.png (accessed on 25 June 2023).
- The Nanodatabase. Available online: https://nanodb.dk/en/ (accessed on 25 June 2023).
- Nanotechnology Products Database. Available online: https://product.statnano.com/ (accessed on 25 June 2023).
- Johnson, J.R.; Delavari, P.; Azar, M. Activities of a nitrofurazone-containing urinary catheter and a silver hydrogel catheter against multidrug-resistant bacteria characteristic of catheter-associated urinary tract infection. Antimicrob. Agents Chemother. 1999, 43, 2990–2995. [Google Scholar] [CrossRef] [Green Version]
- Silverlon Wound Contact Dressing 510(k) Premarket Notification FDA.gov. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpmn/pmn.cfm?ID=K190343 (accessed on 25 June 2023).
- AQUACEL Ag Surgical SP Dressing 510(k) Premarket Notification FDA.gov. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpmn/pmn.cfm?id=K152926 (accessed on 25 June 2023).
- ACTICOAT SILVER COATED DRESSING 510(k) Premarket Notification FDA.gov. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfPMN/pmn.cfm?ID=K955453 (accessed on 25 June 2023).
- Mogilnicka, I.; Bogucki, P.; Ufnal, M. Microbiota and Malodor—Etiology and Management. Int. J. Mol. Sci. 2020, 21, 2886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ong, W.T.J.; Nyam, K.L. Evaluation of silver nanoparticles in cosmeceutical and potential biosafety complications. Saudi. J. Biol. Sci. 2022, 29, 2085–2094. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, F.; Yalamarty, S.S.; Filipczack, N.; Jin, Y.; Li, X. Nano Silver-Induced Toxicity and Associated Mechanisms. Int. J. Nanomed. 2022, 17, 1851–1864. [Google Scholar] [CrossRef]
- Benn, T.; Cavanagh, B.; Hristovski, K.; Posner, J.D.; Westerhoff, P. The Release of Nanosilver from Consumer Products Used in the Home. J. Environ. Qual. 2010, 39, 1875–1882. [Google Scholar] [CrossRef] [Green Version]
- Schäfer, B.; Tentschert, J.; Luch, A. Nanosilver in Consumer Products and Human Health: More Information Required! Environ. Sci. Technol. 2011, 45, 7589–7590. [Google Scholar] [CrossRef]
- Levard, C.; Hotze, E.M.; Lowry, G.V.; Brown, G.E. Environmental Transformations of Silver Nanoparticles: Impact on Stability and Toxicity. Environ. Sci. Technol. 2012, 46, 6900–6914. [Google Scholar] [CrossRef]
- Varner, K.E.; El-Badawy, A.; Feldhake, D.; Venkatapathy, R. State-of-the-Science Review: Everything Nanosilver and More. U.S. EPA: Washington, DC, USA, 2010; EP-C-05-057. [Google Scholar]
- Mueller, N.C.; Nowack, B. Exposure modeling of engineered nanoparticles in the environment. Environ. Sci. Technol. 2008, 15, 4447–4453. [Google Scholar] [CrossRef]
- The Nanodatabase: Ankle Thin Socks with Molecules of Silver. Available online: https://nanodb.dk/en/product/?pid=7138 (accessed on 25 June 2023).
- The Nanodatabase: Platinum Silver Nanocolloid Cream. Available online: https://nanodb.dk/en/product/?pid=2905 (accessed on 25 June 2023).
- Yin, I.X.; Zhang, J.; Zhao, I.S.; Mei, M.L.; Li, Q.; Chu, C.H. The Antibacterial Mechanism of Silver Nanoparticles and Its Application in Dentistry. Int. J. Nanomed. 2020, 15, 2555–2562. [Google Scholar] [CrossRef] [Green Version]
- Galdiero, S.; Falanga, A.; Vitiello, M.; Cantisani, M.; Marra, V.; Galdiero, M. Silver nanoparticles as potential antiviral agents. Molecules 2011, 16, 8894–8918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogers, J.V.; Parkinson, C.V.; Choi, Y.W.; Speshock, J.L.; Hussain, S.M. A Preliminary Assessment of Silver Nanoparticle Inhibition of Monkeypox Virus Plaque Formation. Nanoscale Res. Lett. 2008, 3, 129–133. [Google Scholar] [CrossRef] [Green Version]
- Bruna, T.; Malondonado-Bravo, F.; Jara, P.; Caro, N. Silver Nanoparticles and Their Antibacterial Applications. Int. J. Mol. Sci. 2021, 22, 7202. [Google Scholar] [CrossRef] [PubMed]
- Baptista, P.V.; McCusker, M.P.; Carvalho, A.; Ferreira, D.A.; Mohan, N.M.; Martins, M.; Fernandes, A.R. Nano-Strategies to Fight Multidrug Resistant Bacteria—“A Battle of the Titans”. Front. Microbiol. 2018, 9, 1441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheeseman, S.; Christofferson, A.J.; Kariuki, R.; Cozzolino, D.; Daeneke, T.; Crawford, R.J.; Truong, V.K.; Chapman, J.; Elbourne, A. Antimicrobial Metal Nanomaterials: From Passive to Stimuli-Activated Applications. Adv. Sci. 2020, 7, 1902913. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Nikaido, H. Molecular Basis of Bacterial Outer Membrane Permeability Revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef] [Green Version]
- Papp-Wallace, K.M.; Endimiani, A.; Taracila, M.A.; Bonomo, R.A. Carbapenems: Past, Present, and Future. Antimicrob. Agents Chemother. 2011, 55, 4943–4960. [Google Scholar] [CrossRef] [Green Version]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- Baron, E.J. Classification. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch: Galveston, TX, USA, 1996. [Google Scholar]
- Pitt, T.L.; Barer, M.R. Classification, identification and typing of micro-organisms. Med. Microbiol. 2012, 24–38. [Google Scholar] [CrossRef]
- Silhavy, T.J.; Kahne, D.; Walker, S. The Bacterial Cell Envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Tripathi, N.; Sapra, A. Gram Staining. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Milton, S.R.J.; Kwang-Shin, K. Structure. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch: Galveston, TX, USA, 1996. [Google Scholar]
- Sizar, O.; Leslie, S.W.; Unakal, C.G. Gram-Positive Bacteria. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA.
- Alvarez, L.; Espaillat, A.; Hermoso, J.A.; de Pedro, M.A.; Cava, F. Peptidoglycan Remodeling by the Coordinated Action of Multispecific Enzymes. Microb. Drug Resist. 2014, 20, 190–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irazoki, O.; Hernandez, S.B.; Cava, F. Peptidoglycan Muropeptides: Release, Perception, and Functions as Signaling Molecules. Front. Microbiol. 2019, 10, 500. [Google Scholar] [CrossRef] [Green Version]
- Neuhaus, F.C.; Baddiley, J. A Continuum of Anionic Charge: Structures and Functions of D-Alanyl-Teichoic Acids in Gram-Positive Bacteria. Microbiol. Mol. Biol. Rev. 2003, 67, 686–723. [Google Scholar] [CrossRef] [Green Version]
- Brown, S.; Santa Maria, J.P., Jr.; Walker, S. Wall Teichoic Acids of Gram-Positive Bacteria. Annu. Rev. Microbiol. 2014, 67, 313–336. [Google Scholar] [CrossRef] [Green Version]
- Thomas, K.J., III; Rice, C.V. Equilibrium Binding Behavior of Magnesium to Wall Teichoic Acid. Biochim. Biophys. Acta 2016, 1848, 1981–1987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, Y.; Brown, D.G. Electrostatic Behavior of the Charge-Regulated Bacterial Cell Surface. Langmuir 2008, 24, 5003–5009. [Google Scholar] [CrossRef] [PubMed]
- Gajdás, M.; Ábrók, M.; Lázár, A.; Burián, K. Increasing relevance of Gram-positive cocci in urinary tract infections: A 10-year analysis of their prevalence and resistance trends. Sci. Rep. 2020, 10, 17658. [Google Scholar] [CrossRef]
- Iglewski, B.H. Pseudomonas. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch: Galveston, TX, USA, 1996. [Google Scholar]
- Oliveira, J.; Reygaert, W.C. Gram-Negative Bacteria. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Haftel, A.; Sharman, T. Vibrio vulnificus Infection. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Huang, K.C.; Mukhopadhyay, R.; Wen, B.; Gitai, Z.; Wingreen, N.S. Cell-shape and cell-wall organization in Gram-negative bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 19282–19287. [Google Scholar] [CrossRef]
- Moreillon, P.; Majcherczyk, P.A. Proinflammatory activity of cell-wall constituents from gram-positive bacteria. Scand. J. Infect. Dis. 2003, 35, 632–641. [Google Scholar] [CrossRef]
- Raetz, C.R.H.; Reynolds, C.M.; Trent, M.S.; Bishop, R.E. Lipid a modification systems in gram-negative bacteria. Annu. Rev. Biochem. 2008, 76, 295–329. [Google Scholar] [CrossRef] [Green Version]
- Bertani, B.; Natividad, R. Function and biogenesis of lipopolysaccharides. EcoSal Plus 2018, 8, ESP-0001-2018. [Google Scholar] [CrossRef] [PubMed]
- Farhana, A.; Khan, Y.S. Biochemistry, Lipopolysaccharide. In StatPearls; StatPearls Publishing: Treasure Island, FL, 2023. [Google Scholar]
- Roberts, J.A.; Lipman, J. Pharmacokinetic issues for antibiotics in the critically ill patient. Crit. Care Med. 2009, 37, 840–851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorenzo, F.D.; Duda, K.A.; Lanzetta, R.; Silipo, A.; Castro, C.D.; Molinaro, A. A Journey from Structure to Function of Bacterial lipopolysaccharides. Chem. Rev. 2022, 122, 15767–15821. [Google Scholar] [CrossRef]
- Wassenaar, T.M.; Zimmermann, K. Lipopolysaccharides in Food, Food Supplements, and Probiotics: Should We be Worried? Eur. J. Microbiol. Immunol. 2018, 8, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Dadgostar, P. Antimicrobial Resistance: Implications and Costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef] [Green Version]
- Antibiotic Resistance Threats in the United States. Available online: https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf (accessed on 25 June 2023).
- Hornyak, G.L.; Tibbals, H.F.; Dutta, J.; Moore, J.J. Introduction to Nanoscience and Nanotechnology, 1st ed.; CRC Press: Boca Raton, FL, USA, 2009. [Google Scholar]
- Kibis, L.S.; Stadnichenko, A.I.; Pajetnov, E.M.; Koscheev, S.V.; Zaykovskii, V.I.; Boronin, A.I. The Investigation of Oxidized Silver Nanoparticles Prepared by Thermal Evaporation and Radio-Frequency Sputtering of Metallic Silver under Oxygen. Appl. Surf. Sci. 2010, 257, 404–413. [Google Scholar] [CrossRef]
- Sun, G.; Ye, G.; Wang, K.; Lou, M.; Jia, X.; Xu, F.; Ye, Z. Deposition of Ag Films on Liquid Substrates via Thermal Evaporation for Surface-Enhanced Raman Scattering. ACS Omega 2020, 5, 7440–7445. [Google Scholar] [CrossRef]
- Maślak, E.; Arendowski, A.; Złoch, M.; Walczak-Skierska, J.; Radtke, A.; Piszczek, P.; Pomastowski, P. Silver Nanoparticle Targets Fabricated Using Chemical Vapor Deposition Method for Differentiation of Bacteria Based on Lipidomic Profiles in Laser Desorption/Ionization Mass Spectrometry. Antibiotics 2023, 12, 874. [Google Scholar] [CrossRef]
- Piszczek, P.; Radtke, A. Silver Nanoparticles Fabricated Using Chemical Vapor Deposition and Atomic Layer Deposition Techniques: Properties, Applications and Perspectives: Review. In Noble and Precious Metals—Properties, Nanoscale Effects and Applications, 1st ed.; Seehra, M., Bristow, A., Eds.; IntechOpen: London, UK, 2017. [Google Scholar]
- Rosa Remiro, P.; Martins, J.; Lima, B.; Oishi, M.; Aguiar, M.; Bernardo, A. Atomization of Silver Nanoparticles Suspension as an Alternative for Generating Nanosilver Aerosol. Chem. Ind. Chem. Eng. Q. 2018, 24, 303–307. [Google Scholar] [CrossRef]
- Kumar, N.; Biswas, K.; Kumar, G.R. Green Synthesis of Ag Nanoparticles in Large Quantity by Cryomilling. RSC Adv. 2016, 6, 111380–111388. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Jun, B.-H. Silver Nanoparticles: Synthesis and Application for Nanomedicine. Int. J. Mol. Sci. 2019, 20, 865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, S.; Sato, M.; Sato, Y.; Ando, N.; Takayama, T.; Fujita, M.; Ishihara, M. Synthesis and Application of Silver Nanoparticles (Ag NPs) for the Prevention of Infection in Healthcare Workers. Int. J. Mol. Sci. 2019, 20, 3620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yusuf, M. Silver Nanoparticles: Synthesis and Applications. Handb. Ecomater. 2018, 2343–2356. [Google Scholar] [CrossRef]
- Quan, F.; Mao, A.; Ding, M.; Ran, S.; Wang, J.; Yang, G.; Yan, Y. Combustion Synthesis and Formation Mechanism of Silver Nanoparticles. Int. J. Mater. Res. 2018, 109, 751–755. [Google Scholar] [CrossRef]
- Mulvihill, M.J.; Ling, X.Y.; Henzie, J.; Yang, P. Anisotropic Etching of Silver Nanoparticles for Plasmonic Structures Capable of Single-Particle SERS. J. Am. Chem. Soc. 2010, 132, 268–274. [Google Scholar] [CrossRef]
- Wack, S.; Lunca, P.P.; Adjeroud, N.; Guillot, J.; Pistillo, B.R.; Leturcq, R. Large-Scale Deposition and Growth Mechanism of Silver Nanoparticles by Plasma-Enhanced Atomic Layer Deposition. J. Phys. Chem. C 2019, 123, 27196–27206. [Google Scholar] [CrossRef]
- Paul, S.; Chakraborty, B.B.; Anwar, S.; Paul, S.B.; Choudhury, S. Self-Assembly of Silver Nanoparticles through Functionalization with Coumarin-Thiazole Fused-Ring Thiol. Heliyon 2020, 6, e03674. [Google Scholar] [CrossRef]
- Pillai, Z.S.; Kamat, P.V. What Factors Control the Size and Shape of Silver Nanoparticles in the Citrate Ion Reduction Method? J. Phys. Chem. B 2004, 108, 945–951. [Google Scholar] [CrossRef]
- Chakraborty, I.; Feliu, N.; Roy, S.; Dawson, K.; Parak, W.J. Protein-Mediated Shape Control of Silver Nanoparticles. Bioconjugate Chem. 2018, 29, 1261–1265. [Google Scholar] [CrossRef]
- Nithyaja, B.; Misha, H.; Nampoori, V.P.N. Synthesis of Silver Nanoparticles in DNA Template and Its Influence on Nonlinear Optical Properties. Nanosci. Nanotechnol. 2012, 2, 99–103. [Google Scholar] [CrossRef] [Green Version]
- Iravani, S.; Korbekandi, H.; Mirmohammadi, S.V.; Zolfaghari, B. Synthesis of Silver Nanoparticles: Chemical, Physical and Biological Methods. Res. Pharm. Sci. 2014, 9, 385–406. [Google Scholar]
- Wei, L.; Lu, J.; Xu, H.; Patel, A.; Chen, Z.-S.; Chen, G. Silver Nanoparticles: Synthesis, Properties, and Therapeutic Applications. Drug Discov. Today 2015, 20, 595–601. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.-F.; Liu, Z.-G.; Shen, W.; Gurunathan, S. Silver Nanoparticles: Synthesis, Characterization, Properties, Applications, and Therapeutic Approaches. Int. J. Mol. Sci. 2016, 17, 1534. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, R.; Kumar, A.; Abraham, J. A Biological Approach to the Synthesis of Silver Nanoparticles with Streptomyces Sp JAR1 and Its Antimicrobial Activity. Sci. Pharm. 2013, 81, 607–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esmail, R.; Afshar, A.; Morteza, M.; Abolfazl, A.; Akhondi, E. Synthesis of silver nanoparticles with high efficiency and stability by culture supernatant of Bacillus ROM6 isolated from Zarshouran gold mine and evaluating its antibacterial effects. BMC Microbiol. 2022, 22, 97. [Google Scholar] [CrossRef]
- Hosny, A.M.S.; Kashef, M.T.; Rasmy, S.A.; Aboul-Magd, D.S.; El-Bazza, Z.E. Antimicrobial Activity of Silver Nanoparticles Synthesized Using Honey and Gamma Radiation against Silver-Resistant Bacteria from Wounds and Burns. Adv. Nat. Sci. Nanosci. Nanotechnol. 2017, 8, 45009. [Google Scholar] [CrossRef] [Green Version]
- Mousavi, S.M.; Hashemi, S.A.; Ghasemi, Y.; Atapour, A.; Amani, A.M.; Savar, D.A.; Babapoor, A.; Arjmand, O. Green Synthesis of Silver Nanoparticles toward Bio and Medical Applications: Review Study. Artif. Cells Nanomed. Biotechnol. 2018, 46, S855–S872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, D.; Bhattacharyya, S.; Bhattacharyya, M.; Mandal, P. Green Chemistry Inspired Formation of Bioactive Stable Colloidal Nanosilver and Its Wide-Spectrum Functionalised Properties for Sustainable Industrial Escalation. Results Chem. 2022, 4, 100533. [Google Scholar] [CrossRef]
- Sharma, N.K.; Vishwakarma, J.; Rai, S.; Alomar, T.S.; AlMasoud, N.; Bhattarai, A. Green Route Synthesis and Characterization Techniques of Silver Nanoparticles and Their Biological Adeptness. ACS Omega 2022, 7, 27004–27020. [Google Scholar] [CrossRef]
- Quintero-Quiroz, C.; Acevedo, N.; Zapata-Giraldo, J.; Botero, L.E.; Quintero, J.; Zárate-Triviño, D.; Saldarriaga, J.; Pérez, V.Z. Optimization of Silver Nanoparticle Synthesis by Chemical Reduction and Evaluation of Its Antimicrobial and Toxic Activity. Biomater. Res. 2019, 23, 27. [Google Scholar] [CrossRef] [Green Version]
- Javed, R.; Zia, M.; Naz, S.; Aisida, S.O.; Ain, N.U.; Ao, Q. Role of Capping Agents in the Application of Nanoparticles in Biomedicine and Environmental Remediation: Recent Trends and Future Prospects. J. Nanobiotechnol. 2020, 18, 172. [Google Scholar] [CrossRef] [PubMed]
- Khodashenas, B.; Ghorbani, H.R. Synthesis of Silver Nanoparticles with Different Shapes. Arab. J. Chem. 2019, 12, 1823–1838. [Google Scholar] [CrossRef]
- Tolaymat, T.M.; Badawy, A.M.E.; Genaidy, A.; Scheckel, K.G.; Luxton, T.P.; Suidan, M. An evidence-based environmental perspective of manufactured silver nanoparticle in syntheses and applicationss: A systematic review and critical appraisal of peer-reviewed scientific papers. Sci. Total Environ. 2009, 408, 999–1006. [Google Scholar] [CrossRef] [Green Version]
- Badawy, A.E. Assessment of the Fate and Transport of Silver Nanoparticles in Porous Media; University of Cincinnati: Cincinnati, OH, USA, 2011. [Google Scholar]
- Rabiee, N.; Ahmadi, S.; Iravani, S.; Varma, R.S. Functionalized Silver and Gold Nanomaterials with Diagnostic and Therapeutic Applications. Pharmaceutics 2022, 14, 2182. [Google Scholar] [CrossRef]
- Shruthi, T.S.; Meghana, M.R.; Medha, M.U.; Sanjana, S.; Navya, P.N.; Kumar, D.H. Streptomycin Functionalization on Silver Nanoparticles for Improved Antibacterial Activity. Mater. Today Proc. 2019, 10, 8–15. [Google Scholar] [CrossRef]
- Rezazadeh, N.H.; Buazar, F.; Matroodi, S. Synergistic Effects of Combinatorial Chitosan and Polyphenol Biomolecules on Enhanced Antibacterial Activity of Biofunctionalized Silver Nanoparticles. Sci. Rep. 2020, 10, 19615. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Perveen, S.; Ali, M.; Shah, M.R.; Khan, E.; Sharma, A.S.; Li, H.; Chen, Q. Nano-conjugates of Cefadroxil as Efficient Antibacterial Agent Against Staphylococcus aureus ATCC 11632. J. Clust. Sci. 2020, 31, 811–821. [Google Scholar] [CrossRef]
- Asghar, A.; Tan, Y.C.; Zahoor, M.; Zainal Abidin, S.A.; Yow, Y.Y.; Khan, E.; Lahiri, C. A scaffolded approach to unearth potential antibacterial components from epicarp of Malaysian Nephelium lappaceum L. Sci. Rep. 2021, 11, 13859. [Google Scholar] [CrossRef]
- U.S. Environmental Protection Agency. Exposure Assessment Tools by Chemical Classes—Nanomaterials. Available online: https://www.epa.gov/expobox/exposure-assessment-tools-chemical-classes-nanomaterials (accessed on 19 June 2023).
- Almatroudi, A. Silver Nanoparticles: Synthesis, Characterisation and Biomedical Applications. Open Life Sci. 2020, 15, 819–839. [Google Scholar] [CrossRef]
- Cho, W.-S.; Duffin, R.; Thielbeer, F.; Bradley, M.; Megson, I.L.; MacNee, W.; Poland, C.A.; Tran, C.L.; Donaldson, K. Zeta Potential and Solubility to Toxic Ions as Mechanisms of Lung Inflammation Caused by Metal/Metal Oxide Nanoparticles. Toxicol. Sci. 2012, 126, 469–477. [Google Scholar] [CrossRef] [Green Version]
- Jayawardena, H.S.N.; Liyanage, S.H.; Rathnayake, K.; Patel, U.; Yan, M. Analytical Methods for Characterization of Nanomaterial Surfaces. Anal. Chem. 2021, 93, 1889–1911. [Google Scholar] [CrossRef] [PubMed]
- Mourdikoudis, S.; Pallares, R.M.; Thanh, N.T.K. Characterization techniques for nanoparticles: Comparison and complementarity upon studying nanoparticle properties. Nanoscale 2018, 10, 12871–12934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Y.; Zhu, H.; Shen, Y.; Zhang, W.; Zhang, L. Antibacterial Activity of Silver Nanoparticles of Different Particle Size against Vibrio Natriegens. PLoS ONE 2019, 14, e0222322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nie, P.; Zhao, Y.; Xu, H. Synthesis, Applications, Toxicity and Toxicity Mechanisms of Silver Nanoparticles: A Review. Ecotoxicol. Environ. Saf. 2023, 253, 114636. [Google Scholar] [CrossRef]
- Helmlinger, J.; Sengstock, C.; Groß-Heitfeld, C.; Mayer, C.; Schildhauer, T.A.; Köller, M.; Epple, M. Silver nanoparticles with different size and shape: Equal cytotoxicity, but different antibacterial effects. RSC Adv. 2016, 6, 18490–18501. [Google Scholar] [CrossRef] [Green Version]
- Cheon, J.Y.; Kim, S.J.; Rhee, Y.H.; Kwon, O.H.; Park, W.H. Shape-dependent antimicrobial activities of silver nanoparticles. Int. J. Nanomed. 2019, 14, 2773–2780. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, F.; Salem-Bekhit, M.M.; Khan, F.; Alshehri, S.; Khan, A.; Ghoneim, M.M.; Wu, H.-F.; Taha, E.I.; Elbagory, I. Unique Properties of Surface-Functionalized Nanoparticles for Bio-Application: Functionalization Mechanisms and Importance in Application. Nanomaterials 2022, 12, 1333. [Google Scholar] [CrossRef]
- Menichetti, A.; Mavridi-Printezi, A.; Mordini, D.; Montalti, M. Effect of Size, Shape and Surface Functionalization on the Antibacterial Activity of Silver Nanoparticles. J. Funct. Biomater. 2023, 14, 244. [Google Scholar] [CrossRef]
- Mohsen, E.; El-Borady, O.M.; Mohamed, M.B.; Fahim, I.S. Synthesis and Characterization of Ciprofloxacin Loaded Silver Nanoparticles and Investigation of Their Antibacterial Effect. J. Radiat. Res. Appl. Sci. 2020, 13, 416–425. [Google Scholar] [CrossRef] [Green Version]
- Zharkova, M.S.; Golubeva, O.Y.; Orlov, D.S.; Vladimirova, E.V.; Dmitriev, A.V.; Tossi, A.; Shamova, O.V. Silver Nanoparticles Functionalized With Antimicrobial Polypeptides: Benefits and Possible Pitfalls of a Novel Anti-Infective Tool. Front. Microbiol. 2021, 12, 750556. [Google Scholar] [CrossRef]
- Ellairaja, S.; Krithiga, N.; Ponmariappan, S.; Vasantha, V.S. Novel Pyrimidine Tagged Silver Nanoparticle Based Fluorescent Immunoassay for the Detection of Pseudomonas Aeruginosa. J. Agric. Food Chem. 2017, 65, 1802–1812. [Google Scholar] [CrossRef]
- Guo, H.; Zhang, Z.; Xing, B.; Mukherjee, A.; Musante, C.; White, J.C.; He, L. Analysis of Silver Nanoparticles in Antimicrobial Products Using Surface-Enhanced Raman Spectroscopy (SERS). Environ. Sci. Technol. 2015, 49, 4317–4324. [Google Scholar] [CrossRef]
- Heuer-Jungemann, A.; Feliu, N.; Bakaimi, I.; Hamaly, M.; Alkilany, A.; Chakraborty, I.; Masood, A.; Casula, M.F.; Kostopoulou, A.; Oh, E.; et al. The Role of Ligands in the Chemical Synthesis and Applications of Inorganic Nanoparticles. Chem. Rev. 2019, 119, 4819–4880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rim, K.-T.; Song, S.-W.; Kim, H.-Y. Oxidative DNA Damage from Nanoparticle Exposure and Its Application to Workers’ Health: A Literature Review. Saf. Health Work. 2013, 4, 177–186. [Google Scholar] [CrossRef] [Green Version]
- Franci, G.; Falanga, A.; Galdiero, S.; Palomba, L.; Rai, M.; Morelli, G.; Galdiero, M. Silver Nanoparticles as Potential Antibacterial Agents. Molecules 2015, 20, 8856–8874. [Google Scholar] [CrossRef] [Green Version]
- Bondarenko, O.M.; Sihtmäe, M.; Kuzmičiova, J.; Ragelienė, L.; Kahru, A.; Daugelavičius, R. Plasma membrane is the target of rapid antibacterial action of silver nanoparticles in Escherichia coli and Pseudomonas aeruginosa. Int. J. Nanomed. 2018, 13, 6779–6790. [Google Scholar] [CrossRef] [Green Version]
- Barani, M.; Fathizadeh, H.; Arkaban, H.; Kalantar-Neyestanaki, D.; Akbarizadeh, M.R.; Turki Jalil, A.; Akhavan-Sigari, R. Recent Advances in Nanotechnology for the Management of Klebsiella pneumoniae–Related Infections. Biosensors 2022, 12, 1155. [Google Scholar] [CrossRef] [PubMed]
- Hetta, H.F.; Al-Kadmy, I.M.S.; Khazaal, S.S.; Abbas, S.; Suhail, A.; El-Mokhtar, M.A.; Ellah, N.H.A.; Ahmed, E.A.; Abd-Ellatief, R.B.; El-Masry, E.A.; et al. Antibiofilm and antivirulence potential of silver nanoparticles against multidrug-resistant Acinetobacter baumannii. Sci. Rep. 2021, 11, 10751. [Google Scholar] [CrossRef]
- Sabzi, N.; Moniri, R.; Sehat, M.; Fathizadeh, H.; Nazari-Alam, A. Antimicrobial effect of silver and gold nanoparticles in combination with linezolid on Enterococcus biofilm. Iran. J. Microbiol. 2022, 14, 863–873. [Google Scholar] [CrossRef]
- Yuan, Y.-G.; Peng, Q.-L.; Gurunathan, S. Effects of Silver Nanoparticles on Multiple Drug-Resistant Strains of Staphylococcus aureus and Pseudomonas aeruginosa from Mastitis-Infected Goats: An Alternative Approach for Antimicrobial Therapy. Int. J. Mol. Sci. 2017, 18, 569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, X.; Lang, J.; Chen, P.; Yang, R. Silver Nanoparticles as an Effective Antimicrobial against Otitis Media Pathogens. AIChE J. 2021, 67, e17468. [Google Scholar] [CrossRef] [PubMed]
- Saravanan, M.; Barik, S.K.; MubarakAli, D.; Prakash, P.; Pugazhendhi, A. Synthesis of silver nanoparticles from Bacillus brevis (NCIM 2533) and their antibacterial activity against pathogenic bacteria. Microb. Pathog. 2018, 116, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Mikhailova, E.O. Silver Nanoparticles: Mechanism of Action and Probable Bio-Application. J. Funct. Biomater. 2020, 11, 84. [Google Scholar] [CrossRef] [PubMed]
- Dakal, T.C.; Kumar, A.; Majumdar, R.S.; Yadav, V. Mechanistic Basis of Antimicrobial Actions of Silver Nanoparticles. Front. Microbiol. 2016, 7, 1831. [Google Scholar] [CrossRef] [Green Version]
- Bos, M.P.; Robert, V.; Tommassen, J. Biogenesis of the gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 2007, 61, 191–214. [Google Scholar] [CrossRef] [Green Version]
- Beveridge, T.J. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 1999, 181, 4725–4733. [Google Scholar] [CrossRef]
- Liao, C.; Li, Y.; Tjong, S.C. Bactericidal and Cytotoxic Properties of Silver Nanoparticles. Int. J. Mol. Sci. 2019, 20, 449. [Google Scholar] [CrossRef] [Green Version]
- Pokhrel, L.R.; Jacobs, Z.L.; Dikin, D.; Akula, S.M. Five nanometer size highly positive silver nanoparticles are bactericidal targeting cell wall and adherent fimbriae expression. Sci. Rep. 2022, 12, 6729. [Google Scholar] [CrossRef]
- Przemieniecki, S.W.; Oćwieja, M.; Ciesielski, S.; Halecki, W.; Matras, E.; Gorczyca, A. Chemical Structure of Stabilizing Layers of Negatively Charged Silver Nanoparticles as an Effector of Shifts in Soil Bacterial Microbiome under Short-Term Exposure. Int. J. Environ. Res. Public Health 2022, 19, 14438. [Google Scholar] [CrossRef]
- Nie, X.; Gao, F.; Wang, F.; Liu, C.; You, Y.-Z. Charge-reversal silver clusters for targeted bacterial killing. J. Mater. Chem. B 2021, 9, 4006–4014. [Google Scholar] [CrossRef]
- Durán, N.; Silveira, C.P.; Durán, M.; Martinez, D.S.T. Silver nanoparticle protein corona and toxicity: A mini-review. J. Nanobiotechnol. 2015, 13, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miclăuş, T.; Beer, C.; Chevallier, J.; Scavenius, C.; Bochenkov, V.E.; Enghild, J.J.; Sutherland, D.S. Dynamic protein coronas revealed as a modulator of silver nanoparticle sulphidation in vitro. Nat. Commun. 2016, 7, 11770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skoglund, S.; Lowe, T.A.; Hedberg, J.; Blomberg, E.; Wallinder, I.O.; Wold, S.; Lundin, M. Effect of Laundry Surfactants on Surface Charge and Colloidal Stability of Silver Nanoparticles. Langmuir 2013, 29, 8882–8891. [Google Scholar] [CrossRef]
- Pearson, R.M.; Juettner, V.V.; Hong, S. Biomolecular corona on nanoparticles: A survey of recent literature and its implications in targeted drug delivery. Front. Chem. 2014, 2, 108. [Google Scholar] [CrossRef] [PubMed]
- Spagnoletti, F.N.; Kronberg, F.; Spedalieri, C.; Munarriz, E.; Giacometti, R. Protein corona on biogenic silver nanoparticles provides higher stability and protects cells from toxicity in comparison to chemical nanoparticles. J. Environ. Manag. 2021, 297, 113434. [Google Scholar] [CrossRef] [PubMed]
- Deepak, V.; Umamaheshwaran, P.S.; Guhan, K.; Nanthini, R.A.; Krithiga, B.; Jaithoon, N.M.; Gurunathan, S. Synthesis of gold and silver nanoparticles using purified URAK. Colloids Surf. B Biointerfaces 2011, 86, 353–358. [Google Scholar] [CrossRef]
- Agnihotri, S.; Mukherji, S.; Mukherji, S. Size-controlled silver nanoparticles synthesized over the range 5-100 nm using the same protocol and their antibacterial efficacy. RSC Adv. 2014, 8, 3974–3983. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, G.J.; Wiesenfeld, K.; Nelson, D.C.; Weitz, J.S. Critical cell wall hole size for lysis in Gram-positive bacteria. J. R. Soc. Interface 2013, 10, 20120892. [Google Scholar] [CrossRef] [Green Version]
- Vollmer, W.; Blanot, D.; De Pedro, M.A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008, 32, 149–167. [Google Scholar] [CrossRef] [Green Version]
- Pirahanchi, Y.; Jessu, R.; Aeddula, N.R. Physiology, Sodium Potassium Pump. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Fahmy, H.M.; Mosleh, A.M.; Elghany, A.A.; Shams-Eldin, E.; Abu Serea, E.S.; Ali, S.A.; Shalan, A.E. Coated silver nanoparticles: Synthesis, cytotoxicity, and optical properties. RSC Adv. 2019, 9, 20118–20136. [Google Scholar] [CrossRef]
- Miller, S.I.; Salma, N.R. The gram-negative bacterial periplasm: Size matters. PLoS Biol. 2018, 16, e2004935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritz, D.; Beckwith, J. Roles of thiol-redox pathways in bacteria. Annu. Rev. Microbiol. 2001, 55, 21–48. [Google Scholar] [CrossRef] [PubMed]
- Li, W.R.; Xie, X.B.; Shi, Q.S.; Zeng, H.Y.; Ou-Yang, Y.S.; Chen, Y.B. Antibacterial activity and mechanism of silver nanoparticles on Escherichia coli. Appl. Microbiol. Biotechnol. 2009, 85, 1115–1122. [Google Scholar] [CrossRef] [PubMed]
- Seong, M.; Lee, D.G. Silver nanoparticles against salmonella enterica serotype typhimurium: Role of inner membrane dysfunction. Curr. Microbiol. 2017, 74, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.A.; Das, S.S.; Khatoon, A.; Ansari, M.T.; Mohd, A.; Hasnain, M.S.; Nayak, A.K. Bactericidal activity of silver nanoparticles: A mechanistic review. Mater. Sci. Energy Technol. 2020, 3, 756–769. [Google Scholar] [CrossRef]
- Gunawan, C.; Faiz, M.B.; Mann, R.; Ting, S.R.S.; Sotiriou, G.A.; Marquis, C.P.; Amal, R. Nanosilver Targets the Bacterial Cell Envelope: The Link with Generation of Reactive Oxygen Radicals. ACS Appl. Mat. Interfaces 2020, 12, 5557–5568. [Google Scholar] [CrossRef]
- Adeyemi, O.S.; Shittu, E.O.; Akpor, O.B.; Rotimi, D.; Batiha, G.E. Silver nanoparticles restrict microbial growth by promoting oxidative stress and DNA damage. EXCLI J. 2020, 19, 492–500. [Google Scholar]
- Lipfert, J.; Doniach, S.; Das, R.; Herschlag, D. Understanding nucleic acid-ion interactions. Annu. Rev. Biochem. 2015, 83, 813–841. [Google Scholar] [CrossRef] [Green Version]
- Radzig, M.A.; Nadtochenko, V.A.; Koksharova, O.A.; Kiwi, J.; Lipasova, V.A.; Khmel, I.A. Antibacterial effects of silver nanoparticles on gram-negative bacteria: Influence on the growth and biofilms formation, mechanisms of action. Colloids Surf. B Biointerfaces 2013, 102, 300–306. [Google Scholar] [CrossRef]
- Klueh, U.; Wagner, V.; Kelly, S.; Johnson, A.; Bryers, J.D. Efficacy of silver-coated fabric to prevent bacterial colonization and subsequent device-based biofilm formation. J. Biomed. Mater. Res. 2000, 53, 621–631. [Google Scholar] [CrossRef]
- Mammari, N.; Lamouroux, E.; Boudier, A.; Duval, R.E. Current Knowledge on the Oxidative-Stress-Mediated Antimicrobial Properties of Metal-Based Nanoparticles. Microorganisms 2022, 10, 437. [Google Scholar] [CrossRef]
- Onodera, A.; Nishiumi, F.; Kakiguchi, K.; Tanaka, A.; Tanabe, N.; Honma, A.; Yayama, K.; Yoshioka, Y.; Nakahira, K.; Yonemura, S.; et al. Short-term changes in intracellular ROS localisation after the silver nanoparticles exposure depending on particle size. Toxicol. Rep. 2015, 2, 574–579. [Google Scholar] [CrossRef] [Green Version]
- Beaudrie, C.E.H.; Kandlikar, M.; Ramachandran, G. Assessing Nanoparticle Risks to Human Health, 2nd ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2016; pp. 91–119. [Google Scholar]
- Paiva, C.N.; Bozza, M.T. Are reactive oxygen species always detrimental to pathogens? Antioxid. Redox Signal. 2014, 20, 1000–1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Xiao, S.; Ahn, D.U. Protein oxidation: Basic principles and implications for meat quality. Crit. Rev. Food Sci. Nutr. 2013, 53, 1191–1201. [Google Scholar] [CrossRef]
- Byrgazov, K.; Vesper, O.; Moll, I. Ribosome heterogeneity: Another level of complexity in bacterial translation regulation. Curr. Opin. Microbiol. 2013, 16, 133–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duckett, S. Ernest Duchesne and the concept of fungal antibiotic therapy. Lancet 1999, 354, 2068–2071. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present, future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef]
- Scheffers, D.J.; Pinho, M.G. Bacterial cell wall synthesis: New insights from localization studies. Microbiol. Mol. Biol. Rev. 2005, 69, 585–607. [Google Scholar] [CrossRef] [Green Version]
- Fisher, J.F.; Mobashery, S. Constructing and deconstructing the bacterial cell wall. Protein Sci. 2020, 29, 629–646. [Google Scholar] [CrossRef]
- Buynak, J.D. Cutting and stitching: The cross-linking of peptidoglycan in the assembly of the bacterial cell wall. ACS Chem. Biol. 2007, 2, 602–605. [Google Scholar] [CrossRef] [Green Version]
- Bush, K.; Bradford, P.A. β-Lactams and β-Lactamase Inhibitors: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a025247. [Google Scholar] [CrossRef] [PubMed]
- Pandey, N.; Cascella, M. Beta-Lactam Antibiotics. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Mühlberg, E.; Umstätter, F.; Kleist, C.; Domhan, C.; Mier, W.; Uhl, P. Renaissance of vancomycin: Approaches for breaking antibiotic resistance in multidrug-resistant bacteria. Can. J. Microbiol. 2020, 66, 11–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stone, K.J.; Strominger, J.L. Mechanism of action of bacitracin: Complexation with metal ion and C55-isoprenyl pyrophosphate. Proc. Natl. Acad. Sci. USA 1971, 68, 3223–3227. [Google Scholar] [CrossRef]
- Nguyen, R.; Khanna, N.R.; Safadi, A.O.; Sun, Y. Bacitracin Topical. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Zhu, J.; Wang, S.; Wang, C.; Wang, Z.; Luo, G.; Li, J.; Zhan, Y.; Cai, D.; Chen, S. Microbial synthesis of bacitracin: Recent progress, challenges, and prospects. Synth. Syst. Biotechnol. 2023, 8, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Baran, A.; Kwiatkowska, A.; Potocki, L. Antibiotics and Bacterial Resistance-A Short Story of an Endless Arms Race. Int. J. Mol. Sci. 2023, 24, 5777. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.W. DAPTOMYCIN, its membrane-active mechanism vs. that of other antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183395. [Google Scholar] [CrossRef]
- Mingeot-Leclercq, M.P.; Glupczynski, Y.; Tulkens, P.M. Aminoglycosides: Activity and resistance. Antimicrob. Agents Chemother. 1999, 43, 727–737. [Google Scholar] [CrossRef] [Green Version]
- Markley, J.L.; Wencewicz, T.A. Tetracycline-Inactivating Enzymes. Front. Microbiol. 2018, 9, 1058. [Google Scholar] [CrossRef] [Green Version]
- Tenson, T.; Lovmar, M.; Ehrenberg, M. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J. Mol. Biol. 2003, 330, 1005–1014. [Google Scholar] [CrossRef]
- Bush, N.G.; Evans-Roberts, K.; Maxwell, A. DNA Topoisomerases. EcoSal Plus 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Surette, M.D.; Spanogiannopoulos, P.; Wright, G.D. The Enzymes of the Rifamycin Antibiotic Resistome. Acc. Chem. Res. 2021, 54, 2065–2075. [Google Scholar] [CrossRef] [PubMed]
- Wróbel, A.; Arciszewska, K.; Maliszewski, D.; Drozdowska, D. Trimethoprim and other nonclassical antifolates an excellent template for searching modifications of dihydrofolate reductase enzyme inhibitors. J. Antibiot. 2020, 73, 5–27. [Google Scholar] [CrossRef] [Green Version]
- Kemnic, T.R.; Coleman, M. Trimethoprim Sulfamethoxazole. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Masters, P.A.; O’Bryan, T.A.; Zurlo, J.; Miller, D.Q.; Joshi, N. Trimethoprim-sulfamethoxazole revisited. Arch. Intern. Med. 2003, 163, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Varela, M.F.; Stephen, J.; Lekshmi, M.; Ojha, M.; Wenzel, N.; Sanford, L.M.; Hernandez, A.J.; Parvathi, A.; Kumar, S.H. Bacterial Resistance to Antimicrobial Agents. Antibiotics 2021, 10, 593. [Google Scholar] [CrossRef] [PubMed]
- Aljeldah, M.M. Antimicrobial Resistance and Its Spread Is a Global Threat. Antibiotics 2022, 11, 1082. [Google Scholar] [CrossRef] [PubMed]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.R.M.; Mitra, S.; Emran, T.B.; Dhama, K.; Ripon, M.K.H.; Gajdács, M.; Sahibzada, M.U.K.; et al. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef] [PubMed]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Soucy, S.M.; Huang, J.; Gogarten, J.P. Horizontal gene transfer: Building the web of life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef]
- Rangama, S.; Lidbury, I.D.E.A.; Holden, J.M.; Borsetto, C.; Murphy, A.R.J.; Hawkey, P.M.; Wellington, E.M.H. Mechanisms Involved in the Active Secretion of CTX-M-15 β-Lactamase by Pathogenic Escherichia coli ST131. Antimicrob. Agents Chemother. 2021, 65, e0066321. [Google Scholar] [CrossRef]
- Tooke, C.L.; Hinchliffe, P.; Bragginton, E.C.; Colenso, C.K.; Hirvonen, V.H.A.; Takebayashi, Y.; Spencer, J. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 2019, 431, 3472–3500. [Google Scholar] [CrossRef]
- Mendonça, J.; Guedes, C.; Silva, C.; Sá, S.; Oliveira, M.; Accioly, G.; Baylina, P.; Barata, P.; Pereira, C.; Fernandes, R. New CTX-M Group Conferring β-Lactam Resistance: A Compendium of Phylogenetic Insights from Biochemical, Molecular, and Structural Biology. Biology 2022, 11, 256. [Google Scholar] [CrossRef]
- Bush, K. Past and Present Perspectives on β-Lactamases. Antimicrob. Agents Chemother. 2018, 62, e01076-18. [Google Scholar] [CrossRef] [Green Version]
- Heidary, M.; Samangani, A.E.; Kargari, A.; Nejad, K.A.; Yashmi, I.; Motahar, M.; Taki, E.; Khoshnood, S. Mechanism of action, resistance, synergism, and clinical implications of azithromycin. J. Clin. Lab. Anal. 2022, 36, e24427. [Google Scholar] [CrossRef]
- Rahman, T.; Yarnall, B.; Doyle, D.A. Efflux drug transporters at the forefront of antimicrobial resistance. Eur. Biophys. J. 2017, 46, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Moore-Machacek, A.; Gloe, A.; O’Leary, N.; Reen, F.J. Efflux, Signaling and Warfare in a Polymicrobial World. Antibiotics 2023, 12, 731. [Google Scholar] [CrossRef]
- Zwama, M.; Nishino, K. Ever-Adapting RND Efflux Pumps in Gram-Negative Multidrug-Resistant Pathogens: A Race against Time. Antibiotics 2021, 10, 774. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.M.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Tasneem, U.; Mehmood, K.; Majid, M.; Ullah, S.R.; Andleeb, S. Methicillin resistant Staphylococcus aureus: A brief review of virulence and resistance. J. Pak. Med. Assoc. 2022, 72, 509–515. [Google Scholar] [CrossRef]
- Yu, D.; Guo, D.; Zheng, Y.; Yang, Y. A review of penicillin binding protein and group A Streptococcus with reduced-β-lactam susceptibility. Front. Cell. Infect. Microbiol. 2023, 13, 1117160. [Google Scholar] [CrossRef] [PubMed]
- Monama, M.Z.; Olotu, F.; Bishop, Ö.T. Investigation of Multi-Subunit Mycobacterium tuberculosis DNA-Directed RNA Polymerase and Its Rifampicin Resistant Mutants. Int. J. Mol. Sci. 2023, 24, 3313. [Google Scholar] [CrossRef] [PubMed]
- Delcour, A.H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta 2009, 1794, 808–816. [Google Scholar] [CrossRef] [Green Version]
- Michaux, C.; Ronneau, S.; Giorgio, R.T.; Helaine, S. Antibiotic tolerance and persistence have distinct fitness trade-offs. PLoS Pathog. 2022, 18, e1010963. [Google Scholar] [CrossRef]
- Precit, M.R.; Wolter, D.J.; Griffith, A.; Emerson, J.; Burns, J.L.; Hoffman, L.R. Optimized In Vitro Antibiotic Susceptibility Testing Method for Small-Colony Variant Staphylococcus aureus. Antimicrob. Agents Chemother. 2016, 60, 1725–1735. [Google Scholar] [CrossRef] [Green Version]
- Despotovic, M.; de Nies, L.; Busi, S.B.; Wilmes, P. Reservoirs of antimicrobial resistance in the context of One Health. Curr. Opin. Microbiol. 2023, 73, 10229. [Google Scholar] [CrossRef] [PubMed]
- Jian, Z.; Zeng, L.; Xu, T.; Sun, S.; Yan, S.; Yang, L.; Huang, Y.; Jia, J.; Dou, T. Antibiotic resistance genes in bacteria: Occurrence, spread, and control. J. Basic Microbiol. 2021, 61, 1049–1070. [Google Scholar] [CrossRef]
- Touchon, M.; Moura de Sousa, J.A.; Rocha, E.P. Embracing the enemy: The diversification of microbial gene repertoires by phage-mediated horizontal gene transfer. Curr. Opin. Microbiol. 2017, 38, 66–73. [Google Scholar] [CrossRef]
- Yan, J.; Bassler, B.L. Surviving as a Community: Antibiotic Tolerance and Persistence in Bacterial Biofilms. Cell Host Microbe 2019, 26, 15–21. [Google Scholar] [CrossRef]
- Rather, M.A.; Gupta, K.; Mandal, M. Microbial biofilm: Formation, architecture, antibiotic resistance, and control strategies. Braz. J. Microbiol. 2021, 52, 1701–1718. [Google Scholar] [CrossRef]
- Sauer, K.; Stoodley, P.; Goeres, D.M.; Hall-Stoodley, L.; Burmølle, M.; Stewart, P.S.; Bjarnsholt, T. The biofilm life cycle: Expanding the conceptual model of biofilm formation. Nat. Rev. Microbiol. 2022, 20, 608–620. [Google Scholar] [CrossRef]
- Roell, K.R.; Reif, D.M.; Motsinger-Reif, A.A. An Introduction to Terminology and Methodology of Chemical-Synergy-Perspectobes from Across Disciplines. Front. Pharmacol. 2017, 8, 158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, A.I.; Dias, A.M.; Zille, A. Synergistic Effects Between Metal Nanoparticles and Commercial Antimicrobial Agents: A Review. ACS Appl. Nano Mater. 2022, 5, 3030–3064. [Google Scholar] [CrossRef] [PubMed]
- Jelinkova, P.; Mazumdar, A.; Sur, V.P.; Kociova, S.; Dolezelikova, K.; Jimenez, A.M.J.; Koudelkova, Z.; Mishra, P.K.; Smerkova, K.; Heger, Z.; et al. Nanoparticle-drug conjugates treating bacterial infections. J. Control Release 2019, 307, 166–185. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk Susceptibility Tests, 13th ed.; PCLSI standard M02 Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Wang, Y.-W.; Tang, H.; Wu, D.; Liu, D.; Liu, Y.; Cao, A.; Wang, H. Enhanced bactericidal toxicity of silver nanoparticles by the antibiotic gentamicin. Environ. Sci. Nano 2016, 3, 788–798. [Google Scholar] [CrossRef]
- Panáček, A.; Kvítek, L.; Smékalová, M.; Večeřová, R.; Kolář, M.; Röderová, M.; Dyčka, F.; Šebela, M.; Prucek, R.; Tomanec, O.; et al. Bacterial resistance to silver nanoparticles and how to overcome it. Nat. Nanotechnol. 2018, 13, 65–71. [Google Scholar] [CrossRef]
- Abbaszadegan, A.; Ghahramani, Y.; Gholami, A.; Hemmateenejad, B.; Dorostkar, S.; Nabavizadeh, M.; Sharghi, H. The Effect of Charge at the Surface of Silver Nanoparticles on Antimicrobial Activity against Gram-Positive and Gram-Negative Bacteria: A Preliminary Study. J. Nanomater. 2015, 2015, 720654. [Google Scholar] [CrossRef] [Green Version]
- Patwekar, M.; Patwekar, F.; Alghamdi, S.; Kamal, M.; Allahyani, M.; Almehmadi, M.; Kabrah, A.; Dablool, A.S.; Alsaiari, A.A.; Jawaid, T.; et al. Vancomycin as an Antibacterial Agent Capped with Silver Nanoparticles: An Experimental Potential Analysis. BioMed Res. Int. 2022, 2022, 3682757. [Google Scholar] [CrossRef]
- Khatoon, N.; Alam, H.; Khan, A.; Raza, K.; Sardar, M. Ampicillin Silver Nanoformulations against Multidrug resistant bacteria. Sci. Rep. 2018, 9, 6848. [Google Scholar] [CrossRef] [Green Version]
- Kaur, A.; Preet, S.; Kumar, V.; Kumar, R.; Kumar, R. Synergetic effect of vancomycin loaded silver nanoparticles for enhanced antibacterial activity. Colloids Surf. B Biointerfaces 2019, 176, 62–69. [Google Scholar] [CrossRef]
- Thomas, R.; Jishma, P.; Snigdha, S.; Soumya, K.R.; Mathew, J.; Radhakrishnan, E.K. Enhanced antimicrobial efficacy of biosynthesized silver nanoparticle based antibiotic conjugates. Inorg. Chem. Commun. 2020, 117, 107978. [Google Scholar] [CrossRef]
- Deng, H.; McShan, D.; Zhang, Y.; Sinha, S.S.; Arslan, Z.; Ray, P.C.; Yu, H. Mechanistic Study of the Synergistic Antibacterial Activity of Combined Silver Nanoparticles and Common Antibiotics. Environ. Sci. Technol. 2016, 50, 8840–8880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyle, V.J.; Fancher, M.E.; Ross, R.W., Jr. Rapid, modified Kirby-Bauer susceptibility test with single, high-concentration antimicrobial disks. Antimicrob. Agents Chemother. 1973, 3, 418–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, A.; Mumtaz, S.; Li, C.H.; Hussain, I.; Rotello, V.M. Combatting antibiotic-resistant bacteria using nanomaterials. Chem. Soc. Rev. 2019, 48, 415–427. [Google Scholar] [CrossRef]
- Vallet-Regí, M.; González, B.; Izquierdo-Barba, I. Nanomaterials as Promising Alternative in the Infection Treatment. Int. J. Mol. Sci. 2019, 20, 3806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haji, S.H.; Ali, F.A.; Aka, S.T.H. Synergistic antibacterial activity of silver nanoparticles biosynthesized by carbapenem-resistant Gram-negative bacilli. Sci. Rep. 2022, 12, 15254. [Google Scholar] [CrossRef]
- Santos, L.M.; Stanisic, D.; Menezes, U.J.; Mendonça, M.A.; Barral, T.D.; Seyffert, N.; Azevedo, V.; Durán, N.; Meyer, R.; Tasic, L.; et al. Biogenic Silver Nanoparticles as a Post-surgical Treatment for Corynebacterium pseudotuberculosis Infection in Small Ruminants. Front. Microbiol. 2019, 10, 824. [Google Scholar] [CrossRef] [Green Version]
- Xie, K.; Ziao, Z.; Guo, Y.; Lei, W.; Li, G.; Zhao, S.; Liu, X.; Li, J.; Jiang, W.; Wu, S.; et al. Long-Term Prevention of Bacterial Infection and Enhanced Osteoinductivity of a Hybrid Coating with Selective Silver Toxicity. Adv. Healthc. Mater. 2019, 8, 1801465. [Google Scholar] [CrossRef]
- Thomas, R.; Nair, A.P.; Soumya, K.R.; Mathew, J.; Radhakrishnan, E.K. Antibacterial activity and synergistic effect of biosynthesized agnps with antibiotics against multidrug-resistant biofilm-forming coagulase-negative staphylococci isolated from clinical samples. Appl. Biochem. Biotechnol. 2014, 173, 449–460. [Google Scholar] [CrossRef]
- Sharifi-Rad, J.; Alfatemi, S.M.H.; Sharifi-Rad, M.; Iriti, M. Antimicrobial synergic effect of allicin and silver nanoparticles on skin infection caused by methicillin-resistant Staphylococcus aureus spp. Ann. Med. Health Sci. Res. 2014, 4, 863–868. [Google Scholar]
- Fayaz, A.M.; Balaji, A.M.; Girilal, M.; Yadav, R.; Kalaichelvan, P.T.; Venketesan, R. Biogenic synthesis of silver nanoparticles and their synergistic effect with antibiotics: A study against gram-positive and gram-negative bacteria. Nanomed.-Nanotechnol. Biol. Med. 2010, 6, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, S.Z.H.; Kiran, U.; Ali, M.I.; Jamal, A.; Hameed, A.; Ahmed, S.; Ali, N. Combined efficacy of biologically synthesized silver nanoparticles and different antibiotics against multidrug-resistant bacteria. Int. J. Nanomed. 2013, 8, 3187–3195. [Google Scholar] [CrossRef] [Green Version]
- Thirumurugan, G.; Rao, J.V.L.N.; Dhanaraju, M.D. Elucidating pharmacodynamic interaction of silver nanoparticle—Topical deliverable antibiotics. Sci. Rep. 2016, 6, 29982. [Google Scholar] [CrossRef]
- Smekalova, M.; Aragon, V.; Panacek, A.; Prucek, R.; Zboril, R.; Kvitek, L. Enhanced antibacterial effect of antibiotics in combination with silver nanoparticles against animal pathogens. Vet. J. 2016, 209, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Harshiny, M.; Matheswaran, M.; Arthanareeswaran, G.; Kumaran, S.; Rajasree, S. Enhancement of antibacterial properties of silver nanoparticles-ceftriaxone conjugate through Mukia maderaspatana leaf extract mediated synthesis. Ecotoxicol. Environ. Saf. 2015, 121, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Duran, N.; Marcato, P.D.; De Conti, R.; Alves, O.L.; Costa, F.T.M.; Brocchi, M. Potential use of silver nanoparticles on pathogenic bacteria, their toxicity and possible mechanisms of action. J. Braz. Chem. Soc. 2010, 21, 949–959. [Google Scholar] [CrossRef]
- Li, P.; Li, J.; Wu, C.Z.; Wu, Q.S.; Li, J. Synergistic antibacterial effects of beta-lactam antibiotic combined with silver nanoparticles. Nanotechnology 2005, 16, 1912–1917. [Google Scholar] [CrossRef]
- Saeb, A.T.M.; Alshammari, A.S.; Al-Brahim, H.; Al-Rubeaan, K.A. Production of silver nanoparticles with strong and stable antimicrobial activity against highly pathogenic and multidrug resistant bacteria. Sci. World J. 2014, 2014, 704708. [Google Scholar] [CrossRef] [Green Version]
- Hwang, I.S.; Hwang, J.H.; Choi, H.; Kim, K.J.; Lee, D.G. Synergistic effects between silver nanoparticles and antibiotics and the mechanisms involved. J. Med. Microbiol. 2012, 61, 1719–1726. [Google Scholar] [CrossRef] [Green Version]
- Shahverdi, A.R.; Fakhimi, A.; Shahverdi, H.R.; Minaian, S. Synthesis and effect of silver nanoparticles on the antibacterial activity of different antibiotics against Staphylococcus aureus and Escherichia coli. Nanomed. Nanotechnol. Biol. Med. 2007, 3, 168–171. [Google Scholar] [CrossRef]
- Ghosh, S.; Patil, S.; Ahire, M.; Kitture, R.; Kale, S.; Pardesi, K.; Cameotra, S.S.; Bellare, J.; Dhavale, D.D.; Jabgunde, A.; et al. Synthesis of silver nanoparticles using Dioscorea bulbifera tuber extract and evaluation of its synergistic potential in combination with antimicrobial agents. Int. J. Nanomed. 2012, 7, 483–496. [Google Scholar]
- Hari, N.; Thomas, T.K.; Nair, A.J. Comparative study on the synergistic action of garlic synthesized and citrate capped silver nanoparticles with beta-penem antibiotics. ISRN Nanotechnol. 2013, 2013, 792105. [Google Scholar] [CrossRef] [Green Version]
- Wan, G.; Ruan, L.; Yin, Y.; Yang, T.; Ge, M.; Cheng, X. Effects of silver nanoparticles in combination with antibiotics on the resistant bacteria Acinetobacter baumannii. Int. J. Nanomed. 2016, 11, 3789–3800. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Carrizales, M.; Velasco, K.; Castillo, C.; Flores, A.; Magaña, M.; Martinez-Castanon, G.; Martinez-Gutierrez, F. In Vitro Synergism of Silver Nanoparticles with Antibiotics as an Alternative Treatment in Multiresistant Uropathogens. Antibiotics 2018, 7, 50. [Google Scholar] [CrossRef] [Green Version]
- Salarian, A.A.; Mollamahale, Y.B.; Hami, Z.; Soltani-Rezaee-Rad, M. Cephalexin nanoparticles: Synthesis, cytotoxicity and their synergistic antibacterial study in combination with silver nanoparticles. Mater. Chem. Phys. 2017, 198, 125–130. [Google Scholar] [CrossRef]
- Kora, A.J.; Rastogi, L. Enhancement of Antibacterial Activity of Capped Silver Nanoparticles in Combination with Antibiotics, on Model Gram-Negative and Gram-Positive Bacteria. Bioinorg. Chem. Appl. 2013, 2013, 871097. [Google Scholar] [CrossRef] [Green Version]
- Rogowska, A.; Rafińska, K.; Pomastowski, P.; Walczak, J.; Railean-Plugaru, V.; Buszewska-Forajta, M.; Buszewski, B. Silver nanoparticles functionalized with ampicillin. Electrophoresis 2017, 38, 2757–2764. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Kumar, R. Enhanced bactericidal efficacy of polymer stabilized silver nanoparticles in conjugation with different classes of antibiotics. RSC Adv. 2019, 9, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Mazur, P.; Skiba-Kurek, I.; Mrowiec, P.; Karczewska, E.; Drożdż, R. Synergistic ROS-Associated Antimicrobial Activity of Silver Nanoparticles and Gentamicin Against Staphylococcus epidermidis. Int. J. Nanomed. 2020, 15, 3551–3562. [Google Scholar] [CrossRef]
- McShan, D.; Zhang, Y.; Deng, H.; Ray, P.C.; Yu, H. Synergistic Antibacterial Effect of Silver Nanoparticles Combined with Ineffective Antibiotics on Drug Resistant Salmonella typhimurium DT104. J. Environ. Sci. Health 2015, 33, 369–384. [Google Scholar]
- Panáček, A.; Smékalová, M.; Kilianová, M.; Prucek, R.; Bogdanová, K.; Večeřová, R.; Kolář, M.; Havrdová, M.; Płaza, G.A.; Chojniak, J.; et al. Strong and Nonspecific Synergistic Antibacterial Efficiency of Antibiotics Combined with Silver Nanoparticles at Very Low Concentrations Showing No Cytotoxic Effect. Molecules 2016, 21, 26. [Google Scholar] [CrossRef] [Green Version]
- Panáček, A.; Smékalová, M.; Večeřová, R.; Bogdanová, K.; Röderová, M.; Kolář, M.; Kilianová, M.; Hradilová, Š.; Froning, J.P.; Havrdová, M.; et al. Silver nanoparticles strongly enhance and restore bactericidal activity of inactive antibiotics against multiresistant Enterobacteriaceae. Colloids Surf. 2016, 142, 392–399. [Google Scholar] [CrossRef]
- Wypij, M.; Świecimska, M.; Czarnecka, J.; Dahm, H.; Rai, M.; Golinska, P. Antimicrobial and cytotoxic activity of silver nanoparticles synthesized from two haloalkaliphilic actinobacterial strains alone and in combination with antibiotics. J. Appl. Microbiol. 2018, 124, 1411–1424. [Google Scholar] [CrossRef] [PubMed]
- Katva, S.; Das, S.; Moti, H.S.; Jyoti, A.; Kaushik, S. Antibacterial Synergy of Silver Nanoparticles with Gentamicin and Chloramphenicol against Enterococcus faecalis. Pharmacogn. Mag. 2018, 13, S828–S833. [Google Scholar] [PubMed]
- Naik, M.M.; Prabhu, M.S.; Samant, S.N.; Naik, P.M.; Shirodkar, S. Synergistic Action of Silver Nanoparticles Synthesized from Silver Resistant Estuarine Pseudomonas aeruginosa Strain SN5 with Antibiotics against Antibiotic Resistant Bacterial Human Pathogens. Thalass. Int. J. Mar. Sci. 2017, 33, 73–80. [Google Scholar] [CrossRef]
- Chopade, B.A.; Singh, R.; Wagh, P.; Wadhwani, S.; Gaidhani, S.; Kumbhar, A.; Bellare, J. Synthesis, optimization, and characterization of silver nanoparticles from Acinetobacter calcoaceticus and their enhanced antibacterial activity when combined with antibiotics. Int. J. Nanomed. 2013, 8, 4277–4290. [Google Scholar] [CrossRef] [Green Version]
- Dar, M.A.; Ingle, A.; Rai, M. Enhanced antimicrobial activity of silver nanoparticles synthesized by Cryphonectria sp. Evaluated singly and in combination with antibiotics. Nanomedicine 2013, 9, 105–110. [Google Scholar] [CrossRef]
- Barapatre, A.; Aadil, K.R.; Jha, H. Synergistic antibacterial and antibiofilm activity of silver nanoparticles biosynthesized by lignin degrading fungus. Front. Microbiol. 2016, 3, 8. [Google Scholar] [CrossRef] [Green Version]
- Ranpariya, B.; Salunke, G.; Karmakar, S.; Babiya, K.; Sutar, S.; Kadoo, N.; Kumbhakar, P.; Ghosh, S. Antimicrobial Synergy of Silver-Platinum Nanohybrids With Antibiotics. Front. Microbiol. 2021, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Saratale, G.D.; Saratale, R.G.; Benelli, G.; Kumar, G.; Pugazhendhi, A.; Kim, D.-S.; Shin, H.-S. Anti-diabetic Potential of Silver Nanoparticles Synthesized with Argyreia nervosa Leaf Extract High Synergistic Antibacterial Activity with Standard Antibiotics Against Foodborne Bacteria. J. Clust. Sci. 2017, 28, 1709–1727. [Google Scholar] [CrossRef]
- Rastogi, L.; Kora, A.J.; Sashidhar, R.B. Antibacterial effects of gum kondagogu reduced/stabilized silver nanoparticles in combination with various antibiotics: A mechanistic approach. Appl. Nanosci. 2015, 5, 535–543. [Google Scholar] [CrossRef] [Green Version]
- Halawani, E.M.; Hassan, A.M.; Gad El-Rab, S.M.F. Nanoformulation of Biogenic Cefotaxime-Conjugated-Silver Nanoparticles for Enhanced Antibacterial Efficacy Against Multidrug- Resistant Bacteria and Anticancer Studies. Int. J. Nanomed. 2020, 15, 1889–1901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abo-Shama, U.H.; El-Gendy, H.; Mousa, W.S.; Hamouda, R.A.; Yousuf, W.E.; Hetta, H.F.; Abdeen, E.E. Synergistic and Antagonistic Effects of Metal Nanoparticles in Combination with Antibiotics Against Some Reference Strains of Pathogenic Microorganisms. Infect. Drug Resist. 2020, 13, 351–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, P.; Wang, F.-Q.; Li, C.-T.; Yan, Z.-F. An Enhancement of Antibacterial Activity and Synergistic Effect of Biosynthesized Silver Nanoparticles by Eurotium cristatum with Various Antibiotics. Biotechnol. Bioprocess Eng. 2020, 25, 450–458. [Google Scholar] [CrossRef]
- Jyoti, K.; Baunthiyal, M.; Singh, A. Characterization of silver nanoparticles synthesized using Urtica dioica Linn. leaves and their synergistic effects with antibiotics. J. Radiat. Res. Appl. Sci. 2016, 9, 217–227. [Google Scholar] [CrossRef] [Green Version]
- Patra, J.K.; Baek, K.-H. Antibacterial Activity and Synergistic Antibacterial Potential of Biosynthesized Silver Nanoparticles against Foodborne Pathogenic Bacteria along with its Anticandidal and Antioxidant Effects. Front. Microbiol. 2017, 8, 1. [Google Scholar] [CrossRef] [Green Version]
- Sabir, D. Synergistic Effect of Silver Nanoparticles Combined with Different Antibiotics against Multidrug-Resistant Acinetobacter Baumannii Strain H72721. In Proceedings of the 3rd International Conference of Natural Science 2018-Biotechnology, Chamchamal, Iraq, 3 May 2018; pp. 7–11. [Google Scholar]
- Malawong, S.; Thammawithan, S.; Sirithongsuk, P.; Daduang, S.; Klaynongsruang, S.; Wong, P.T.; Patramanon, R. Silver Nanoparticles Enhance Antimicrobial Efficacy of Antibiotics and Restore That Efficacy against the Melioidosis Pathogen. Antibiotics 2021, 10, 839. [Google Scholar] [CrossRef]
- Vazquez-Munoz, R.; Meza-Villezcas, A.; Fournier, P.G.J.; Soria-Castro, E.; Juarez-Moreno, K.; Gallego-Hernandez, A.L.; Bogdanchikova, N.; Vazquez-Duhalt, R.; Huerta-Saquero, A. Enhancement of antibiotics antimicrobial activity due to the silver nanoparticles impact on the cell membrane. PLoS ONE 2019, 14, e0224904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moorthy, K.; Chang, K.C.; Yang, H.H.; Su, W.M.; Chiang, C.K.; Yuan, Z. Recent developments in detection and therapeutic approaches for antibiotic-resistant bacterial infections. J. Food Drug Anal. 2023, 31, 1–19. [Google Scholar] [CrossRef]
- Makabenta, J.M.V.; Nabawy, A.; Li, C.H.; Schmidt-Malan, S.; Patel, R.; Rotello, V.M. Nanomaterial-based therapeutics for antibiotic-resistant bacterial infections. Nat. Rev. Microbiol. 2021, 19, 23–36. [Google Scholar] [CrossRef]
- Modi, S.K.; Gaur, S.; Sengupta, M.; Singh, M.S. Mechanistic insights into nanoparticle-surface bacterial membrane interactions in overcoming antibiotic resistance. Front. Microbiol. 2023, 14, 1135579. [Google Scholar] [CrossRef]
- Gupta, A.; Saleh, N.M.; Das, R.; Landis, R.F.; Bigdeli, A.; Motamedchaboki, K.; Rosa Campos, A.; Pomeroy, K.; Mahmoudi, M.; Rotello, V.M. Synergistic antimicrobial therapy using nanoparticles and antibiotics for the treatment of multidrug-resistant bacterial infection. Nano Futures 2017, 1, 015004. [Google Scholar] [CrossRef]
- Ellis, T.; Chiappi, M.; Garcia-Trenco, A.; Al-Eijji, M.; Sarkar, S.; Georgiou, T.K.; Shaffer, M.S.P.; Tetley, T.D.; Schwander, S.; Ryan, M.P.; et al. Multimetallic Microparticles Increase the Potency of Rifampicin against Intracellular Mycobacterium tuberculosis. ACS Nano 2018, 12, 5228–5240. [Google Scholar] [CrossRef] [PubMed]
- Geng, Z.; Cao, Z.; Liu, J. Recent advances in targeted antibacterial therapy basing on nanomaterials. Exploration 2023, 3, 20210117. [Google Scholar] [CrossRef]
- Cooper, G.M. The Cell: A Molecular Approach. 2nd edition. Sunderland (MA): Sinauer Associates; 2000. Summary. Available online: https://www.ncbi.nlm.nih.gov/books/NBK9943/ (accessed on 30 June 2023).
- National Primary Drinking Water Regulations. Available online: https://www.epa.gov/ground-water-and-drinking-water/national-primary-drinking-water-regulations (accessed on 18 July 2023).
- Secondary Drinking Water Standards: Guidance for Nuisance Chemicals. Available online: https://www.epa.gov/sdwa/secondary-drinking-water-standards-guidance-nuisance-chemicals (accessed on 30 June 2023).
- World Health Organization. Guidelines for Drinking-Water Quality, 2nd ed.; World Health Organization: Geneva, Switzerland, 1996; Volume 2. [Google Scholar]
- AshaRani, P.V.; Mun, G.L.K.; Hande, M.P.; Valiyaveettil, S. Cytotoxicity and genotoxicity of silver nanoparticles in human cells. ACS Nano 2009, 3, 279–290. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, L.; Chen, Q.; Chen, C. Cytotoxic potential of silver nanoparticles. Yonsei Med. J. 2014, 55, 283–291. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Huang, Z.; Yin, S.; Zhou, X.; Jiang, Y.; Shao, L. The lysosome-mitochondrion crosstalk engaged in silver nanoparticles-disturbed mitochondrial homeostasis. Sci. Total Environ. 2023, 889, 164078. [Google Scholar] [CrossRef] [PubMed]
- Lansdown, A.B. Silver in health care: Antimicrobial effects and safety in use. Curr. Probl. Dermatol. 2006, 33, 17–34. [Google Scholar] [PubMed] [Green Version]
- Campagnolo, L.; Massimiani, M.; Vecchione, L.; Piccirilli, D.; Toschi, N.; Magrini, A.; Bonanno, E.; Scimeca, M.; Castagnozzi, L.; Buonanno, G.; et al. Silver nanoparticles inhaled during pregnancy reach and affect the placenta and the foetus. Nanotoxicology 2017, 11, 687–698. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Zhao, L.; Bai, R.; Liu, Y.; Han, L.; Xu, Z.; Chen, F.; Autrup, H.; Long, D.; Chen, C. Silver nanoparticles induced oxidative and endoplasmic reticulum stresses in mouse tissues: Implications for the development of acute toxicity after intravenous administration. Toxicol. Res. 2016, 5, 602–608. [Google Scholar] [CrossRef] [Green Version]
- Quan, J.H.; Gao, F.F.; Lee, M.; Yuk, J.M.; Cha, G.H.; Chu, J.Q.; Wang, H.; Lee, Y.H. Involvement of endoplasmic reticulum stress response and IRE1-mediated ASK1/JNK/Mcl-1 pathways in silver nanoparticle-induced apoptosis of human retinal pigment epithelial cells. Toxicology 2020, 442, 152540. [Google Scholar] [CrossRef]
- Dąbrowska-Bouta, B.; Sulkowski, G.; Gewartowska, M.; Strużyńska, L. Endoplasmic Reticulum Stress Underlies Nanosilver-Induced Neurotoxicity in Immature Rat Brain. Int. J. Mol. Sci. 2022, 23, 13013. [Google Scholar] [CrossRef] [PubMed]
- Maurer, L.L.; Yang, X.; Schindler, A.J.; Taggart, R.K.; Jiang, C.; Hsu-Kim, H.; Sherwood, D.R.; Meyer, J.N. Intracellular trafficking pathways in silver nanoparticle uptake and toxicity in Caenorhabditis elegans. Nanotoxicology 2016, 10, 831–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caballero-Díaz, E.; Pfeiffer, C.; Kastl, L.; Rivera-Gil, P.; Simonet, B.; Valcárcel, M.; Jiménez-Lamana, J.; Laborda, F.; Parak, W.J. The toxicity of silver nanoparticles depends on their uptake by cells and thus on their surface chemistry. Part. Part. Syst. Charact. 2013, 30, 1079–1085. [Google Scholar] [CrossRef]
- Greulich, C.; Diendorf, J.; Simon, T.; Eggeler, G.; Epple, M.; Köller, M. Uptake and intracellular distribution of silver nanoparticles in human mesenchymal stem cells. Acta Biomater. 2011, 7, 347–354. [Google Scholar] [CrossRef]
- Ahamed, M.; Alsalhi, M.S.; Siddiqui, M.K. Silver nanoparticle applications and human health. Clin. Chim. Acta 2010, 411, 1841–1848. [Google Scholar] [CrossRef]
- Garcia, T.X.; Costa, G.M.; França, L.R.; Hofmann, M.C. Sub-acute intravenous administration of silver nanoparticles in male mice alters Leydig cell function and testosterone levels. Reprod. Toxicol. 2014, 45, 59–70. [Google Scholar] [CrossRef] [Green Version]
- Brouillard, C.; Bursztejn, A.C.; Latarche, C.; Cuny, J.F.; Truchetet, F.; Goullé, J.P.; Schmutz, J. Silver absorption and toxicity evaluation of silver wound dressings in 40 patients with chronic wounds. J. Eur. Acad. Dermatol. Venerol. 2018, 32, 2295–2299. [Google Scholar] [CrossRef]
- Tang, J.; Xiong, L.; Zhou, G.; Wang, S.; Wang, J.; Liu, L.; Li, J.; Yuan, F.; Lu, S.; Wan, Z.; et al. Silver nanoparticles crossing through and distribution in the blood-brain barrier in vitro. J. Nanosci. Nanotechnol. 2010, 10, 6313–6317. [Google Scholar] [CrossRef]
- Tiwari, D.K.; Jin, T.; Behari, J. Dose-dependent in-vivo toxicity assessment of silver nanoparticle in Wistar rats. Toxicol. Mech. Methods 2011, 21, 13–24. [Google Scholar] [CrossRef]
- Sung, J.H.; Ji, J.H.; Park, J.D.; Yoon, J.U.; Kim, D.S.; Jeon, K.S.; Song, M.Y.; Jeong, J.; Han, B.S.; Han, J.H.; et al. Subchronic inhalation toxicity of silver nanoparticles. Toxicol. Sci. 2009, 108, 452–461. [Google Scholar] [CrossRef]
- Stebounova, L.V.; Adamcakova-Dodd, A.; Kim, J.S.; Park, H.; O’Shaughnessy, P.T.; Grassian, V.H.; Thorne, P.S. Nanosilver induces minimal lung toxicity or inflammation in a subacute murine inhalation model. Part. Fibre Toxicol. 2011, 8, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hadrup, N.; Lam, H.R. Oral toxicity of silver ions, silver nanoparticles and colloidal silver--a review. Regul. Toxicol. Pharmacol. 2014, 68, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Loeschner, K.; Hadrup, N.; Qvortrup, K.; Larsen, A.; Gao, X.; Vogel, U.; Mortensen, A.; Lam, H.R.; Larsen, E.H. Distribution of silver in rats following 28 days of repeated oral exposure to silver nanoparticles or silver acetate. Part. Fibre Toxicol. 2011, 8, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Recordati, C.; De Maglie, M.; Cella, C.; Argentiere, S.; Paltrinieri, S.; Bianchessi, S.; Losa, M.; Fiordaliso, F.; Corbelli, A.; Milite, G.; et al. Repeated oral administration of low doses of silver in mice: Tissue distribution and effects on central nervous system. Part. Fibre Toxicol. 2021, 18, 23. [Google Scholar] [CrossRef]
- Schneider, M.; Stracke, F.; Hansen, S.; Schaefer, U.F. Nanoparticles and their interactions with the dermal barrier. Dermato-endocrinology 2009, 1, 197–206. [Google Scholar] [CrossRef] [Green Version]
- Kraeling, M.E.K.; Topping, V.D.; Keltner, Z.M.; Belgrave, K.R.; Bailey, K.D.; Gao, X.; Yourick, J.J. In vitro percutaneous penetration of silver nanoparticles in pig and human skin. Regul. Toxicol. Pharmacol. 2018, 95, 314–322. [Google Scholar] [CrossRef]
- Choi, J.E.; Kim, S.; Ahn, J.H.; Youn, P.; Kang, J.S.; Park, K.; Yi, J.; Ryu, D.Y. Induction of oxidative stress and apoptosis by silver nanoparticles in the liver of adult zebrafish. Aquat. Toxicol. 2010, 100, 151–159. [Google Scholar] [CrossRef]
- Ferdous, Z.; Nemmar, A. Health Impact of Silver Nanoparticles: A Review of the Biodistribution and Toxicity Following Various Routes of Exposure. Int. J. Mol. Sci. 2020, 21, 2375. [Google Scholar] [CrossRef] [Green Version]
- Lansdown, A.B. A pharmacological and toxicological profile of silver as an antimicrobial agent in medical devices. Adv. Pharmacol. Sci. 2010, 2010, 910686. [Google Scholar] [CrossRef] [Green Version]
- Thirumoorthy, N.; Manisenthil Kumar, K.T.; Shyam Sundar, A.; Panayappan, L.; Chatterjee, M. Metallothionein: An overview. World J. Gastroenterol. 2007, 13, 993–996. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Lai, W.; Cui, M.; Liang, L.; Lin, Y.; Fang, Q.; Liu, Y.; Xie, L. An Evaluation of Blood Compatibility of Silver Nanoparticles. Sci. Rep. 2016, 6, 25518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, A.; Casey, A.; Byrne, G.; Chambers, G.; Howe, O. Silver nanoparticles induce pro-inflammatory gene expression and inflammasome activation in human monocytes. J. Appl. Toxicol. 2016, 36, 1311–1320. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q.; Fang, L.; Ling, J.; Ding, C.Z.; Kang, B.; Huang, C.Z. Nanotoxicity of silver nanoparticles to red blood cells: Size dependent adsorption, uptake, and hemolytic activity. Chem. Res. Toxicol. 2015, 28, 501–509. [Google Scholar] [CrossRef]
- Bian, Y.; Kim, K.; Ngo, T.; Kim, I.; Bae, O.N.; Lim, K.M.; Chung, J.H. Silver nanoparticles promote procoagulant activity of red blood cells: A potential risk of thrombosis in susceptible population. Part. Fibre Toxicol. 2019, 16, 9. [Google Scholar] [CrossRef] [PubMed]
- Rosales, C. Neutrophil: A Cell with Many Roles in Inflammation or Several Cell Types? Front. Physiol. 2018, 9, 113. [Google Scholar] [CrossRef] [PubMed]
- Fraser, J.A.; Kemp, S.; Young, L.; Ross, M.; Prach, M.; Hutchinson, G.R.; Malone, E. Silver nanoparticles promote the emergence of heterogeneic human neutrophil sub-populations. Sci. Rep. 2018, 8, 7506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poon, V.K.; Burd, A. In vitro cytotoxity of silver: Implication for clinical wound care. Burns 2004, 30, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Shah, J.M.; Omar, E.; Pai, D.R.; Sood, S. Cellular events and biomarkers of wound healing. Indian J. Plast. Surg. 2012, 45, 220–228. [Google Scholar]
- Bastos, V.; Ferreira de Oliveira, J.M.; Brown, D.; Jonhston, H.; Malheiro, E.; Daniel-da-Silva, A.L.; Duarte, I.F.; Santos, C.; Oliveira, H. The influence of Citrate or PEG coating on silver nanoparticle toxicity to a human keratinocyte cell line. Toxicol. Lett. 2016, 249, 29–41. [Google Scholar] [CrossRef]
- Larese, F.F.; D’Agostin, F.; Crosera, M.; Adami, G.; Renzi, N.; Bovenzi, M.; Maina, G. Human skin penetration of silver nanoparticles through intact and damaged skin. Toxicology 2009, 255, 33–37. [Google Scholar] [CrossRef]
- Rulemaking History for OTC Colloidal Silver Drug Products. Available online: https://www.fda.gov/drugs/historical-status-otc-rulemakings/rulemaking-history-otc-colloidal-silver-drug-products (accessed on 19 July 2023).
- Akter, M.; Sikder, M.T.; Rahman, M.M.; Ullah, A.K.M.A.; Hossain, K.F.B.; Banik, S.; Hosokawa, T.; Saito, T.; Kurasaki, M. A systematic review on silver nanoparticles-induced cytotoxicity: Physicochemical properties and perspectives. J. Adv. Res. 2017, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]

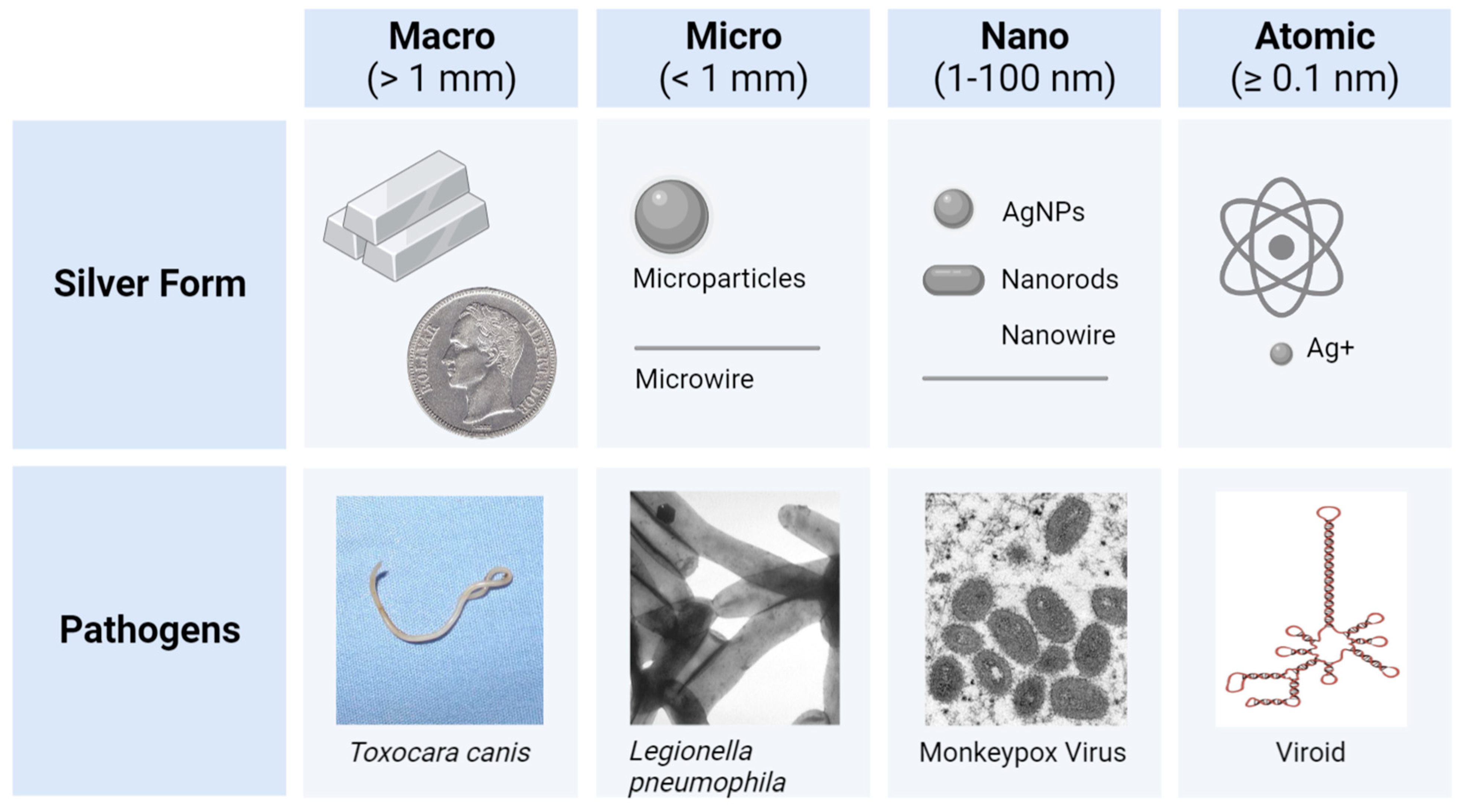
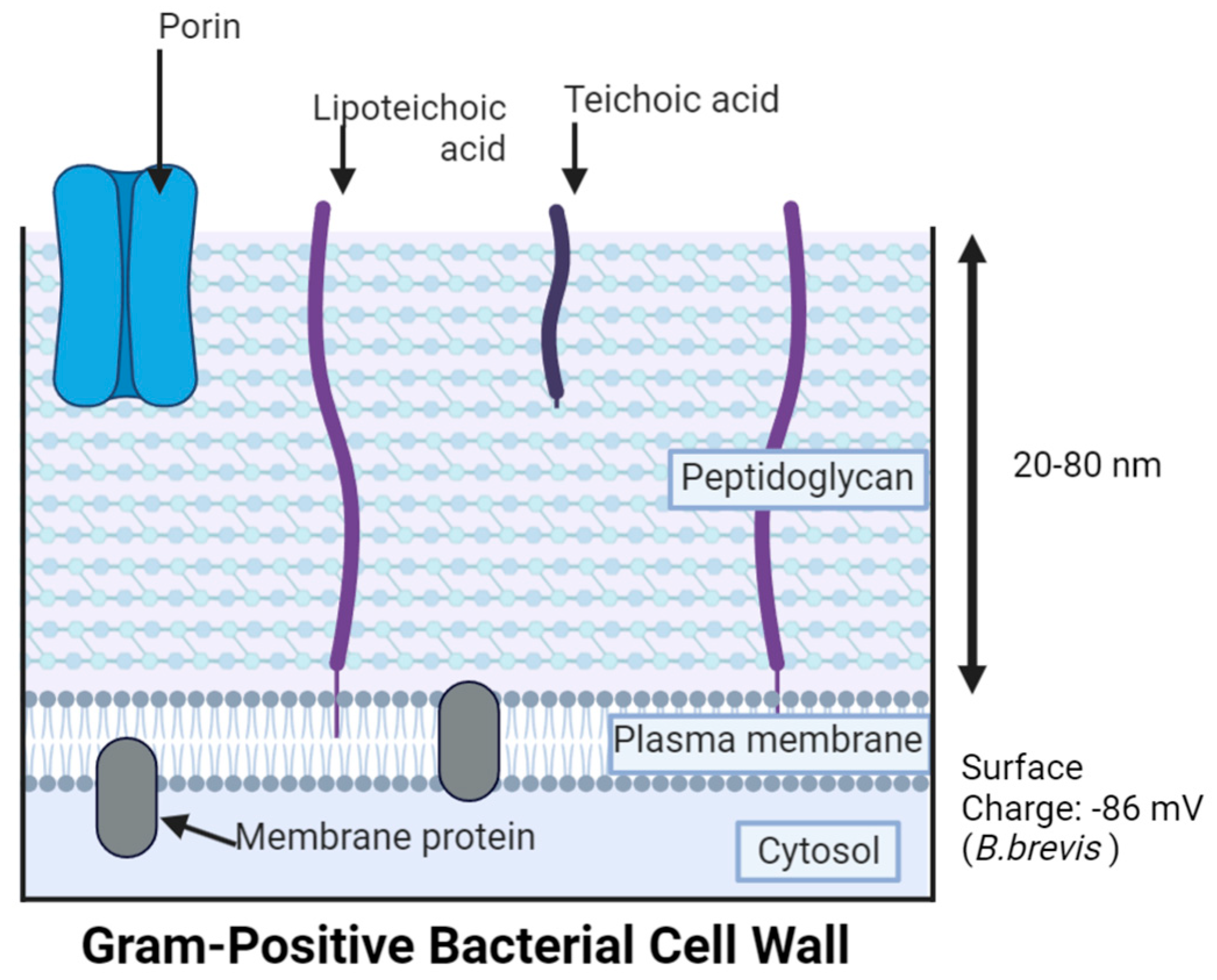
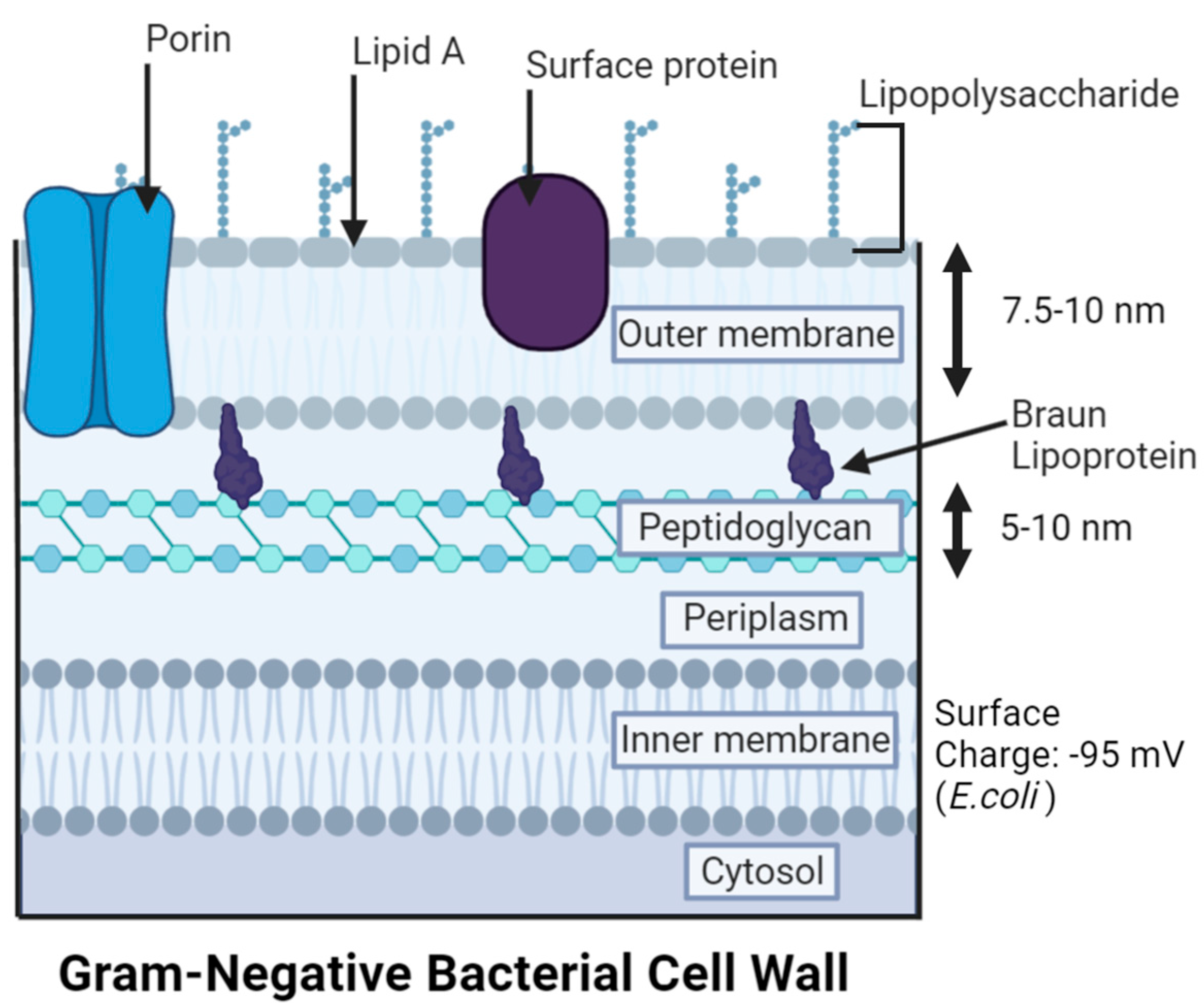
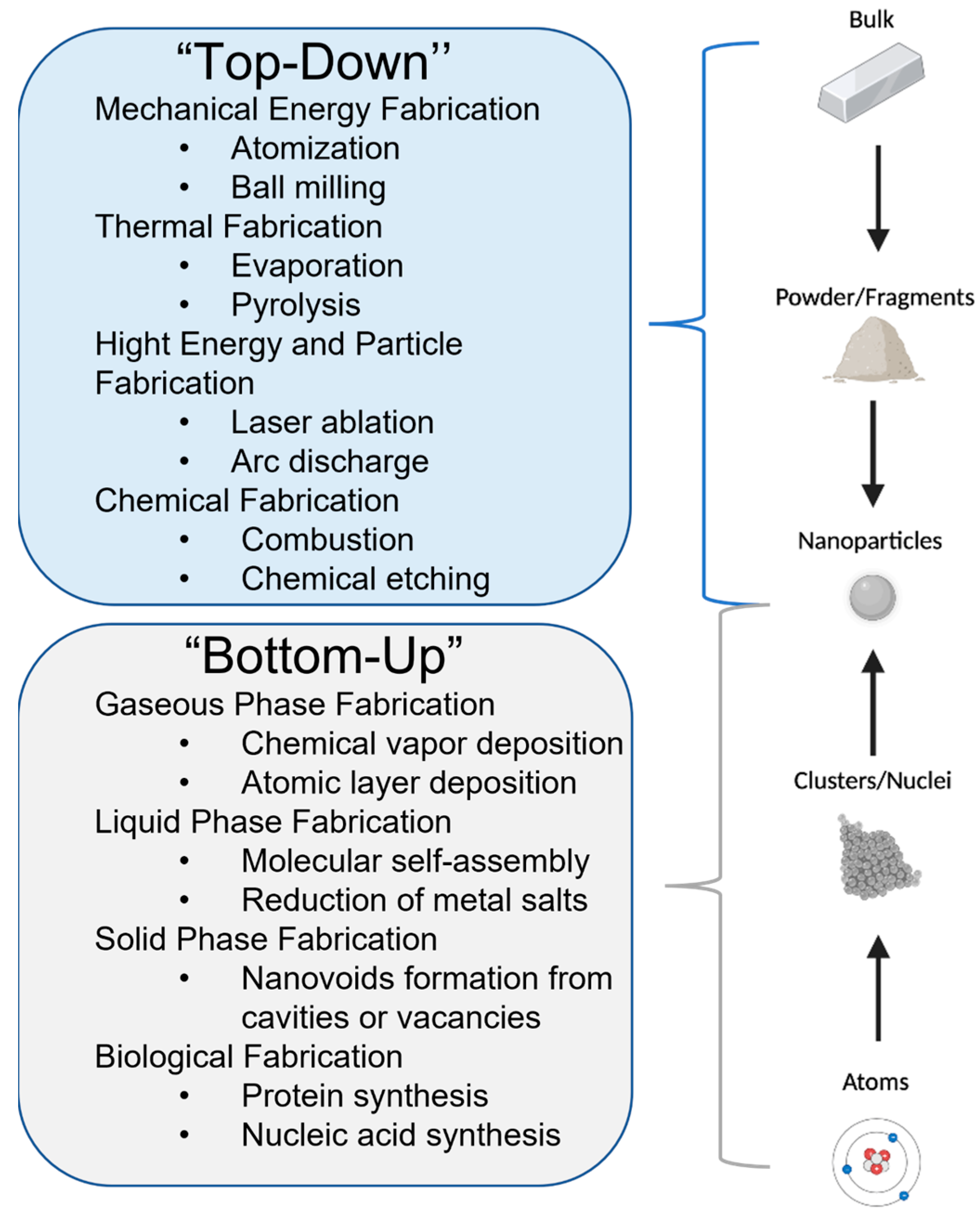
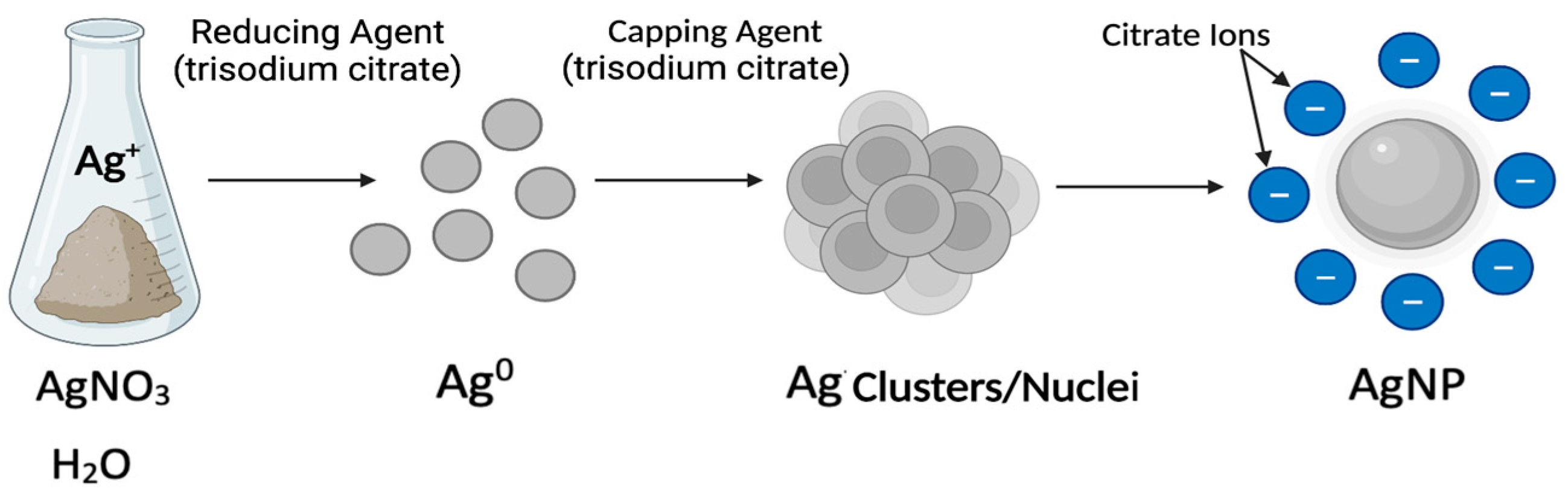
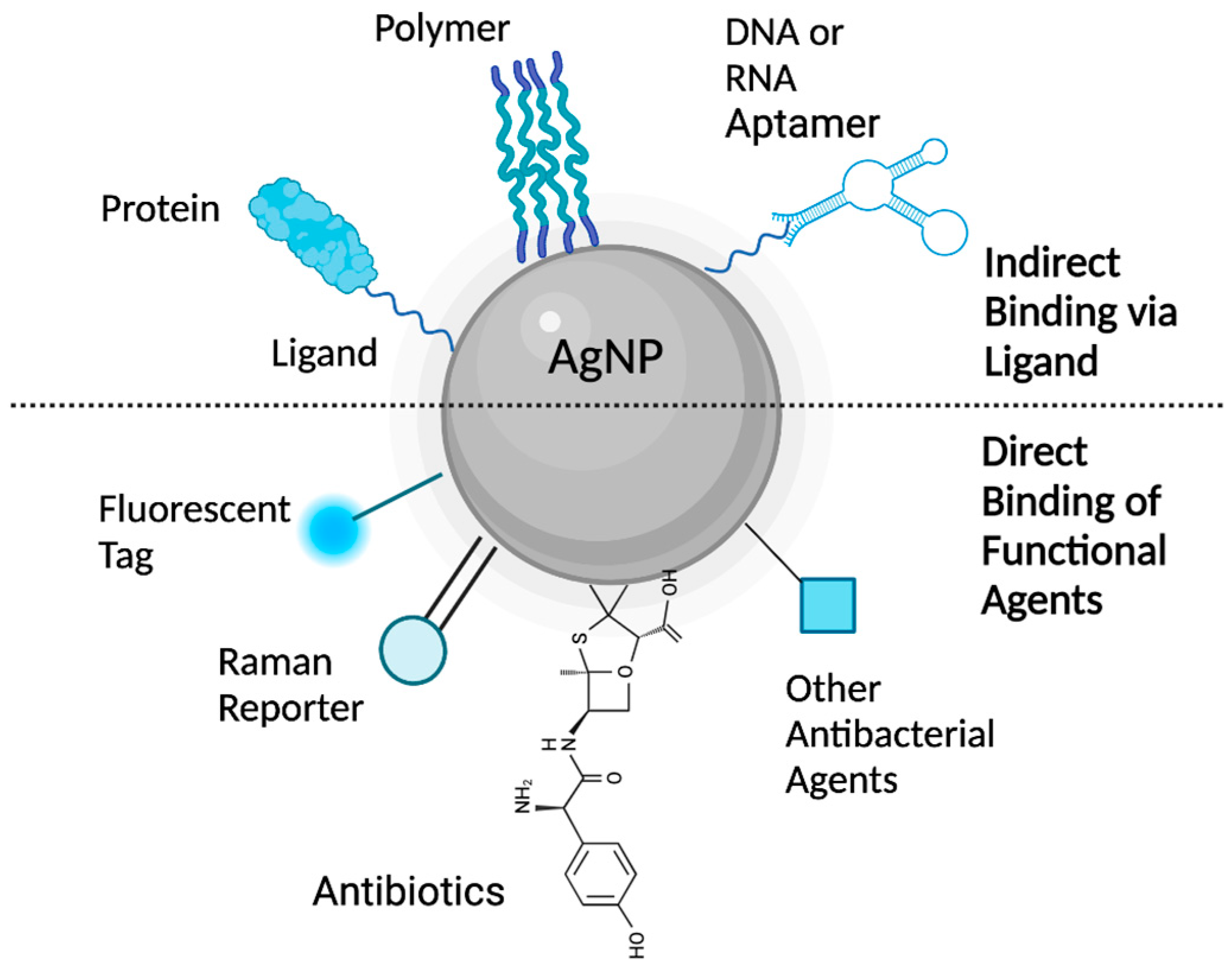
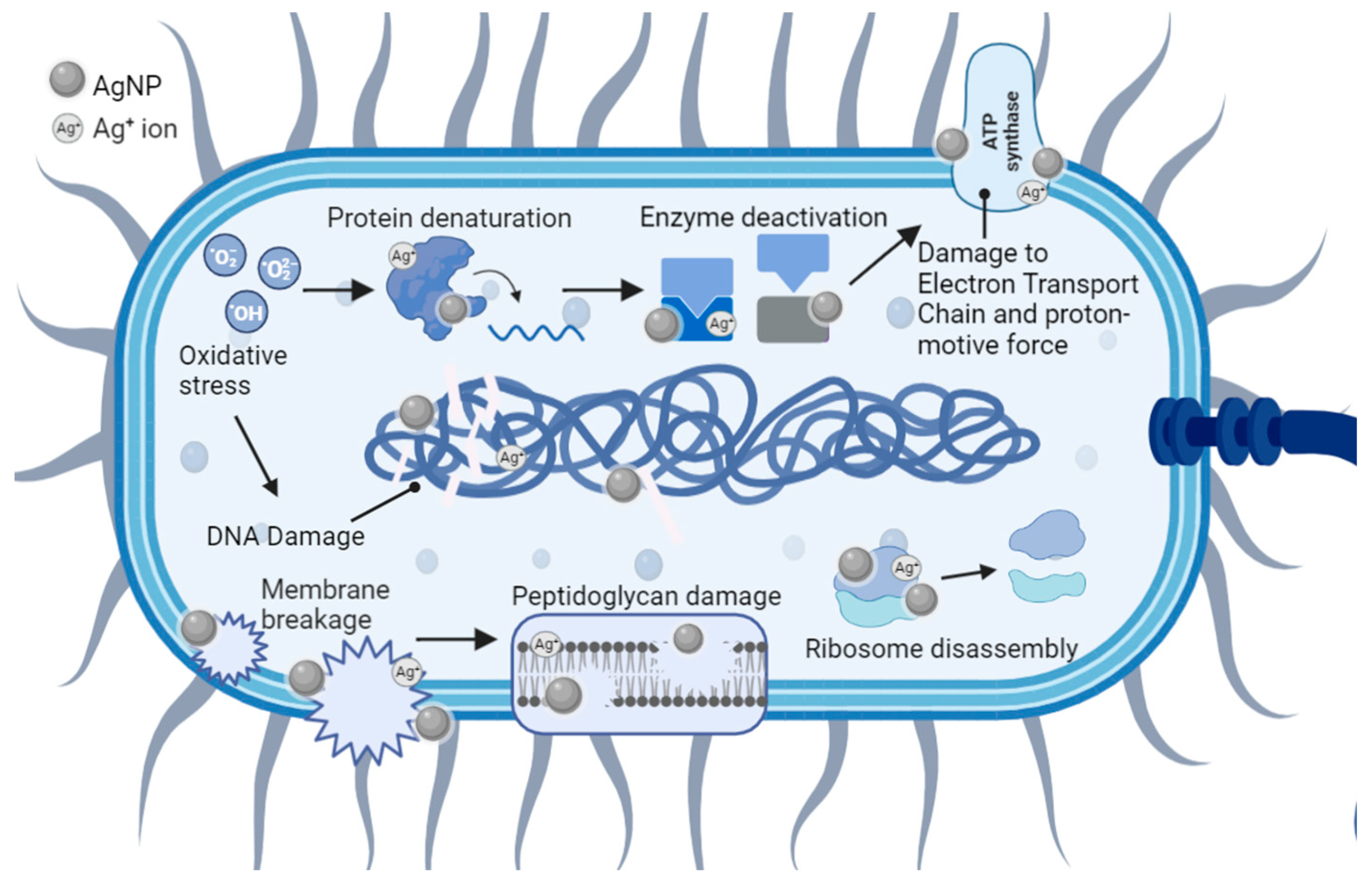
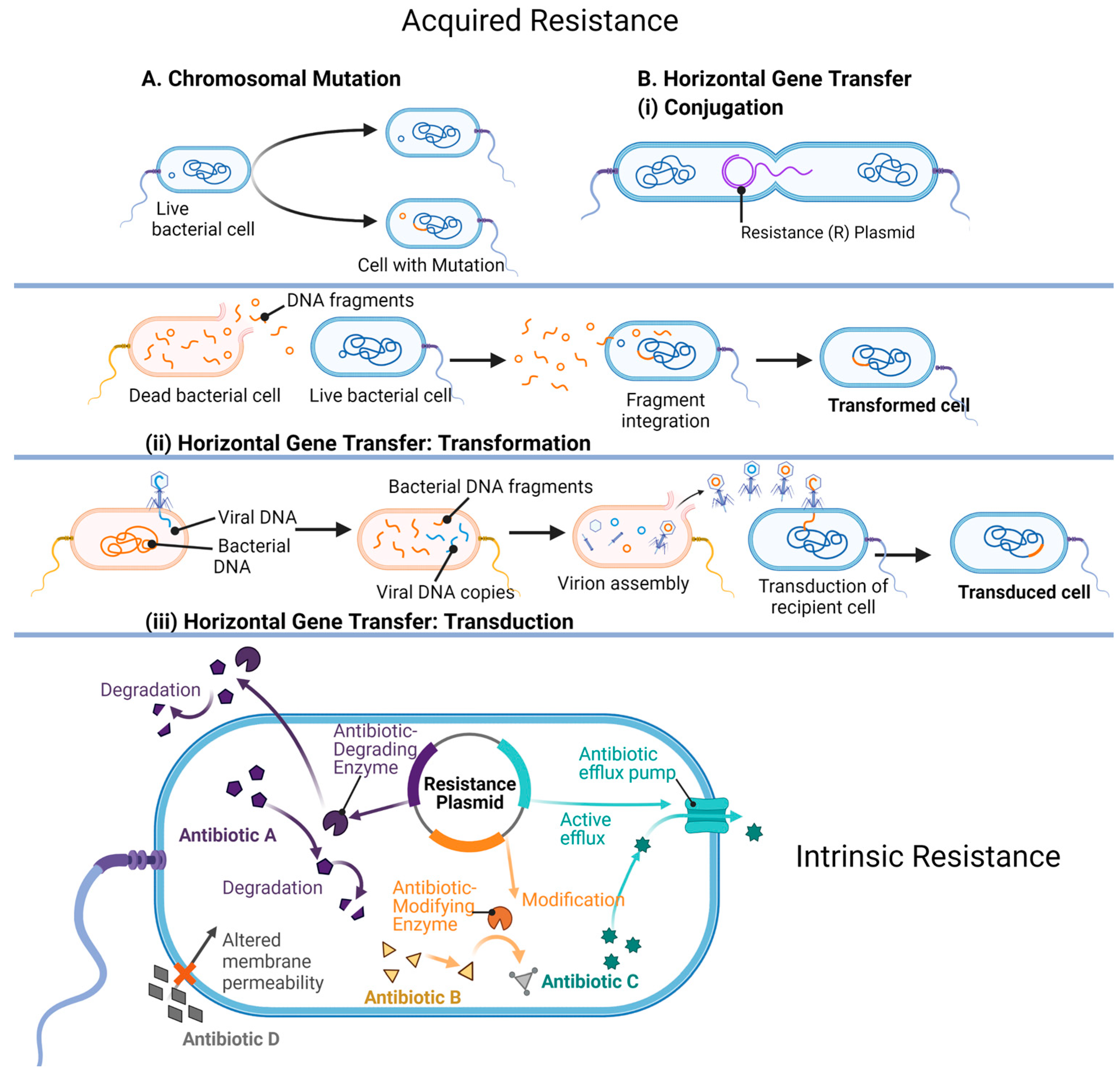

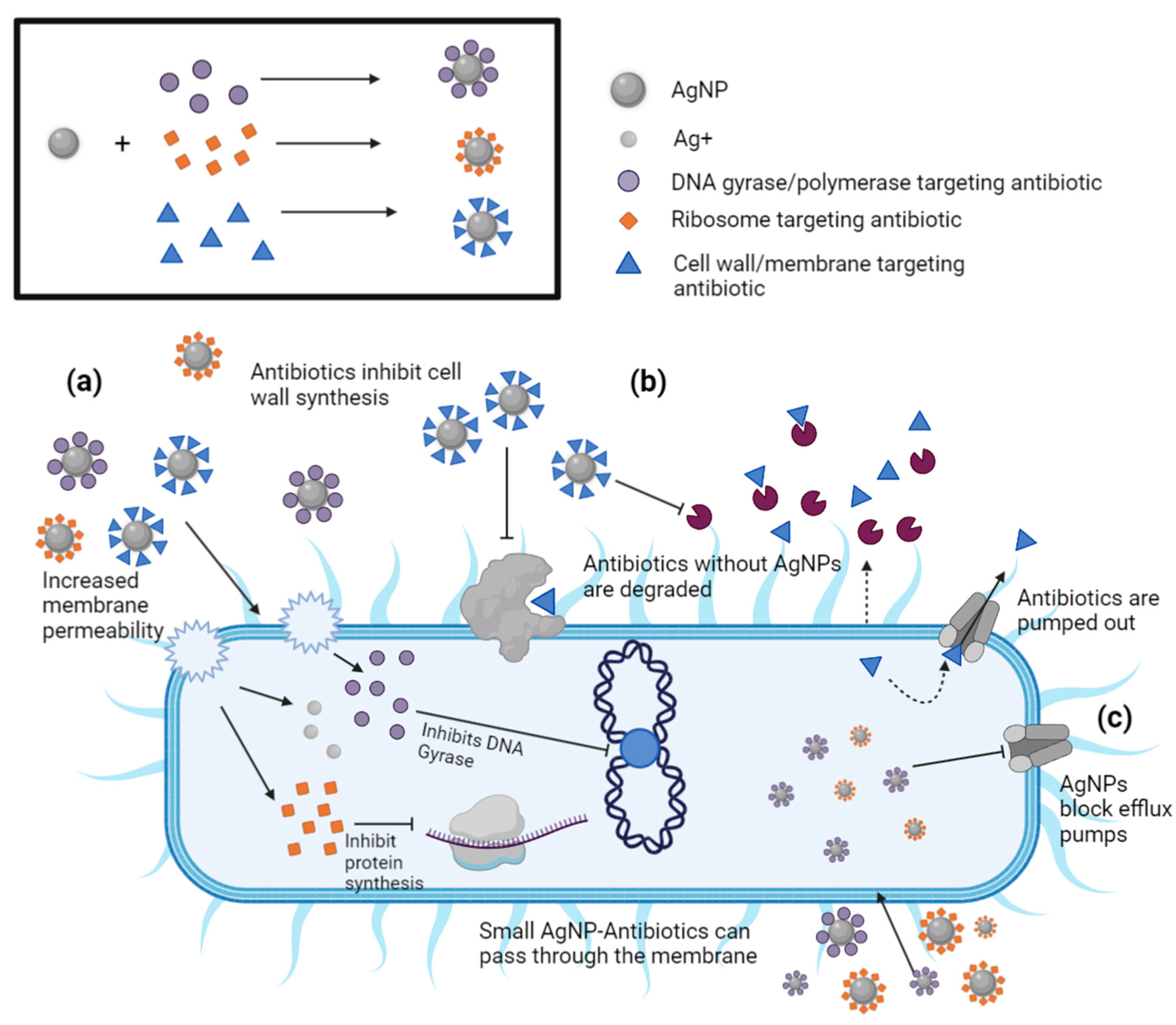
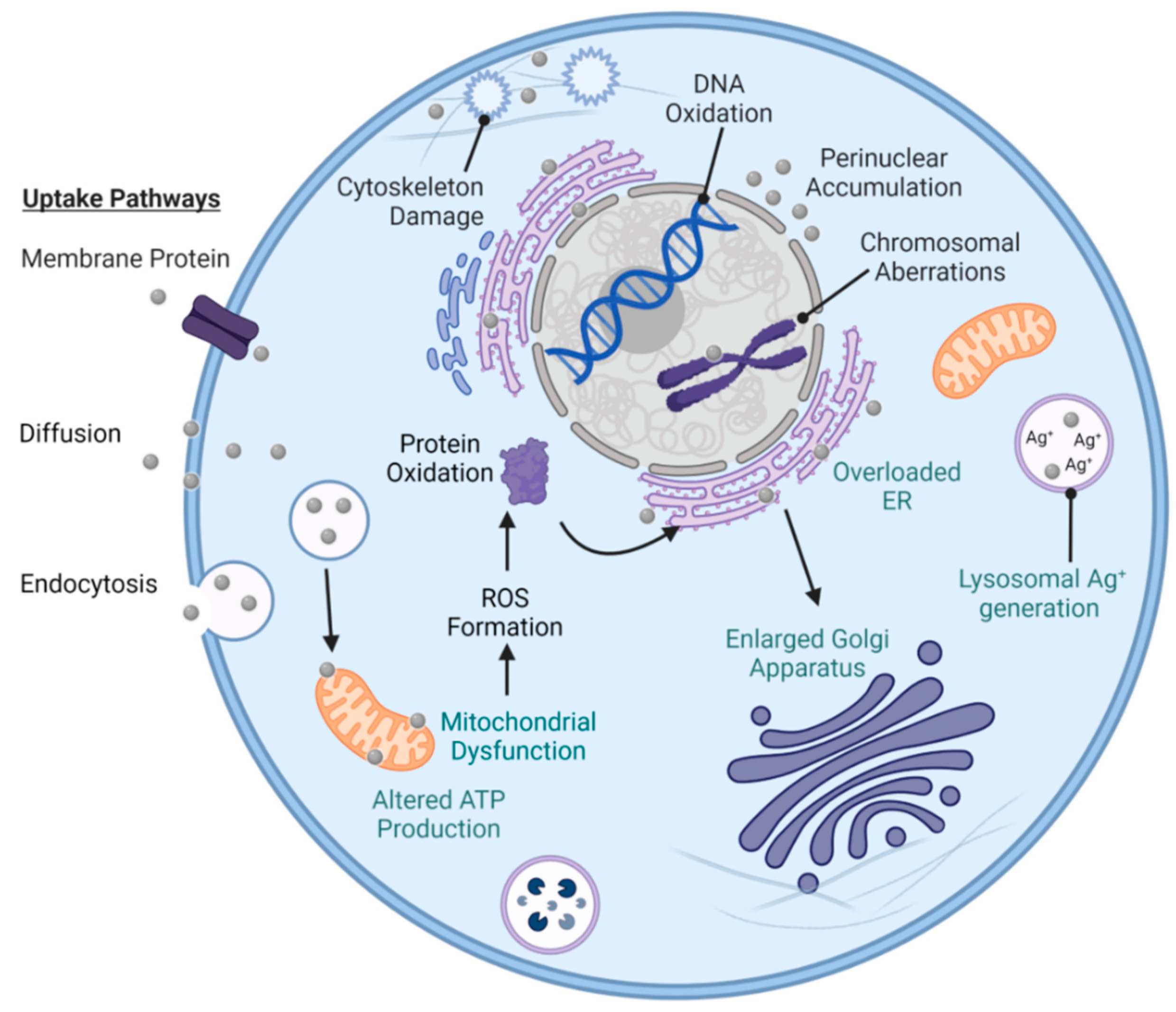
| Silver B.C.E. [1,4,7] | Silver Pre-Industrialization [1,7] | Silver during and Post Industrialization [8] | |
|---|---|---|---|
| Knowledge |
|
|
|
| Applications |
|
|
|
| Processes | Components | Advantages | Disadvantages |
|---|---|---|---|
| Chemical [116,123,124] PubMed: “silver nanoparticles chemical fabrication” n = 1459 |
|
|
|
| Biological [116,117,120,121,122] PubMed: “silver nanoparticles biological fabrication” n = 940 |
|
|
|
| Physical [115,116,125] PubMed: “silver nanoparticles physical fabrication” n = 290 |
|
|
|
| PCC Properties | Characterization Techniques |
|---|---|
| Size distribution and agglomeration | UV-Vis absorption spectrophotometry, dynamic light scattering (DLS), X-ray diffraction, scanning electron microscopy (SEM), and transmission electron microscopy (TEM) |
| Shape | SEM, TEM, STM, and atomic force microscopy (AFM) |
| Surface area and surface-to-volume ratio | TEM and Brunauer–Emmett–Teller measurements |
| Chemical composition and purity | UV-Vis absorption spectrophotometry, Raman spectroscopy, surface-enhanced Raman spectroscopy (SERS), XPS, flame atomic absorption spectroscopy (FAAS), inductively coupled plasma–optical emission spectroscopy (ICP-OES), ICP–mass spectrometry (ICP-MS), and scanning probe microscopy (SPM) |
| Surface functionalization | Fourier transform-infrared spectroscopy (FT-IR), Raman spectroscopy, X-ray photoelectron spectroscopy (XPS), thermogravimetric analysis (TGA), nuclear magnetic resonance (NMR), and X-ray diffraction spectroscopy (XRD) |
| Solubility and surface charge | Solubility tests, zeta potential measurements, electrophoretic mobility, contact angle measurements, hydrophobic interaction chromatography (HIC), atomic force microscopy (AFM), and scanning ion conductance microscopy (SICM) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaiser, K.G.; Delattre, V.; Frost, V.J.; Buck, G.W.; Phu, J.V.; Fernandez, T.G.; Pavel, I.E. Nanosilver: An Old Antibacterial Agent with Great Promise in the Fight against Antibiotic Resistance. Antibiotics 2023, 12, 1264. https://doi.org/10.3390/antibiotics12081264
Kaiser KG, Delattre V, Frost VJ, Buck GW, Phu JV, Fernandez TG, Pavel IE. Nanosilver: An Old Antibacterial Agent with Great Promise in the Fight against Antibiotic Resistance. Antibiotics. 2023; 12(8):1264. https://doi.org/10.3390/antibiotics12081264
Chicago/Turabian StyleKaiser, Kyra G., Victoire Delattre, Victoria J. Frost, Gregory W. Buck, Julianne V. Phu, Timea G. Fernandez, and Ioana E. Pavel. 2023. "Nanosilver: An Old Antibacterial Agent with Great Promise in the Fight against Antibiotic Resistance" Antibiotics 12, no. 8: 1264. https://doi.org/10.3390/antibiotics12081264
APA StyleKaiser, K. G., Delattre, V., Frost, V. J., Buck, G. W., Phu, J. V., Fernandez, T. G., & Pavel, I. E. (2023). Nanosilver: An Old Antibacterial Agent with Great Promise in the Fight against Antibiotic Resistance. Antibiotics, 12(8), 1264. https://doi.org/10.3390/antibiotics12081264