Sensing of Antibiotic–Bacteria Interactions
Abstract
:1. Introduction
2. Sensing the Antibiotic Mechanism of Action
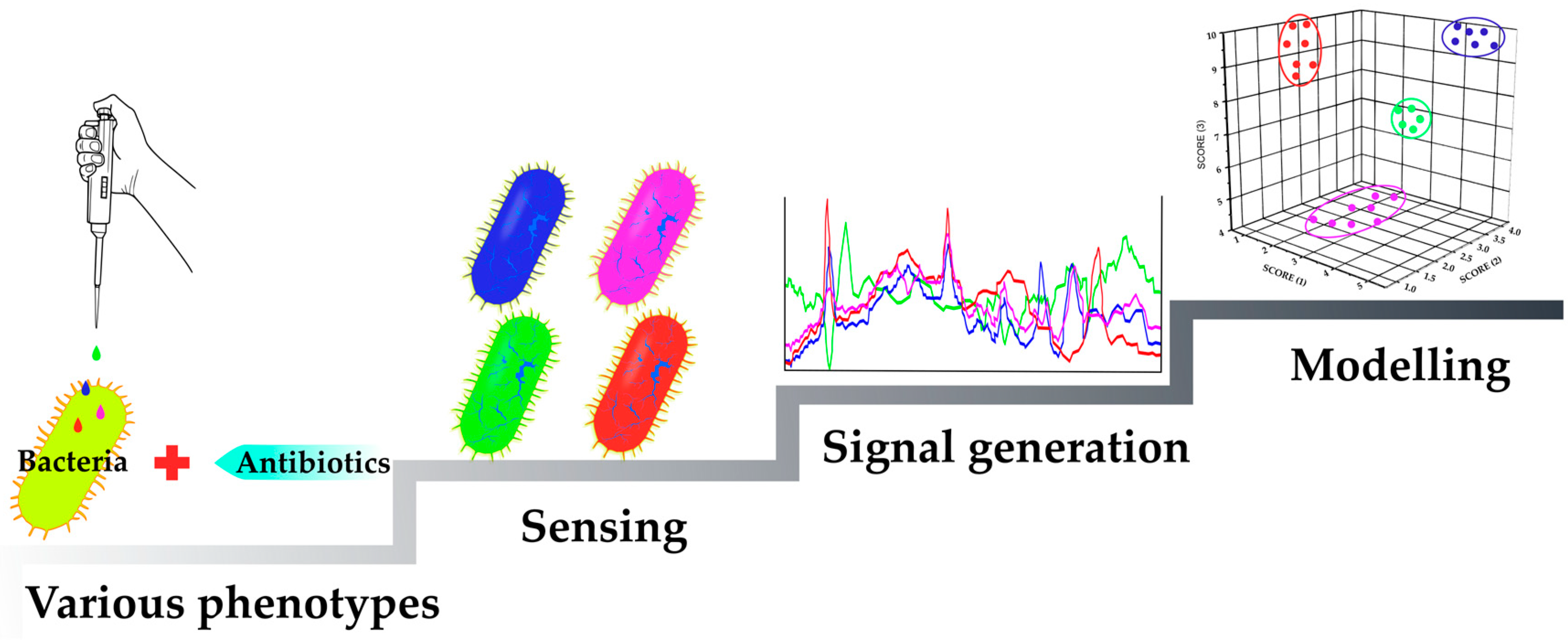
2.1. Mechanism-Independent Methods
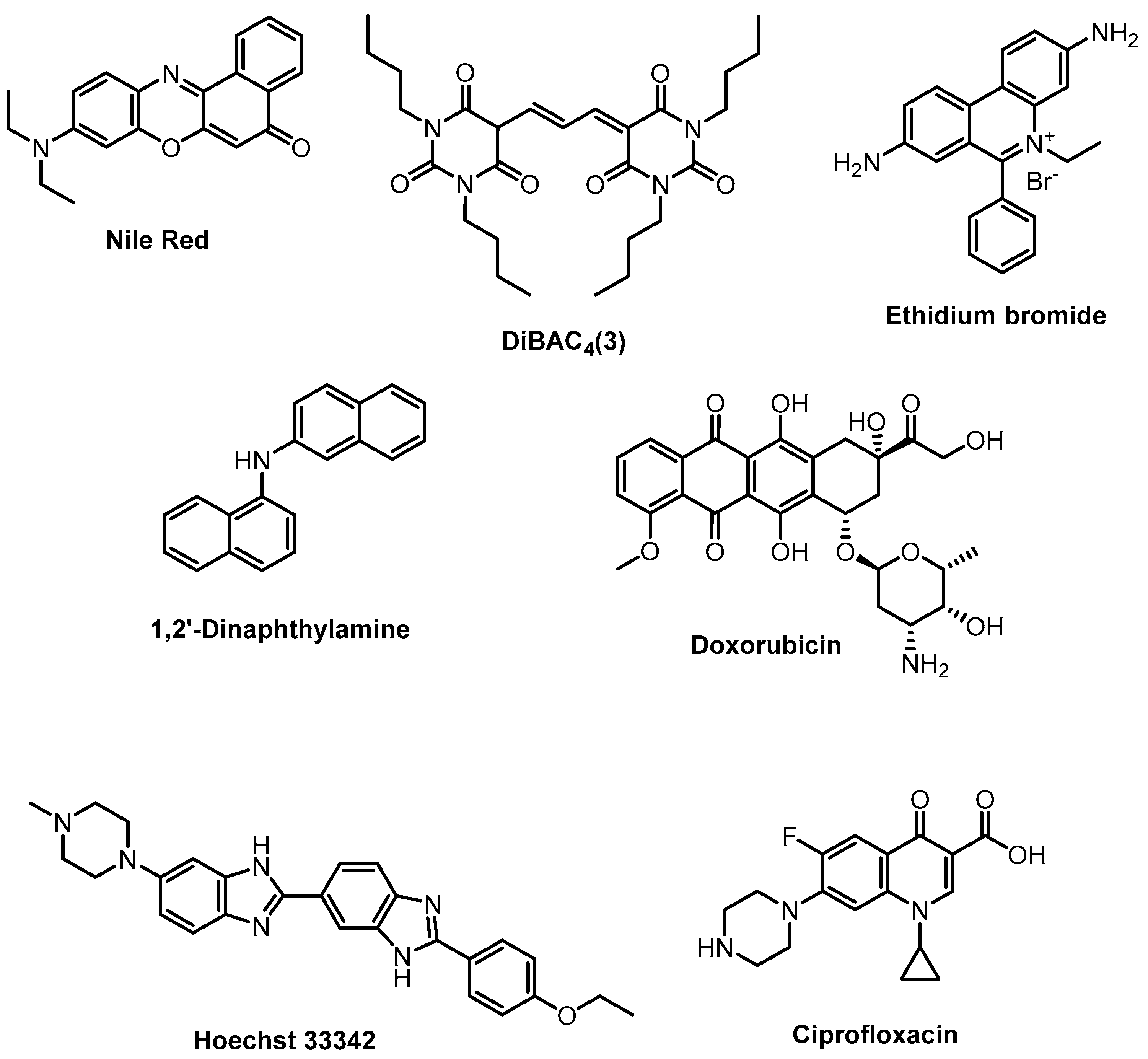
2.1.1. Sensing Phenotypes: Fluorescent Stains
2.1.2. Sensing Phenotypes: Fluorescent Array Sensors
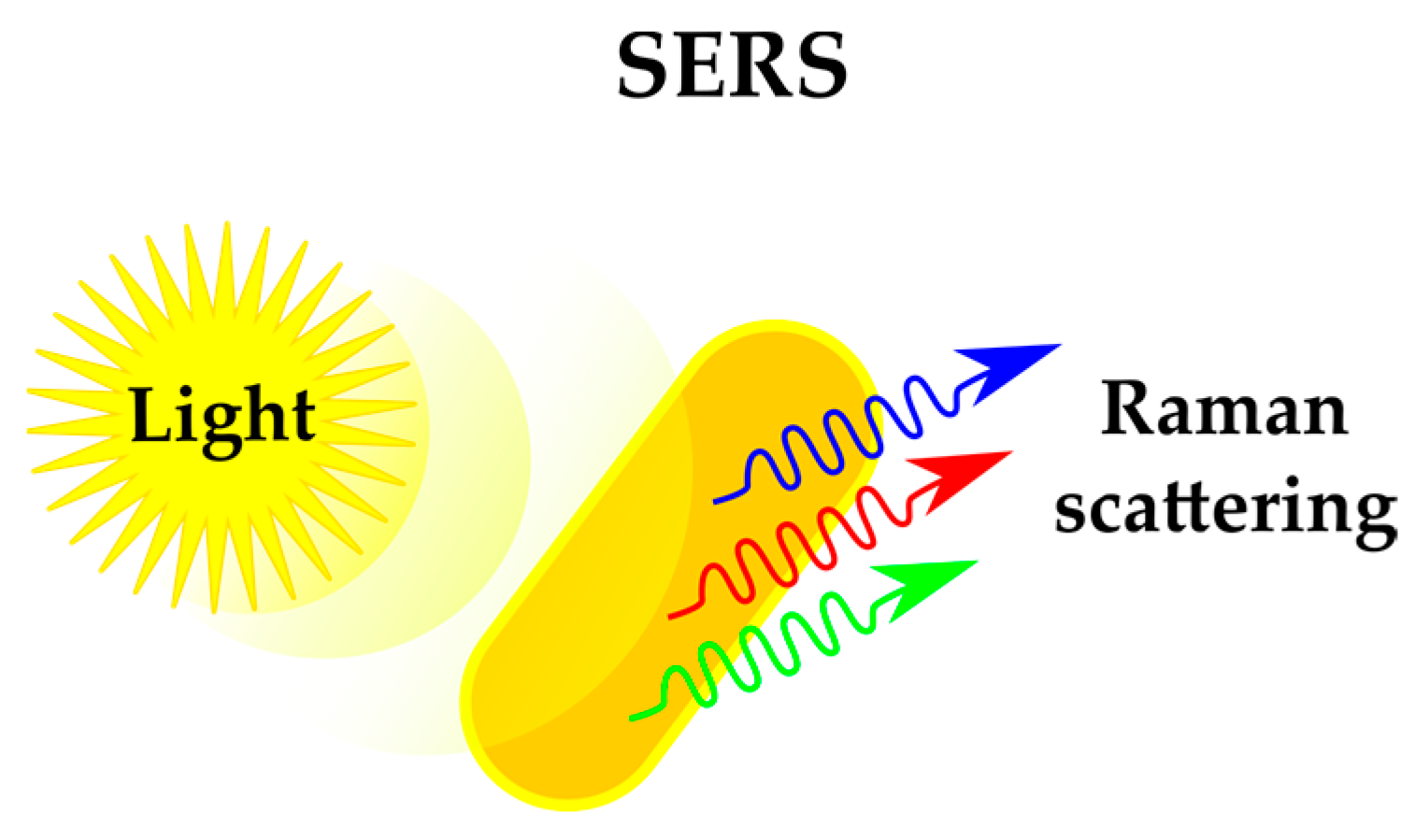
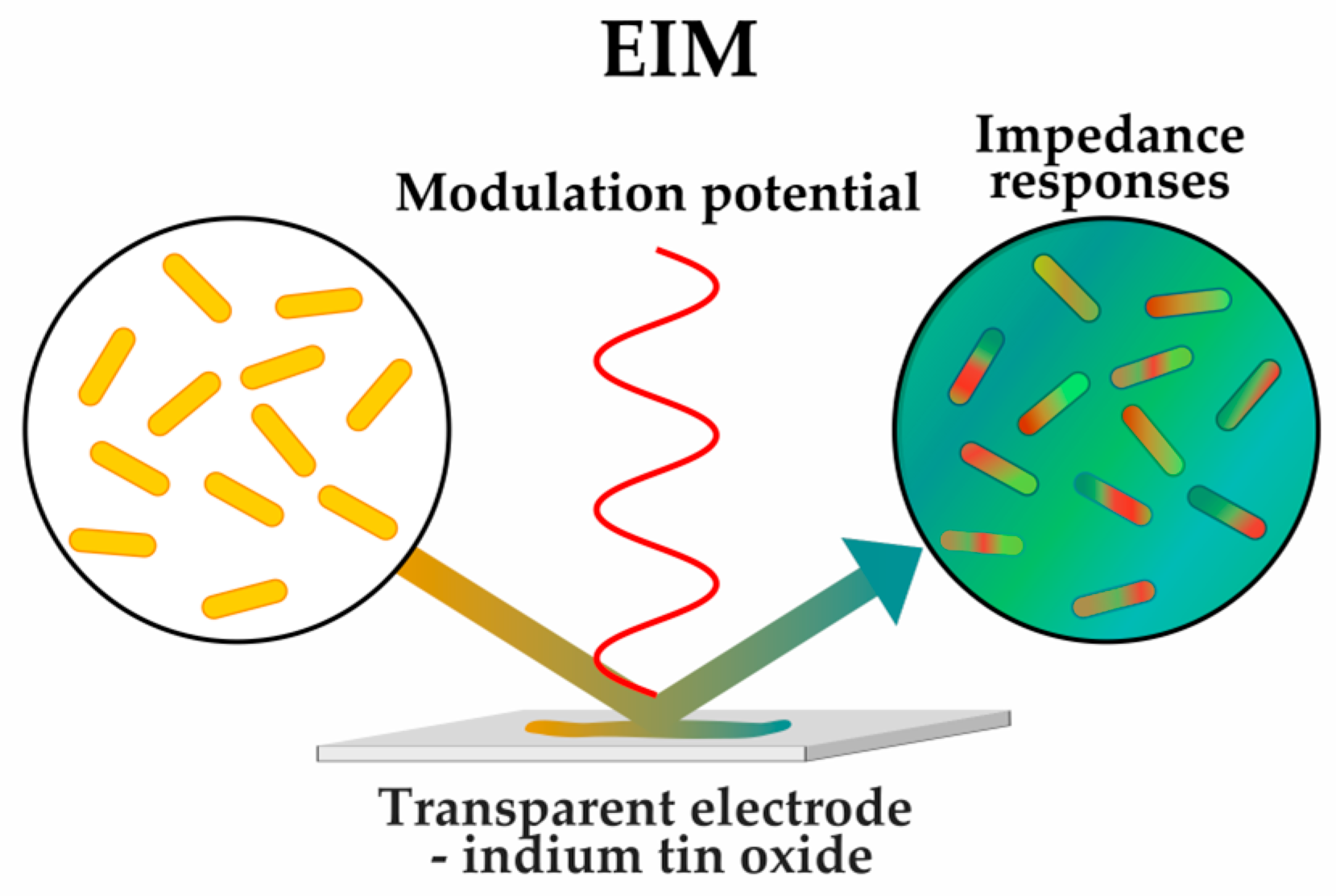
2.1.3. Sensing Phenotypes: Label-Free Methods
2.2. Narrow MoA Elucidation Techniques
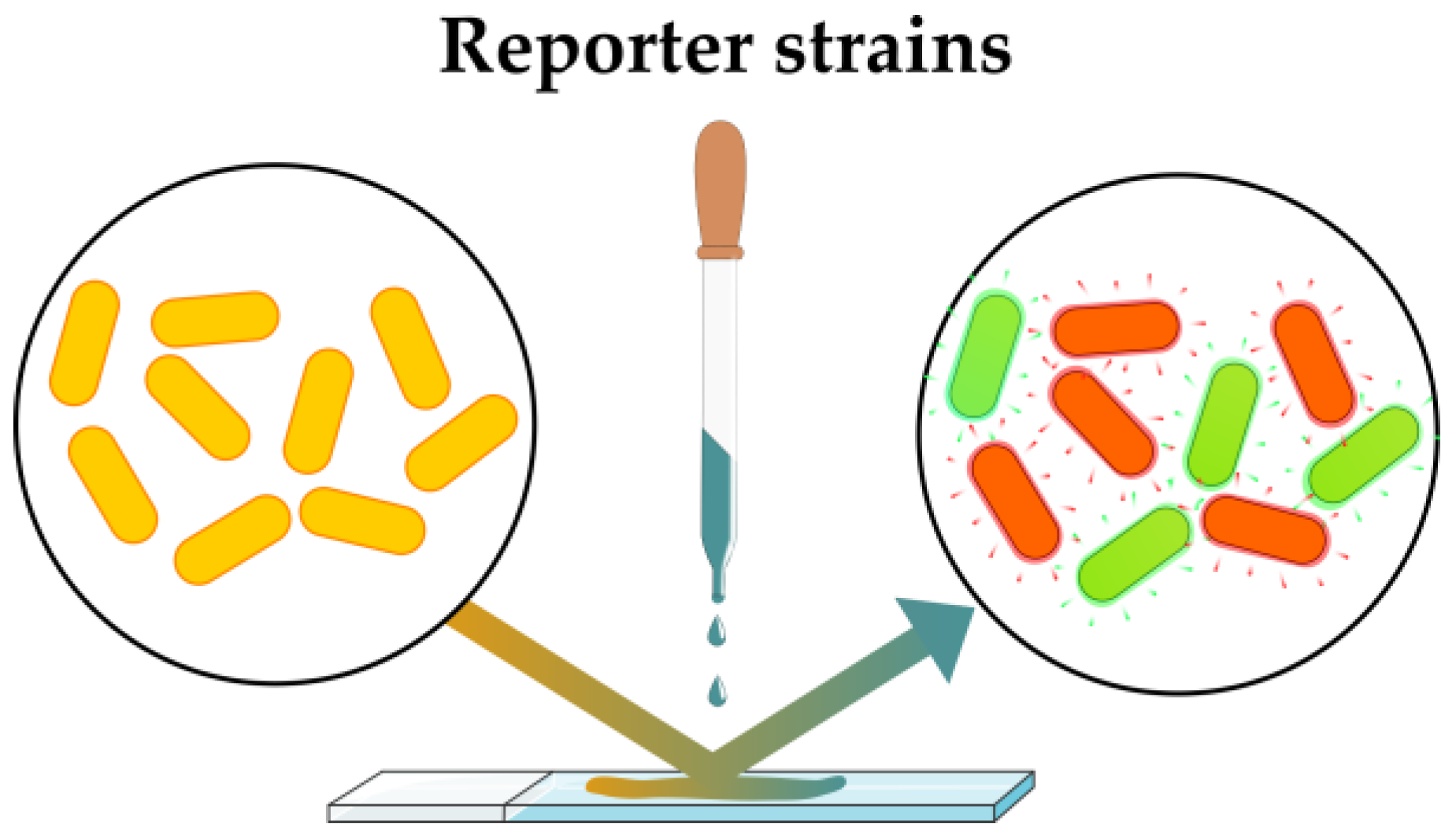
2.2.1. Sensing Artificial Phenotypes: Reporter Strains
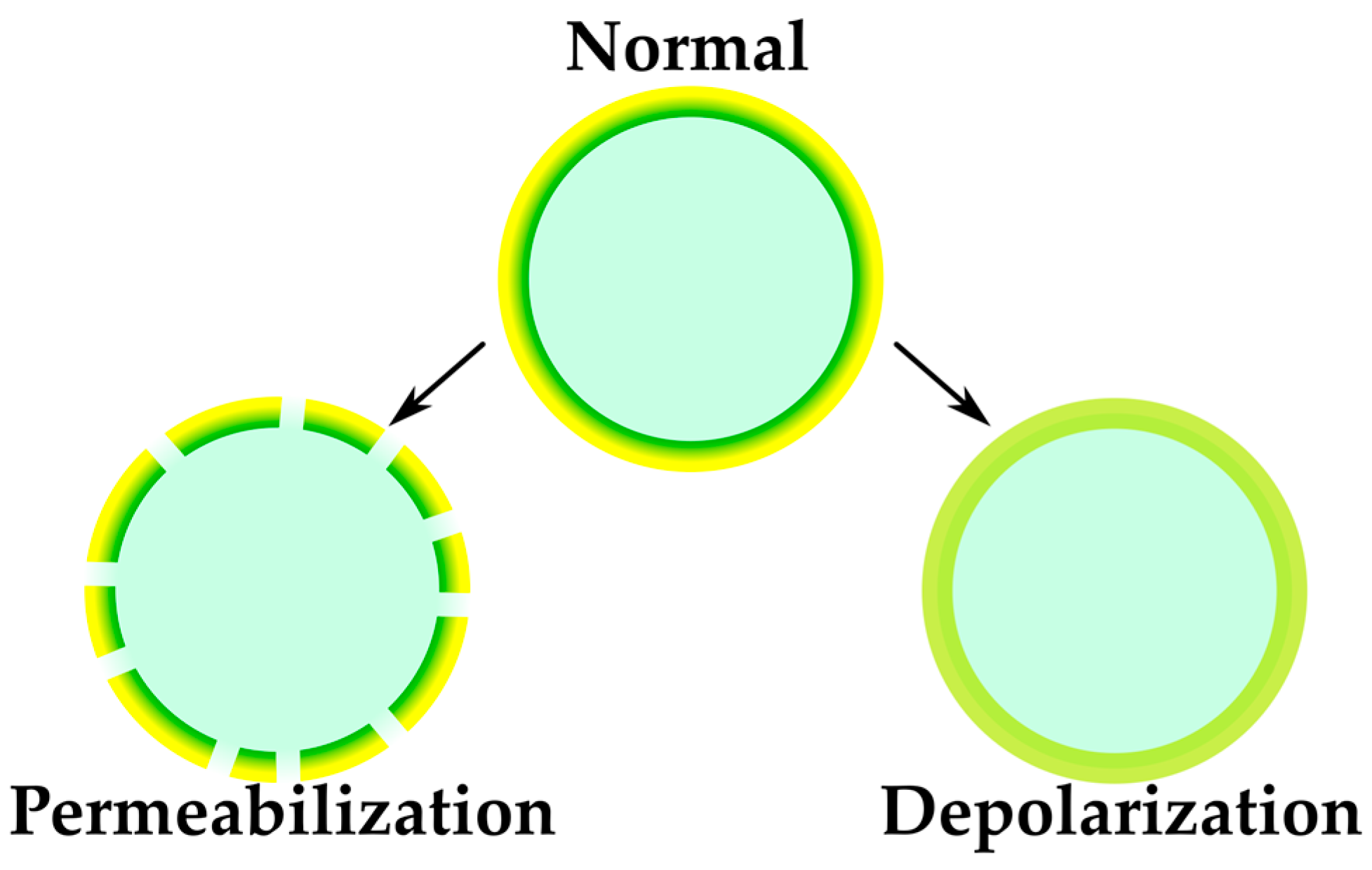
2.2.2. Sensing of Membrane-Targeting Antibiotics and Other Membrane-Related Effects
2.2.3. Peptidoglycan Targeting
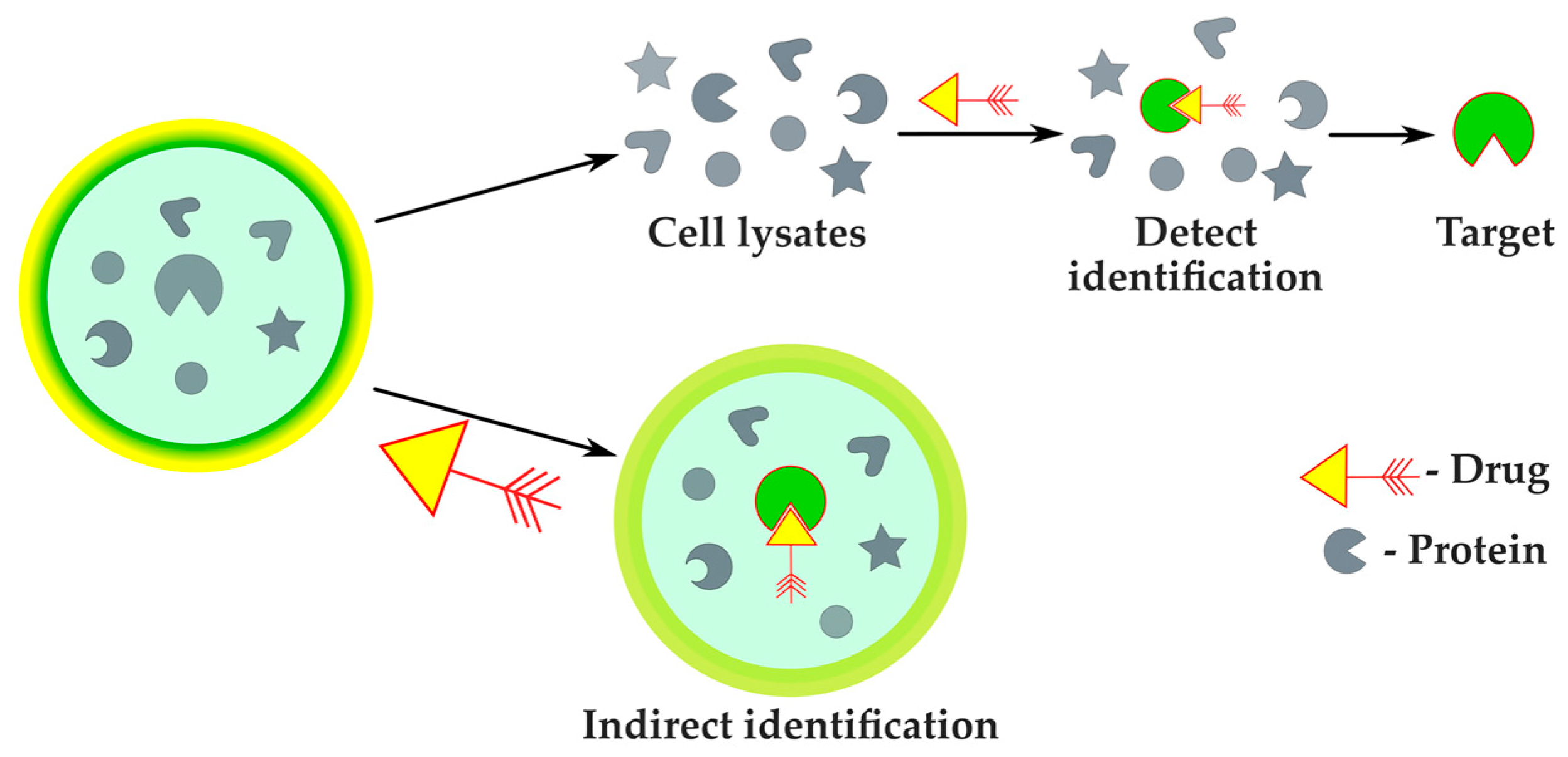
2.2.4. Protein Target Identification
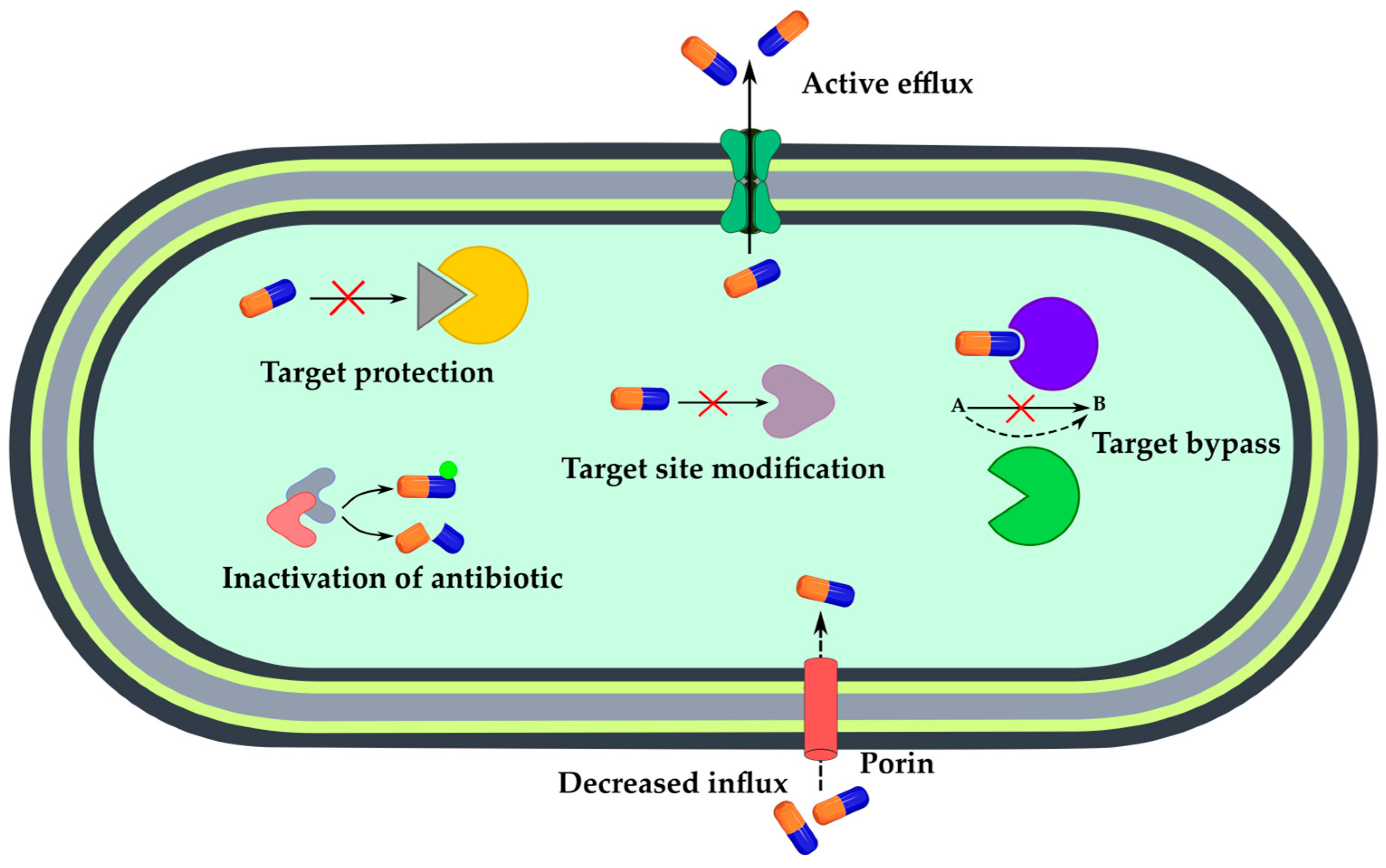
3. Sensing Bacterial Resistance
3.1. Non-Specific Sensors of Bacterial Growth
3.2. Mechanism-Specific Sensors
3.2.1. Enzymatic Inactivation of Antibiotics
3.2.2. Active Efflux or Decreased Influx
4. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Baranova, A.A.; Alferova, V.A.; Korshun, V.A.; Tyurin, A.P. Modern Trends in Natural Antibiotic Discovery. Life 2023, 13, 1073. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. The Science of Antibiotic Discovery. Cell 2020, 181, 29–45. [Google Scholar] [CrossRef]
- Miethke, M.; Pieroni, M.; Weber, T.; Brönstrup, M.; Hammann, P.; Halby, L.; Arimondo, P.B.; Glaser, P.; Aigle, B.; Bode, H.B.; et al. Towards the Sustainable Discovery and Development of New Antibiotics. Nat. Rev. Chem. 2021, 5, 726–749. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Antimicrobial Stewardship Programmes in Health-Care Facilities in Low- and Middle-Income Countries: A WHO Practical Toolkit; World Health Organization: Geneva, Switzerland, 2019; ISBN 978-92-4-151548-1.
- World Health Organization. WHO Policy Guidance on Integrated Antimicrobial Stewardship Activities; World Health Organization: Geneva, Switzerland, 2021; ISBN 978-92-4-002553-0.
- Wenzel, M. Do We Really Understand How Antibiotics Work? Future Microbiol. 2020, 15, 1307–1311. [Google Scholar] [CrossRef]
- Gray, D.A.; Wenzel, M. Multitarget Approaches against Multiresistant Superbugs. ACS Infect. Dis. 2020, 6, 1346–1365. [Google Scholar] [CrossRef] [PubMed]
- Farha, M.A.; French, S.; Brown, E.D. Systems-Level Chemical Biology to Accelerate Antibiotic Drug Discovery. Acc. Chem. Res. 2021, 54, 1909–1920. [Google Scholar] [CrossRef]
- Da Cunha, B.R.; Zoio, P.; Fonseca, L.P.; Calado, C.R.C. Technologies for High-Throughput Identification of Antibiotic Mechanism of Action. Antibiotics 2021, 10, 565. [Google Scholar] [CrossRef]
- Farha, M.A.; Brown, E.D. Strategies for Target Identification of Antimicrobial Natural Products. Nat. Prod. Rep. 2016, 33, 668–680. [Google Scholar] [CrossRef]
- Moser, C.; Lerche, C.J.; Thomsen, K.; Hartvig, T.; Schierbeck, J.; Jensen, P.Ø.; Ciofu, O.; Høiby, N. Antibiotic Therapy as Personalized Medicine—General Considerations and Complicating Factors. APMIS 2019, 127, 361–371. [Google Scholar] [CrossRef]
- Zhang, H.-W.; Lv, C.; Zhang, L.-J.; Guo, X.; Shen, Y.-W.; Nagle, D.G.; Zhou, Y.-D.; Liu, S.-H.; Zhang, W.-D.; Luan, X. Application of Omics- and Multi-Omics-Based Techniques for Natural Product Target Discovery. Biomed. Pharmacother. 2021, 141, 111833. [Google Scholar] [CrossRef]
- Ortmayr, K.; De La Cruz Moreno, R.; Zampieri, M. Expanding the Search for Small-Molecule Antibacterials by Multidimensional Profiling. Nat. Chem. Biol. 2022, 18, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y. Label-Free Drug Discovery. Front. Pharmacol. 2014, 5, 52. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, S.; Sievers, S.; Waldmann, H. Morphological Profiling of Small Molecules. Cell Chem. Biol. 2021, 28, 300–319. [Google Scholar] [CrossRef]
- Hudson, M.A.; Lockless, S.W. Elucidating the Mechanisms of Action of Antimicrobial Agents. mBio 2022, 13, e02240-21. [Google Scholar] [CrossRef] [PubMed]
- Cushnie, T.P.T.; O’Driscoll, N.H.; Lamb, A.J. Morphological and Ultrastructural Changes in Bacterial Cells as an Indicator of Antibacterial Mechanism of Action. Cell. Mol. Life Sci. 2016, 73, 4471–4492. [Google Scholar] [CrossRef]
- Nonejuie, P.; Burkart, M.; Pogliano, K.; Pogliano, J. Bacterial Cytological Profiling Rapidly Identifies the Cellular Pathways Targeted by Antibacterial Molecules. Proc. Natl. Acad. Sci. USA 2013, 110, 16169–16174. [Google Scholar] [CrossRef] [PubMed]
- Peters, C.E.; Lamsa, A.; Liu, R.B.; Quach, D.; Sugie, J.; Brumage, L.; Pogliano, J.; Lopez-Garrido, J.; Pogliano, K. Rapid Inhibition Profiling Identifies a Keystone Target in the Nucleotide Biosynthesis Pathway. ACS Chem. Biol. 2018, 13, 3251–3258. [Google Scholar] [CrossRef]
- Montaño, E.T.; Nideffer, J.F.; Sugie, J.; Enustun, E.; Shapiro, A.B.; Tsunemoto, H.; Derman, A.I.; Pogliano, K.; Pogliano, J. Bacterial Cytological Profiling Identifies Rhodanine-Containing PAINS Analogs as Specific Inhibitors of Escherichia coli Thymidylate Kinase in vivo. J. Bacteriol. 2021, 203, e0010521. [Google Scholar] [CrossRef]
- Kepplinger, B.; Mardiana, L.; Cowell, J.; Morton-Laing, S.; Dashti, Y.; Wills, C.; Marrs, E.C.L.; Perry, J.D.; Gray, J.; Goodfellow, M.; et al. Discovery, Isolation, Heterologous Expression and Mode-of-Action Studies of the Antibiotic Polyketide Tatiomicin from Amycolatopsis Sp. DEM30355. Sci. Rep. 2022, 12, 15579. [Google Scholar] [CrossRef]
- Martin, J.K.; Sheehan, J.P.; Bratton, B.P.; Moore, G.M.; Mateus, A.; Li, S.H.-J.; Kim, H.; Rabinowitz, J.D.; Typas, A.; Savitski, M.M.; et al. A Dual-Mechanism Antibiotic Kills Gram-Negative Bacteria and Avoids Drug Resistance. Cell 2020, 181, 1518–1532.e14. [Google Scholar] [CrossRef]
- Nonejuie, P.; Trial, R.M.; Newton, G.L.; Lamsa, A.; Ranmali Perera, V.; Aguilar, J.; Liu, W.-T.; Dorrestein, P.C.; Pogliano, J.; Pogliano, K. Application of Bacterial Cytological Profiling to Crude Natural Product Extracts Reveals the Antibacterial Arsenal of Bacillus Subtilis. J. Antibiot. 2016, 69, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Quach, D.T.; Sakoulas, G.; Nizet, V.; Pogliano, J.; Pogliano, K. Bacterial Cytological Profiling (BCP) as a Rapid and Accurate Antimicrobial Susceptibility Testing Method for Staphylococcus Aureus. eBioMedicine 2016, 4, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Htoo, H.H.; Brumage, L.; Chaikeeratisak, V.; Tsunemoto, H.; Sugie, J.; Tribuddharat, C.; Pogliano, J.; Nonejuie, P. Bacterial Cytological Profiling as a Tool To Study Mechanisms of Action of Antibiotics That Are Active against Acinetobacter Baumannii. Antimicrob. Agents Chemother. 2019, 63, e02310-18. [Google Scholar] [CrossRef] [PubMed]
- Lamsa, A.; Lopez-Garrido, J.; Quach, D.; Riley, E.P.; Pogliano, J.; Pogliano, K. Rapid Inhibition Profiling in Bacillus Subtilis to Identify the Mechanism of Action of New Antimicrobials. ACS Chem. Biol. 2016, 11, 2222–2231. [Google Scholar] [CrossRef]
- Coram, M.A.; Wang, L.; Godinez, W.J.; Barkan, D.T.; Armstrong, Z.; Ando, D.M.; Feng, B.Y. Morphological Characterization of Antibiotic Combinations. ACS Infect. Dis. 2022, 8, 66–77. [Google Scholar] [CrossRef]
- Tyurin, A.P.; Alferova, V.A.; Paramonov, A.S.; Shuvalov, M.V.; Kudryakova, G.K.; Rogozhin, E.A.; Zherebker, A.Y.; Brylev, V.A.; Chistov, A.A.; Baranova, A.A.; et al. Gausemycins A,B: Cyclic Lipoglycopeptides from Streptomyces sp. Angew. Chem. Int. Ed. 2021, 60, 18694–18703. [Google Scholar] [CrossRef]
- Ouyang, X.; Hoeksma, J.; Lubbers, R.J.M.; Siersma, T.K.; Hamoen, L.W.; Den Hertog, J. Classification of Antimicrobial Mechanism of Action Using Dynamic Bacterial Morphology Imaging. Sci. Rep. 2022, 12, 11162. [Google Scholar] [CrossRef]
- Zoffmann, S.; Vercruysse, M.; Benmansour, F.; Maunz, A.; Wolf, L.; Blum Marti, R.; Heckel, T.; Ding, H.; Truong, H.H.; Prummer, M.; et al. Machine Learning-Powered Antibiotics Phenotypic Drug Discovery. Sci. Rep. 2019, 9, 5013. [Google Scholar] [CrossRef]
- Von Chamier, L.; Laine, R.F.; Jukkala, J.; Spahn, C.; Krentzel, D.; Nehme, E.; Lerche, M.; Hernández-Pérez, S.; Mattila, P.K.; Karinou, E.; et al. Democratising Deep Learning for Microscopy with ZeroCostDL4Mic. Nat. Commun. 2021, 12, 2276. [Google Scholar] [CrossRef]
- Spahn, C.; Gómez-de-Mariscal, E.; Laine, R.F.; Pereira, P.M.; Von Chamier, L.; Conduit, M.; Pinho, M.G.; Jacquemet, G.; Holden, S.; Heilemann, M.; et al. DeepBacs for Multi-Task Bacterial Image Analysis Using Open-Source Deep Learning Approaches. Commun. Biol. 2022, 5, 688. [Google Scholar] [CrossRef]
- Samernate, T.; Htoo, H.H.; Sugie, J.; Chavasiri, W.; Pogliano, J.; Chaikeeratisak, V.; Nonejuie, P. High-Resolution Bacterial Cytological Profiling Reveals Intrapopulation Morphological Variations upon Antibiotic Exposure. Antimicrob. Agents Chemother. 2023, 67, e01307-22. [Google Scholar] [CrossRef]
- Sridhar, S.; Forrest, S.; Warne, B.; Maes, M.; Baker, S.; Dougan, G.; Bartholdson Scott, J. High-Content Imaging to Phenotype Antimicrobial Effects on Individual Bacteria at Scale. mSystems 2021, 6, e00028-21. [Google Scholar] [CrossRef] [PubMed]
- Krentzel, D.; Shorte, S.L.; Zimmer, C. Deep Learning in Image-Based Phenotypic Drug Discovery. Tr. Cell Biol. 2023, 33, 538–554. [Google Scholar] [CrossRef]
- Klymchenko, A.S. Solvatochromic and Fluorogenic Dyes as Environment-Sensitive Probes: Design and Biological Applications. Acc. Chem. Res. 2017, 50, 366–375. [Google Scholar] [CrossRef]
- Basu Roy, S.; Nabawy, A.; Chattopadhyay, A.N.; Geng, Y.; Makabenta, J.M.; Gupta, A.; Rotello, V.M. A Polymer-Based Multichannel Sensor for Rapid Cell-Based Screening of Antibiotic Mechanisms and Resistance Development. ACS Appl. Mater. Interfaces 2022, 14, 27515–27522. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jin, K.; Chen, H.; Zhang, L.; Zhang, G.; Jiang, Y.; Zou, H.; Wang, W.; Qi, G.; Qu, X. A Machine Learning Approach-Based Array Sensor for Rapidly Predicting the Mechanisms of Action of Antibacterial Compounds. Nanoscale 2022, 14, 3087–3096. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Zhang, D.; Yang, K.; Zhang, X.; Zhu, Y.-G. Perspective on Surface-Enhanced Raman Spectroscopic Investigation of Microbial World. Anal. Chem. 2019, 91, 15345–15354. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.S.; Landry, Z.; Pereira, F.C.; Wagner, M.; Berry, D.; Huang, W.E.; Taylor, G.T.; Kneipp, J.; Popp, J.; Zhang, M.; et al. Raman Microspectroscopy for Microbiology. Nat. Rev. Methods Primers 2021, 1, 80. [Google Scholar] [CrossRef]
- Mehta, M.; Liu, Y.; Waterland, M.; Holmes, G. Monitoring the Mode of Action of Synthetic and Natural Biocides against Aeromonas Hydrophila by Raman Spectroscopy and Chemometrics. J. Leather Sci. Eng. 2021, 3, 22. [Google Scholar] [CrossRef]
- Wang, W.; Rahman, A.; Huang, Q.; Vikesland, P.J. Surface-Enhanced Raman Spectroscopy Enabled Evaluation of Bacterial Inactivation. Water Res. 2022, 220, 118668. [Google Scholar] [CrossRef]
- Liu, W.; Wei, L.; Wang, D.; Zhu, C.; Huang, Y.; Gong, Z.; Tang, C.; Fan, M. Phenotyping Bacteria through a Black-Box Approach: Amplifying Surface-Enhanced Raman Spectroscopy Spectral Differences among Bacteria by Inputting Appropriate Environmental Stress. Anal. Chem. 2022, 94, 6791–6798. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro Da Cunha, B.; Fonseca, L.P.; Calado, C.R.C. Metabolic Fingerprinting with Fourier-Transform Infrared (FTIR) Spectroscopy: Towards a High-Throughput Screening Assay for Antibiotic Discovery and Mechanism-of-Action Elucidation. Metabolites 2020, 10, 145. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro Da Cunha, B.; Fonseca, L.P.; Calado, C.R.C. Simultaneous Elucidation of Antibiotic Mechanism of Action and Potency with High-Throughput Fourier-Transform Infrared (FTIR) Spectroscopy and Machine Learning. Appl. Microbiol. Biotechnol. 2021, 105, 1269–1286. [Google Scholar] [CrossRef]
- Ribeiro Da Cunha, B.; Aleixo, S.M.; Fonseca, L.P.; Calado, C.R.C. Fast Identification of Off-target Liabilities in Early Antibiotic Discovery with Fourier-transform Infrared Spectroscopy. Biotechnol. Bioeng. 2021, 118, 4465–4476. [Google Scholar] [CrossRef] [PubMed]
- Von Gundlach, A.R.; Garamus, V.M.; Gorniak, T.; Davies, H.A.; Reischl, M.; Mikut, R.; Hilpert, K.; Rosenhahn, A. Small Angle X-Ray Scattering as a High-Throughput Method to Classify Antimicrobial Modes of Action. Biochim. Biophys. Acta Biomembr. 2016, 1858, 918–925. [Google Scholar] [CrossRef]
- Von Gundlach, A.R.; Garamus, V.M.; Willey, T.M.; Ilavsky, J.; Hilpert, K.; Rosenhahn, A. Use of Small-Angle X-Ray Scattering to Resolve Intracellular Structure Changes of Escherichia coli Cells Induced by Antibiotic Treatment. J. Appl. Crystallogr. 2016, 49, 2210–2216. [Google Scholar] [CrossRef]
- Von Gundlach, A.; Ashby, M.P.; Gani, J.; Lopez-Perez, P.M.; Cookson, A.R.; Ann Huws, S.; Rumancev, C.; Garamus, V.M.; Mikut, R.; Rosenhahn, A.; et al. BioSAXS Measurements Reveal That Two Antimicrobial Peptides Induce Similar Molecular Changes in Gram-Negative and Gram-Positive Bacteria. Front. Pharmacol. 2019, 10, 1127. [Google Scholar] [CrossRef]
- Hilpert, K.; Gani, J.; Rumancev, C.; Simpson, N.; Lopez-Perez, P.M.; Garamus, V.M.; Von Gundlach, A.R.; Markov, P.; Scocchi, M.; Mikut, R.; et al. Rational Designed Hybrid Peptides Show up to a 6-Fold Increase in Antimicrobial Activity and Demonstrate Different Ultrastructural Changes as the Parental Peptides Measured by BioSAXS. Front. Pharmacol. 2021, 12, 769739. [Google Scholar] [CrossRef]
- Rumancev, C.; Rosenhahn, A.; Hilpert, K. BioSAXS–an Emerging Method to Accelerate, Enrich and de-Risk Antimicrobial Drug Development. Front. Pharmacol. 2022, 13, 947005. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, S.; Yang, Y.; Jiang, J.; Tao, N. Imaging Single Bacterial Cells with Electro-Optical Impedance Microscopy. ACS Sens. 2021, 6, 348–354. [Google Scholar] [CrossRef]
- Haddad, G.; Takakura, T.; Bellali, S.; Fontanini, A.; Ominami, Y.; Khalil, J.B.; Raoult, D. A Preliminary Investigation into Bacterial Viability Using Scanning Electron Microscopy–Energy-Dispersive X-Ray Analysis: The Case of Antibiotics. Front. Microbiol. 2022, 13, 967904. [Google Scholar] [CrossRef] [PubMed]
- Sergiev, P.V.; Osterman, I.A.; Golovina, A.Y.; Andreyanova, E.S.; Laptev, I.G.; Pletnev, P.I.; Evfratov, S.A.; Marusich, E.I.; Leonov, S.V.; Ivanenkov, Y.A.; et al. Application of Reporter Strains for Screening of New Antibiotics. Biochem. Moscow Suppl. Ser. B 2016, 10, 293–299. [Google Scholar] [CrossRef]
- Peach, K.C.; Bray, W.M.; Winslow, D.; Linington, P.F.; Linington, R.G. Mechanism of Action-Based Classification of Antibiotics Using High-Content Bacterial Image Analysis. Mol. BioSyst. 2013, 9, 1837. [Google Scholar] [CrossRef] [PubMed]
- Osterman, I.A.; Komarova, E.S.; Shiryaev, D.I.; Korniltsev, I.A.; Khven, I.M.; Lukyanov, D.A.; Tashlitsky, V.N.; Serebryakova, M.V.; Efremenkova, O.V.; Ivanenkov, Y.A.; et al. Sorting Out Antibiotics’ Mechanisms of Action: A Double Fluorescent Protein Reporter for High-Throughput Screening of Ribosome and DNA Biosynthesis Inhibitors. Antimicrob. Agents. Chemother. 2016, 60, 7481–7489. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-H.; Wang, B.-W.; Pan, M.; Zeng, Y.-N.; Rego, H.; Javid, B. Rifampicin Can Induce Antibiotic Tolerance in Mycobacteria via Paradoxical Changes in rpoB Transcription. Nat. Commun. 2018, 9, 4218. [Google Scholar] [CrossRef]
- Wenzel, M.; Rautenbach, M.; Vosloo, J.A.; Siersma, T.; Aisenbrey, C.H.M.; Zaitseva, E.; Laubscher, W.E.; Van Rensburg, W.; Behrends, J.C.; Bechinger, B.; et al. The Multifaceted Antibacterial Mechanisms of the Pioneering Peptide Antibiotics Tyrocidine and Gramicidin S. mBio 2018, 9, e00802-18. [Google Scholar] [CrossRef] [PubMed]
- Grein, F.; Müller, A.; Scherer, K.M.; Liu, X.; Ludwig, K.C.; Klöckner, A.; Strach, M.; Sahl, H.-G.; Kubitscheck, U.; Schneider, T. Ca2+-Daptomycin Targets Cell Wall Biosynthesis by Forming a Tripartite Complex with Undecaprenyl-Coupled Intermediates and Membrane Lipids. Nat. Commun. 2020, 11, 1455. [Google Scholar] [CrossRef]
- Abramovitch, R.B. Mycobacterium tuberculosis Reporter Strains as Tools for Drug Discovery and Development. IUBMB Life 2018, 70, 818–825. [Google Scholar] [CrossRef]
- Yuan, T.; Werman, J.M.; Sampson, N.S. The Pursuit of Mechanism of Action: Uncovering Drug Complexity in TB Drug Discovery. RSC Chem. Biol. 2021, 2, 423–440. [Google Scholar] [CrossRef]
- Wex, K.W.; Saur, J.S.; Handel, F.; Ortlieb, N.; Mokeev, V.; Kulik, A.; Niedermeyer, T.H.J.; Mast, Y.; Grond, S.; Berscheid, A.; et al. Bioreporters for Direct Mode of Action-Informed Screening of Antibiotic Producer Strains. Cell Chem. Biol. 2021, 28, 1242–1252.e4. [Google Scholar] [CrossRef]
- Volynkina, I.A.; Zakalyukina, Y.V.; Alferova, V.A.; Belik, A.R.; Yagoda, D.K.; Nikandrova, A.A.; Buyuklyan, Y.A.; Udalov, A.V.; Golovin, E.V.; Kryakvin, M.A.; et al. Mechanism-Based Approach to New Antibiotic Producers Screening among Actinomycetes in the Course of the Citizen Science Project. Antibiotics 2022, 11, 1198. [Google Scholar] [CrossRef] [PubMed]
- Benfield, A.H.; Henriques, S.T. Mode-of-Action of Antimicrobial Peptides: Membrane Disruption vs. Intracellular Mechanisms. Front. Med. Technol. 2020, 2, 610997. [Google Scholar] [CrossRef]
- Morris, M.A.; Vallmitjana, A.; Grein, F.; Schneider, T.; Arts, M.; Jones, C.R.; Nguyen, B.T.; Hashemian, M.H.; Malek, M.; Gratton, E.; et al. Visualizing the Mode of Action and Supramolecular Assembly of Teixobactin Analogues in Bacillus subtilis. Chem. Sci. 2022, 13, 7747–7754. [Google Scholar] [CrossRef]
- McAuley, S.; Huynh, A.; Czarny, T.L.; Brown, E.D.; Nodwell, J.R. Membrane Activity Profiling of Small Molecule B. Subtilis Growth Inhibitors Utilizing Novel Duel-Dye Fluorescence Assay. Med. Chem. Commun. 2018, 9, 554–561. [Google Scholar] [CrossRef]
- Epand, R.M.; Walker, C.; Epand, R.F.; Magarvey, N.A. Molecular Mechanisms of Membrane Targeting Antibiotics. Biochim. Biophys. Acta Biomembr. 2016, 1858, 980–987. [Google Scholar] [CrossRef]
- Parasassi, T.; Gratton, E. Membrane Lipid Domains and Dynamics as Detected by Laurdan Fluorescence. J. Fluoresc. 1995, 5, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Zou, G.; Hari, T.P.A.; Wilt, I.K.; Zhu, W.; Galle, N.; Faizi, H.A.; Hendricks, G.L.; Tori, K.; Pan, W.; et al. A Selective Membrane-Targeting Repurposed Antibiotic with Activity against Persistent Methicillin-Resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2019, 116, 16529–16534. [Google Scholar] [CrossRef] [PubMed]
- Danylchuk, D.I.; Jouard, P.-H.; Klymchenko, A.S. Targeted Solvatochromic Fluorescent Probes for Imaging Lipid Order in Organelles under Oxidative and Mechanical Stress. J. Am. Chem. Soc. 2021, 143, 912–924. [Google Scholar] [CrossRef]
- Oshchepkov, A.S.; Silva, E.C.D.; Morozov, B.S.; Aparin, I.O.; Kataev, E.A.; Klymchenko, A.S. Fluorescent Macrocycle-Dye-Anchor Conjugates for Sensing Phospholipids in Biomembranes. Sens. Actuators B Chem. 2023, 390, 133911. [Google Scholar] [CrossRef]
- Xu, M.; Kelley, S.P.; Glass, T.E. A Multi-Component Sensor System for Detection of Amphiphilic Compounds. Angew. Chem. Int. Ed. 2018, 57, 12741–12744. [Google Scholar] [CrossRef]
- Colom, A.; Derivery, E.; Soleimanpour, S.; Tomba, C.; Molin, M.D.; Sakai, N.; González-Gaitán, M.; Matile, S.; Roux, A. A Fluorescent Membrane Tension Probe. Nat. Chem. 2018, 10, 1118–1125. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Zhang, T.; Jiang, J.; Wu, C.; Zhu, G.; You, M.; Chen, X.; Zhang, L.; Cui, C.; Yu, R.; et al. Cell Membrane-Anchored Biosensors for Real-Time Monitoring of the Cellular Microenvironment. J. Am. Chem. Soc. 2014, 136, 13090–13093. [Google Scholar] [CrossRef] [PubMed]
- Vyšniauskas, A.; Kuimova, M.K. A Twisted Tale: Measuring Viscosity and Temperature of Microenvironments Using Molecular Rotors. Int. Rev. Phys. Chem. 2018, 37, 259–285. [Google Scholar] [CrossRef]
- Rütten, A.; Kirchner, T.; Musiol-Kroll, E.M. Overview on Strategies and Assays for Antibiotic Discovery. Pharmaceuticals 2022, 15, 1302. [Google Scholar] [CrossRef]
- Steenhuis, M.; Abdallah, A.M.; De Munnik, S.M.; Kuhne, S.; Sterk, G.; Van Den Berg Van Saparoea, B.; Westerhausen, S.; Wagner, S.; Van Der Wel, N.N.; Wijtmans, M.; et al. Inhibition of Autotransporter Biogenesis by Small Molecules. Mol. Microbiol. 2019, 112, 81–98. [Google Scholar] [CrossRef]
- Steenhuis, M.; Ten Hagen-Jongman, C.M.; Van Ulsen, P.; Luirink, J. Stress-Based High-Throughput Screening Assays to Identify Inhibitors of Cell Envelope Biogenesis. Antibiotics 2020, 9, 808. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Zhu, Y.; Liang, Y.; Luo, Y.; Lou, J.; Hu, X.; Meng, Q.; Zhu, T.; Yu, Z. Development of Whole-Cell Biosensors for Screening of Peptidoglycan-Targeting Antibiotics in a Gram-Negative Bacterium. Appl. Environ. Microbiol. 2022, 88, e00846-22. [Google Scholar] [CrossRef]
- Purcell, R.H.; Hall, R.A. Adhesion G Protein–Coupled Receptors as Drug Targets. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 429–449. [Google Scholar] [CrossRef]
- Fihn, C.A.; Carlson, E.E. Targeting a Highly Conserved Domain in Bacterial Histidine Kinases to Generate Inhibitors with Broad Spectrum Activity. Curr. Opin. Microbiol. 2021, 61, 107–114. [Google Scholar] [CrossRef]
- Bem, A.E.; Velikova, N.; Pellicer, M.T.; Baarlen, P.V.; Marina, A.; Wells, J.M. Bacterial Histidine Kinases as Novel Antibacterial Drug Targets. ACS Chem. Biol. 2015, 10, 213–224. [Google Scholar] [CrossRef]
- Chen, H.; Yu, C.; Wu, H.; Li, G.; Li, C.; Hong, W.; Yang, X.; Wang, H.; You, X. Recent Advances in Histidine Kinase-Targeted Antimicrobial Agents. Front. Chem. 2022, 10, 866392. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Li, C.; Chen, P.; Yang, H. An Update of Label-Free Protein Target Identification Methods for Natural Active Products. Theranostics 2022, 12, 1829–1854. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.-Y.; Corson, T.W. Small Molecule Target Identification Using Photo-Affinity Chromatography. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2019; Volume 622, pp. 347–374. ISBN 978-0-12-818119-5. [Google Scholar]
- Nishiya, Y.; Hamada, T.; Abe, M.; Takashima, M.; Tsutsumi, K.; Okawa, K. A New Efficient Method of Generating Photoaffinity Beads for Drug Target Identification. Bioorg. Med. Chem. Lett. 2017, 27, 834–840. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Van Voorhis, W.C.; Quinn, R.J. Development of a Target Identification Approach Using Native Mass Spectrometry. Sci. Rep. 2021, 11, 2387. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Ou, M.; Zheng, Q.; Ding, Y. Unmodified Methodologies in Target Discovery for Small Molecule Drugs: A Rising Star. Chin. Chem. Lett. 2022, 33, 4980–4988. [Google Scholar] [CrossRef]
- Santa Maria, J.P.; Park, Y.; Yang, L.; Murgolo, N.; Altman, M.D.; Zuck, P.; Adam, G.; Chamberlin, C.; Saradjian, P.; Dandliker, P.; et al. Linking High-Throughput Screens to Identify MoAs and Novel Inhibitors of Mycobacterium tuberculosis Dihydrofolate Reductase. ACS Chem. Biol. 2017, 12, 2448–2456. [Google Scholar] [CrossRef]
- Gosschalk, J.E.; Chang, C.; Sue, C.K.; Siegel, S.D.; Wu, C.; Kattke, M.D.; Yi, S.W.; Damoiseaux, R.; Jung, M.E.; Ton-That, H.; et al. A Cell-Based Screen in Actinomyces oris to Identify Sortase Inhibitors. Sci. Rep. 2020, 10, 8520. [Google Scholar] [CrossRef]
- Darby, E.M.; Trampari, E.; Siasat, P.; Gaya, M.S.; Alav, I.; Webber, M.A.; Blair, J.M.A. Molecular Mechanisms of Antibiotic Resistance Revisited. Nat. Rev. Microbiol. 2023, 21, 280–295. [Google Scholar] [CrossRef]
- Schaenzer, A.J.; Wright, G.D. Antibiotic Resistance by Enzymatic Modification of Antibiotic Targets. Trends Mol. Med. 2020, 26, 768–782. [Google Scholar] [CrossRef]
- Wilson, D.N.; Hauryliuk, V.; Atkinson, G.C.; O’Neill, A.J. Target Protection as a Key Antibiotic Resistance Mechanism. Nat. Rev. Microbiol. 2020, 18, 637–648. [Google Scholar] [CrossRef]
- Huang, L.; Wu, C.; Gao, H.; Xu, C.; Dai, M.; Huang, L.; Hao, H.; Wang, X.; Cheng, G. Bacterial Multidrug Efflux Pumps at the Frontline of Antimicrobial Resistance: An Overview. Antibiotics 2022, 11, 520. [Google Scholar] [CrossRef] [PubMed]
- The EUCAST Guideline on Detection of Resistance Mechanisms v 2.0 (2017-07-11). Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_170711.pdf (accessed on 1 July 2023).
- US Centers for Disease Control and Prevention (CDC). Available online: https://www.cdc.gov/drugresistance/about.html (accessed on 1 July 2023).
- 2017(WHO/EMP/IAU/2017.12); Geneva: World Health Organization Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis. World Health Organization: Geneva, Switzerland, 2017.
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef] [PubMed]
- CLSI Guideline M45; Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria, 3rd ed. Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2016; ISBN 1-56238-918-1.
- CLSI Standard M02; Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Disk Susceptibility Tests, 13th ed. Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2018; ISBN 1-56238-835-5.
- CLSI Standard M11 (ISBN 978-1-68440-021-8 [Print]; ISBN 978-1-68440-022-5 [Electronic]); Methods for Antimicrobial Susceptibility Testing of Anaerobic Bacteria, 9th ed. Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2018; ISBN 978-1-68440-022-5.
- CLSI Standard M07; Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th ed. Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2018; ISBN 1-56238-837-1.
- CLSI Standard M24; Susceptibility Testing of Mycobacteria, Nocardia spp., and Other Aerobic Actinomycetes, 3rd ed. Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2018; ISBN 978-1-68440-026-3.
- ISO 20776-1; Susceptibility Testing of Infectious Agents and Evaluation of Performance of Antimicrobial Susceptibility Test Devices—Part 1: Broth Microdilution Reference Method for Testing the in Vitro Activity of Antimicrobial Agents against Rapidly Growing Aerobic Bacteria Involved in Infectious Diseases. International Organization for Standardization: Geneva, Switzerland, 2019.
- ISO 20776-2; Susceptibility Testing of Infectious Agents and Evaluation of Performance of Antimicrobial Susceptibility Test Devices—Part 2: Evaluation of Performance of Antimicrobial Susceptibility Test Devices against Reference Broth Micro-Dilution. International Organization for Standardization: Geneva, Switzerland, 2022.
- Shi, X.; Kadiyala, U.; VanEpps, J.S.; Yau, S.-T. Culture-Free Bacterial Detection and Identification from Blood with Rapid, Phenotypic, Antibiotic Susceptibility Testing. Sci. Rep. 2018, 8, 3416. [Google Scholar] [CrossRef] [PubMed]
- Kittel, M.; Findeisen, P.; Ghebremedhin, B.; Miethke, T.; Grundt, A.; Ahmad-Nejad, P.; Neumaier, M. Rapid Susceptibility Testing of Multi-Drug Resistant Escherichia coli and Klebsiella by Glucose Metabolization Monitoring. Clin. Chem. Lab. 2019, 57, 1271–1279. [Google Scholar] [CrossRef] [PubMed]
- Kuss, S.; Couto, R.A.S.; Evans, R.M.; Lavender, H.; Tang, C.C.; Compton, R.G. Versatile Electrochemical Sensing Platform for Bacteria. Anal. Chem. 2019, 91, 4317–4322. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Cai, X.; Du, J.; Lian, J.; Hui, P.; Gu, M.; Li, F.; Wang, J.; Chen, W. Bioinspired Plasmonic Nanosensor for On-Site Antimicrobial Susceptibility Testing in Urine Samples. ACS Nano 2022, 16, 19229–19239. [Google Scholar] [CrossRef]
- Fonseca E Silva, D.; Andrade, F.F.; Gomes, R.; Silva-Dias, A.; Martins-Oliveira, I.; Pérez-Viso, B.; Ramos, M.H.; Rodrigues, A.G.; Cantón, R.; Pina-Vaz, C. Ultra-Rapid Flow Cytometry Assay for Colistin MIC Determination in Enterobacterales, Pseudomonas aeruginosa and Acinetobacter baumannii. Clin. Microbiol. Infect. 2020, 26, 1559.e1–1559.e4. [Google Scholar] [CrossRef]
- Sawada, T.; Katayama, M.; Takatani, S.; Ohiro, Y. Early Detection of Drug-Resistant Streptococcus Pneumoniae and Haemophilus Influenzae by Quantitative Flow Cytometry. Sci. Rep. 2021, 11, 2873. [Google Scholar] [CrossRef]
- Ekelund, O.; Klokkhammer Hetland, M.A.; Høyland Löhr, I.; Schön, T.; Somajo, S. Rapid High-Resolution Detection of Colistin Resistance in Gram-Negative Bacteria Using Flow Cytometry: A Comparison with Broth Microdilution, a Commercial Screening Test and WGS. J. Antimicrob. Chemother. 2021, 76, 3183–3191. [Google Scholar] [CrossRef]
- Novelli-Rousseau, A.; Espagnon, I.; Filiputti, D.; Gal, O.; Douet, A.; Mallard, F.; Josso, Q. Culture-Free Antibiotic-Susceptibility Determination From Single-Bacterium Raman Spectra. Sci. Rep. 2018, 8, 3957. [Google Scholar] [CrossRef]
- Zhang, M.; Hong, W.; Abutaleb, N.S.; Li, J.; Dong, P.; Zong, C.; Wang, P.; Seleem, M.N.; Cheng, J. Rapid Determination of Antimicrobial Susceptibility by Stimulated Raman Scattering Imaging of D2O Metabolic Incorporation in a Single Bacterium. Adv. Sci. 2020, 7, 2001452. [Google Scholar] [CrossRef] [PubMed]
- Bi, L.; Wang, X.; Cao, X.; Liu, L.; Bai, C.; Zheng, Q.; Choo, J.; Chen, L. SERS-Active Au@Ag Core-Shell Nanorod (Au@AgNR) Tags for Ultrasensitive Bacteria Detection and Antibiotic-Susceptibility Testing. Talanta 2020, 220, 121397. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Song, Y.; Xu, X.; Peng, D.; Wang, J.; Qie, X.; Lin, K.; Yu, M.; Ge, M.; Wang, Y.; et al. Development of a Fast Raman-Assisted Antibiotic Susceptibility Test (FRAST) for the Antibiotic Resistance Analysis of Clinical Urine and Blood Samples. Anal. Chem. 2021, 93, 5098–5106. [Google Scholar] [CrossRef] [PubMed]
- Van Belkum, A.; Burnham, C.-A.D.; Rossen, J.W.A.; Mallard, F.; Rochas, O.; Dunne, W.M. Innovative and Rapid Antimicrobial Susceptibility Testing Systems. Nat. Rev. Microbiol. 2020, 18, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Dietvorst, J.; Vilaplana, L.; Uria, N.; Marco, M.-P.; Muñoz-Berbel, X. Current and Near-Future Technologies for Antibiotic Susceptibility Testing and Resistant Bacteria Detection. TrAC Trends Anal. Chem. 2020, 127, 115891. [Google Scholar] [CrossRef]
- Pitruzzello, G.; Conteduca, D.; Krauss, T.F. Nanophotonics for Bacterial Detection and Antimicrobial Susceptibility Testing. Nanophotonics 2020, 9, 4447–4472. [Google Scholar] [CrossRef]
- Dina, N.E.; Tahir, M.A.; Bajwa, S.Z.; Amin, I.; Valev, V.K.; Zhang, L. SERS-Based Antibiotic Susceptibility Testing: Towards Point-of-Care Clinical Diagnosis. Biosens. Bioelectron. 2023, 219, 114843. [Google Scholar] [CrossRef]
- Haddad, G.; Fontanini, A.; Bellali, S.; Takakura, T.; Ominami, Y.; Hisada, A.; Hadjadj, L.; Rolain, J.-M.; Raoult, D.; Bou Khalil, J.Y. Rapid Detection of Imipenem Resistance in Gram-Negative Bacteria Using Tabletop Scanning Electron Microscopy: A Preliminary Evaluation. Front. Microbiol. 2021, 12, 658322. [Google Scholar] [CrossRef]
- Yu, H.; Jing, W.; Iriya, R.; Yang, Y.; Syal, K.; Mo, M.; Grys, T.E.; Haydel, S.E.; Wang, S.; Tao, N. Phenotypic Antimicrobial Susceptibility Testing with Deep Learning Video Microscopy. Anal. Chem. 2018, 90, 6314–6322. [Google Scholar] [CrossRef]
- Song, D.; Lei, Y. Mini-Review: Recent Advances in Imaging-Based Rapid Antibiotic Susceptibility Testing. Sens. Actuators Rep. 2021, 3, 100053. [Google Scholar] [CrossRef]
- Xu, X.; Chen, S.; Yu, Y.; Virtanen, P.; Wu, J.; Hu, Q.; Koskiniemi, S.; Zhang, Z. All-Electrical Antibiotic Susceptibility Testing within 30 Min Using Silicon Nano Transistors. Sens. Actuators B Chem. 2022, 357, 131458. [Google Scholar] [CrossRef]
- Gao, X.; Li, M.; Zhao, M.; Wang, X.; Wang, S.; Liu, Y. Metabolism-Triggered Colorimetric Sensor Array for Fingerprinting and Antibiotic Susceptibility Testing of Bacteria. Anal. Chem. 2022, 94, 6957–6966. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J.; Liu, H.; Du, X.; Yang, L.; Zeng, J. Single-Probe-Based Colorimetric and Photothermal Dual-Mode Identification of Multiple Bacteria. Anal. Chem. 2023, 95, 3037–3044. [Google Scholar] [CrossRef] [PubMed]
- Tahir, M.A.; Dina, N.E.; Cheng, H.; Valev, V.K.; Zhang, L. Surface-Enhanced Raman Spectroscopy for Bioanalysis and Diagnosis. Nanoscale 2021, 13, 11593–11634. [Google Scholar] [CrossRef]
- Zhang, W.; He, S.; Hong, W.; Wang, P. A Review of Raman-Based Technologies for Bacterial Identification and Antimicrobial Susceptibility Testing. Photonics 2022, 9, 133. [Google Scholar] [CrossRef]
- Kim, S.; Masum, F.; Jeon, J.S. Recent Developments of Chip-Based Phenotypic Antibiotic Susceptibility Testing. BioChip J. 2019, 13, 43–52. [Google Scholar] [CrossRef]
- Zhang, K.; Qin, S.; Wu, S.; Liang, Y.; Li, J. Microfluidic Systems for Rapid Antibiotic Susceptibility Tests (ASTs) at the Single-Cell Level. Chem. Sci. 2020, 11, 6352–6361. [Google Scholar] [CrossRef]
- Qin, N.; Zhao, P.; Ho, E.A.; Xin, G.; Ren, C.L. Microfluidic Technology for Antibacterial Resistance Study and Antibiotic Susceptibility Testing: Review and Perspective. ACS Sens. 2021, 6, 3–21. [Google Scholar] [CrossRef]
- Postek, W.; Pacocha, N.; Garstecki, P. Microfluidics for Antibiotic Susceptibility Testing. Lab Chip 2022, 22, 3637–3662. [Google Scholar] [CrossRef]
- Li, H.; Hsieh, K.; Wong, P.K.; Mach, K.E.; Liao, J.C.; Wang, T.-H. Single-Cell Pathogen Diagnostics for Combating Antibiotic Resistance. Nat. Rev. Methods Primers 2023, 3, 6. [Google Scholar] [CrossRef]
- Scheler, O.; Makuch, K.; Debski, P.R.; Horka, M.; Ruszczak, A.; Pacocha, N.; Sozański, K.; Smolander, O.-P.; Postek, W.; Garstecki, P. Droplet-Based Digital Antibiotic Susceptibility Screen Reveals Single-Cell Clonal Heteroresistance in an Isogenic Bacterial Population. Sci. Rep. 2020, 10, 3282. [Google Scholar] [CrossRef] [PubMed]
- Pitruzzello, G.; Baumann, C.G.; Johnson, S.; Krauss, T.F. Single-Cell Motility Rapidly Quantifying Heteroresistance in Populations of Escherichia coli and Salmonella typhimurium. Small Sci. 2022, 2, 2100123. [Google Scholar] [CrossRef]
- Shan, H.; Zhu, G.; Zhang, Y.; Ke, L.; Yang, X.; Qiao, A.; Wei, B.; Wang, Y.; Fan, Y.; Du, M. Multiplex PCR-ASE Functionalized Microfluidic Diagnostic Platform for the Detection of Clarithromycin Resistance Mutations in Helicobacter Pylori. Sens. Actuators B Chem. 2023, 387, 133808. [Google Scholar] [CrossRef]
- Smith, K.P.; Kirby, J.E. The Inoculum Effect in the Era of Multidrug Resistance: Minor Differences in Inoculum Have Dramatic Effect on MIC Determination. Antimicrob. Agents Chemother. 2018, 62, e00433-18. [Google Scholar] [CrossRef]
- Postek, W.; Gargulinski, P.; Scheler, O.; Kaminski, T.S.; Garstecki, P. Microfluidic Screening of Antibiotic Susceptibility at a Single-Cell Level Shows the Inoculum Effect of Cefotaxime on E. coli. Lab Chip 2018, 18, 3668–3677. [Google Scholar] [CrossRef]
- Postek, W.; Garstecki, P. Droplet Microfluidics for High-Throughput Analysis of Antibiotic Susceptibility in Bacterial Cells and Populations. Acc. Chem. Res. 2022, 55, 605–615. [Google Scholar] [CrossRef]
- Zhang, P.; Kaushik, A.M.; Hsieh, K.; Li, S.; Lewis, S.; Mach, K.E.; Liao, J.C.; Carroll, K.C.; Wang, T. A Cascaded Droplet Microfluidic Platform Enables High-Throughput Single Cell Antibiotic Susceptibility Testing at Scale. Small Methods 2022, 6, 2101254. [Google Scholar] [CrossRef]
- Pacocha, N.; Zapotoczna, M.; Makuch, K.; Bogusławski, J.; Garstecki, P. You Will Know by Its Tail: A Method for Quantification of Heterogeneity of Bacterial Populations Using Single-Cell MIC Profiling. Lab Chip 2022, 22, 4317–4326. [Google Scholar] [CrossRef]
- Song, K.; Yu, Z.; Zu, X.; Huang, L.; Fu, D.; Yao, J.; Hu, Z.; Xue, Y. Microfluidic Chip for Detection of Drug Resistance at the Single-Cell Level. Micromachines 2022, 14, 46. [Google Scholar] [CrossRef]
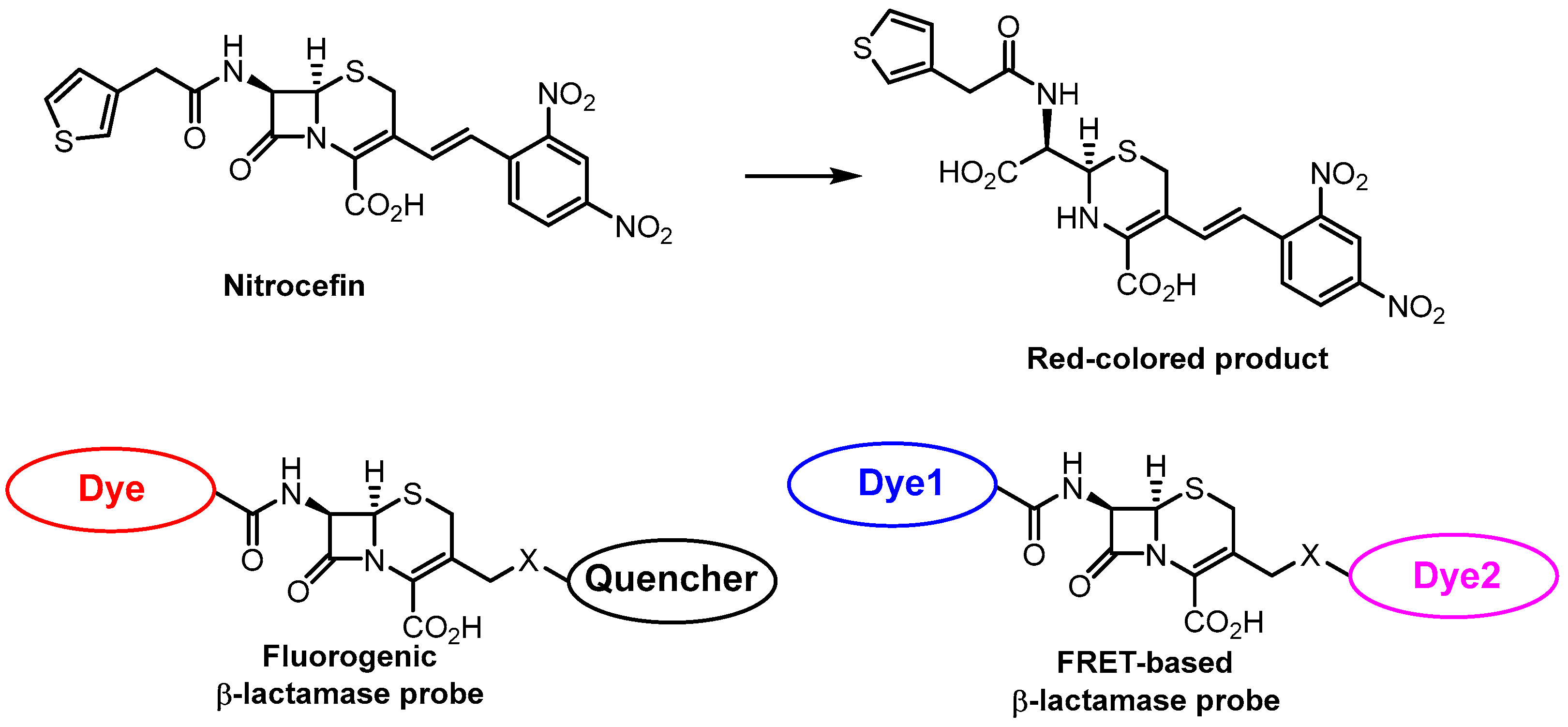
- O’Callaghan, C.H.; Morris, A.; Kirby, S.M.; Shingler, A.H. Novel Method for Detection of β-Lactamases by Using a Chromogenic Cephalosporin Substrate. Antimicrob. Agents Chemother. 1972, 1, 283–288. [Google Scholar] [CrossRef]
- Ding, Y.; Li, Z.; Xu, C.; Qin, W.; Wu, Q.; Wang, X.; Cheng, X.; Li, L.; Huang, W. Fluorogenic Probes/Inhibitors of Β-Lactamase and Their Applications in Drug-Resistant Bacteria. Angew. Chem. Int. Ed. 2021, 60, 24–40. [Google Scholar] [CrossRef] [PubMed]
- Dai, T.; Xie, J.; Buonomo, J.A.; Moreno, A.; Banaei, N.; Bertozzi, C.R.; Rao, J. Bioluminogenic Probe for Rapid, Ultrasensitive Detection of β-Lactam-Resistant Bacteria. Anal. Chem. 2023, 95, 7329–7335. [Google Scholar] [CrossRef]
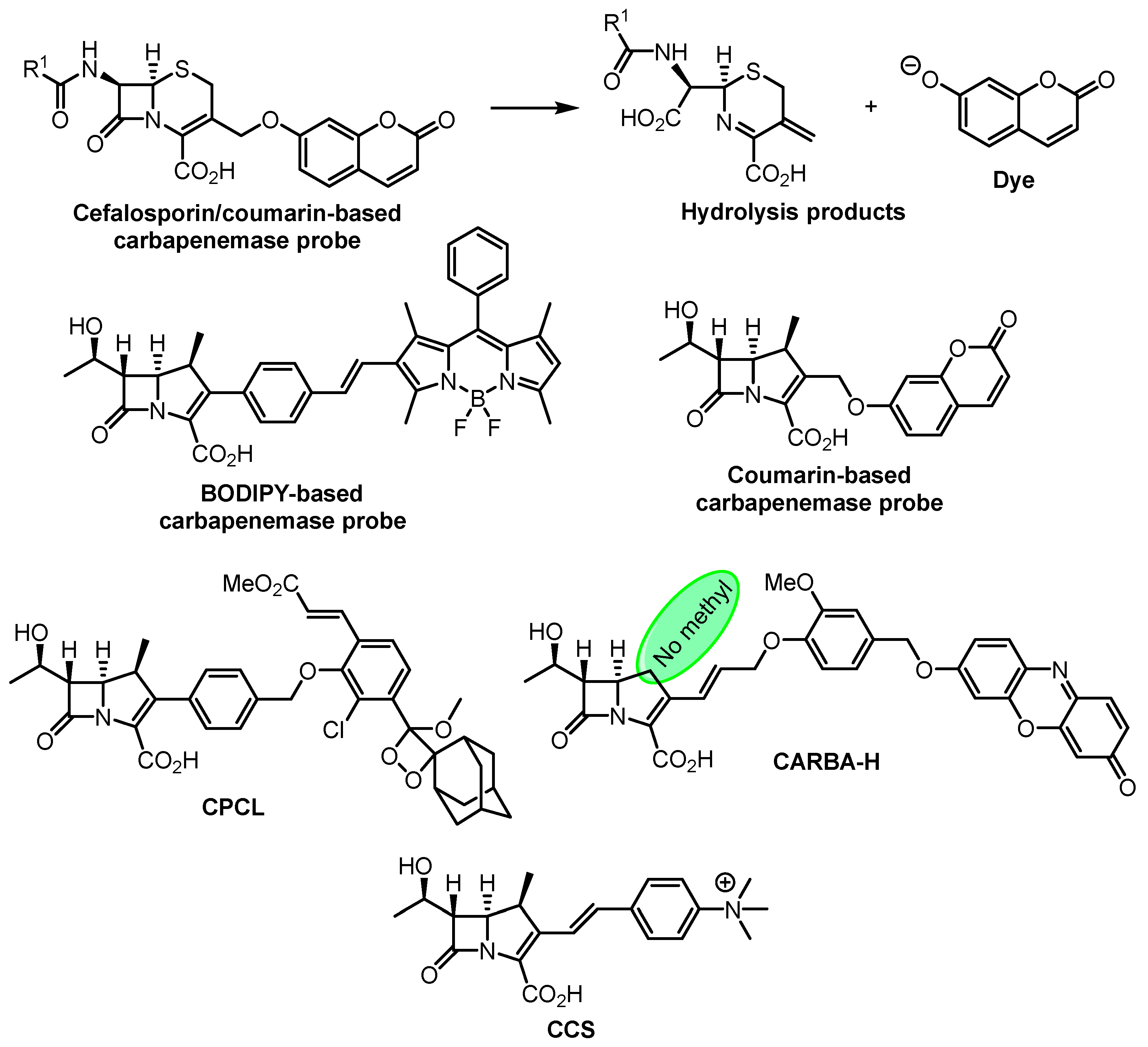
- Shi, H.; Cheng, Y.; Lee, K.H.; Luo, R.F.; Banaei, N.; Rao, J. Engineering the Stereochemistry of Cephalosporin for Specific Detection of Pathogenic Carbapenemase-Expressing Bacteria. Angew. Chem. Int. Ed. 2014, 53, 8113–8116. [Google Scholar] [CrossRef] [PubMed]
- Mao, W.; Xia, L.; Xie, H. Detection of Carbapenemase-Producing Organisms with a Carbapenem-Based Fluorogenic Probe. Angew. Chem. Int. Ed. 2017, 56, 4468–4472. [Google Scholar] [CrossRef] [PubMed]
- Mao, W.; Wang, Y.; Qian, X.; Xia, L.; Xie, H. A Carbapenem-Based Off–On Fluorescent Probe for Specific Detection of Metallo-β-Lactamase Activities. ChemBioChem 2019, 20, 511–515. [Google Scholar] [CrossRef]
- Das, S.; Ihssen, J.; Wick, L.; Spitz, U.; Shabat, D. Chemiluminescent Carbapenem-Based Molecular Probe for Detection of Carbapenemase Activity in Live Bacteria. Chem. Eur. J. 2020, 26, 3647–3652. [Google Scholar] [CrossRef]
- Ma, C.-W.; Ng, K.K.-H.; Yam, B.H.-C.; Ho, P.-L.; Kao, R.Y.-T.; Yang, D. Rapid Broad Spectrum Detection of Carbapenemases with a Dual Fluorogenic-Colorimetric Probe. J. Am. Chem. Soc. 2021, 143, 6886–6894. [Google Scholar] [CrossRef]
- Li, W.; Wang, J.; Li, C.; Zong, Z.; Zhao, J.; Gao, H.; Liu, D. Achieving Ultrasensitive Chromogenic Probes for Rapid, Direct Detection of Carbapenemase-Producing Bacteria in Sputum. JACS Au 2023, 3, 227–238. [Google Scholar] [CrossRef]
- Rybenkov, V.V.; Zgurskaya, H.I.; Ganguly, C.; Leus, I.V.; Zhang, Z.; Moniruzzaman, M. The Whole Is Bigger than the Sum of Its Parts: Drug Transport in the Context of Two Membranes with Active Efflux. Chem. Rev. 2021, 121, 5597–5631. [Google Scholar] [CrossRef]
- Blair, J.M.A.; Piddock, L.J.V. How to Measure Export via Bacterial Multidrug Resistance Efflux Pumps. mBio 2016, 7, e00840-16. [Google Scholar] [CrossRef]
- Six, D.A.; Krucker, T.; Leeds, J.A. Advances and Challenges in Bacterial Compound Accumulation Assays for Drug Discovery. Curr. Opin. Chem. Biol. 2018, 44, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Cama, J.; Voliotis, M.; Metz, J.; Smith, A.; Iannucci, J.; Keyser, U.F.; Tsaneva-Atanasova, K.; Pagliara, S. Single-Cell Microfluidics Facilitates the Rapid Quantification of Antibiotic Accumulation in Gram-Negative Bacteria. Lab Chip 2020, 20, 2765–2775. [Google Scholar] [CrossRef] [PubMed]
- Prajapati, J.D.; Kleinekathöfer, U.; Winterhalter, M. How to Enter a Bacterium: Bacterial Porins and the Permeation of Antibiotics. Chem. Rev. 2021, 121, 5158–5192. [Google Scholar] [CrossRef]
- Masi, M.; Réfregiers, M.; Pos, K.M.; Pagès, J.-M. Mechanisms of Envelope Permeability and Antibiotic Influx and Efflux in Gram-Negative Bacteria. Nat. Microbiol. 2017, 2, 17001. [Google Scholar] [CrossRef] [PubMed]
- Vergalli, J.; Bodrenko, I.V.; Masi, M.; Moynié, L.; Acosta-Gutiérrez, S.; Naismith, J.H.; Davin-Regli, A.; Ceccarelli, M.; Van Den Berg, B.; Winterhalter, M.; et al. Porins and Small-Molecule Translocation across the Outer Membrane of Gram-Negative Bacteria. Nat. Rev. Microbiol. 2020, 18, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Richter, R.; Kamal, M.A.M.; García-Rivera, M.A.; Kaspar, J.; Junk, M.; Elgaher, W.A.M.; Srikakulam, S.K.; Gress, A.; Beckmann, A.; Grißmer, A.; et al. A Hydrogel-Based in vitro Assay for the Fast Prediction of Antibiotic Accumulation in Gram-Negative Bacteria. Mater. Today Bio 2020, 8, 100084. [Google Scholar] [CrossRef] [PubMed]
- Peveler, W.J.; Landis, R.F.; Yazdani, M.; Day, J.W.; Modi, R.; Carmalt, C.J.; Rosenberg, W.M.; Rotello, V.M. A Rapid and Robust Diagnostic for Liver Fibrosis Using a Multichannel Polymer Sensor Array. Adv. Mater. 2018, 30, 1800634. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Liu, W.; Wang, D.; Gong, Z.; Fan, M. Boosting Bacteria Differentiation Efficiency with Multidimensional Surface-enhanced Raman Scattering: The Example of Bacillus cereus. Luminescence 2022, 37, 1145–1151. [Google Scholar] [CrossRef]
- Liu, L.; Ma, W.; Wang, X.; Li, S. Recent Progress of Surface-Enhanced Raman Spectroscopy for Bacteria Detection. Biosensors 2023, 13, 350. [Google Scholar] [CrossRef]
- Lluka, T.; Stokes, J.M. Antibiotic Discovery in the Artificial Intelligence Era. Ann. N. Y. Acad. Sci. 2023, 1519, 74–93. [Google Scholar] [CrossRef]
- Yang, J.; Lu, S.; Chen, B.; Hu, F.; Li, C.; Guo, C. Machine Learning-Assisted Optical Nano-Sensor Arrays in Microorganism Analysis. TrAC Trends Anal. Chem. 2023, 159, 116945. [Google Scholar] [CrossRef]
- Sahayasheela, V.J.; Lankadasari, M.B.; Dan, V.M.; Dastager, S.G.; Pandian, G.N.; Sugiyama, H. Artificial Intelligence in Microbial Natural Product Drug Discovery: Current and Emerging Role. Nat. Prod. Rep. 2022, 39, 2215–2230. [Google Scholar] [CrossRef] [PubMed]
- Serafim, M.S.M.; Kronenberger, T.; Oliveira, P.R.; Poso, A.; Honório, K.M.; Mota, B.E.F.; Maltarollo, V.G. The Application of Machine Learning Techniques to Innovative Antibacterial Discovery and Development. Expert Opin. Drug Discov. 2020, 15, 1165–1180. [Google Scholar] [CrossRef] [PubMed]
- Madhukar, N.S.; Khade, P.K.; Huang, L.; Gayvert, K.; Galletti, G.; Stogniew, M.; Allen, J.E.; Giannakakou, P.; Elemento, O. A Bayesian Machine Learning Approach for Drug Target Identification Using Diverse Data Types. Nat. Commun. 2019, 10, 5221. [Google Scholar] [CrossRef]
- Amiri Souri, E.; Laddach, R.; Karagiannis, S.N.; Papageorgiou, L.G.; Tsoka, S. Novel Drug-Target Interactions via Link Prediction and Network Embedding. BMC Bioinform. 2022, 23, 121. [Google Scholar] [CrossRef]
- Wan, F.; Hong, L.; Xiao, A.; Jiang, T.; Zeng, J. NeoDTI: Neural Integration of Neighbor Information from a Heterogeneous Network for Discovering New Drug–Target Interactions. Bioinformatics 2019, 35, 104–111. [Google Scholar] [CrossRef]
- Santiago, M.; Lee, W.; Fayad, A.A.; Coe, K.A.; Rajagopal, M.; Do, T.; Hennessen, F.; Srisuknimit, V.; Müller, R.; Meredith, T.C.; et al. Genome-Wide Mutant Profiling Predicts the Mechanism of a Lipid II Binding Antibiotic. Nat. Chem. Biol. 2018, 14, 601–608. [Google Scholar] [CrossRef]







| Method | Advantages | Disadvantages | Development Points | References |
|---|---|---|---|---|
| Mechanism-independent approaches | ||||
| Fluorescent stains | Informative due to vast variety of dyes | High resolution required demands sophisticated instrumentation | Instrumentation and acquisition upgrades | [29,35] |
| Single-cell imaging | [33,34] | |||
| Versatile for various mechanisms | Data processing for enhanced resolution | [30,31,32,35] | ||
| Fluorescent array sensors | Versatile for various mechanisms | Construction of array sensors is synthetically complicated | Development of novel array sensors | [37,38] |
| Label-free phenotyping | Alternative physicochemical methods provide insights in various cellular stress responses | Each method of phenotyping requires specific instrumentation, preventing combination of approaches | Raman scattering (SERS) | [41,42,43] |
| Infrared spectroscopy (FTIR) | [44,45,46] | |||
| Small-angle X-ray scattering (SAXS) | [47,48,49,50] | |||
| Impedance microscopy (EIM) | [52] | |||
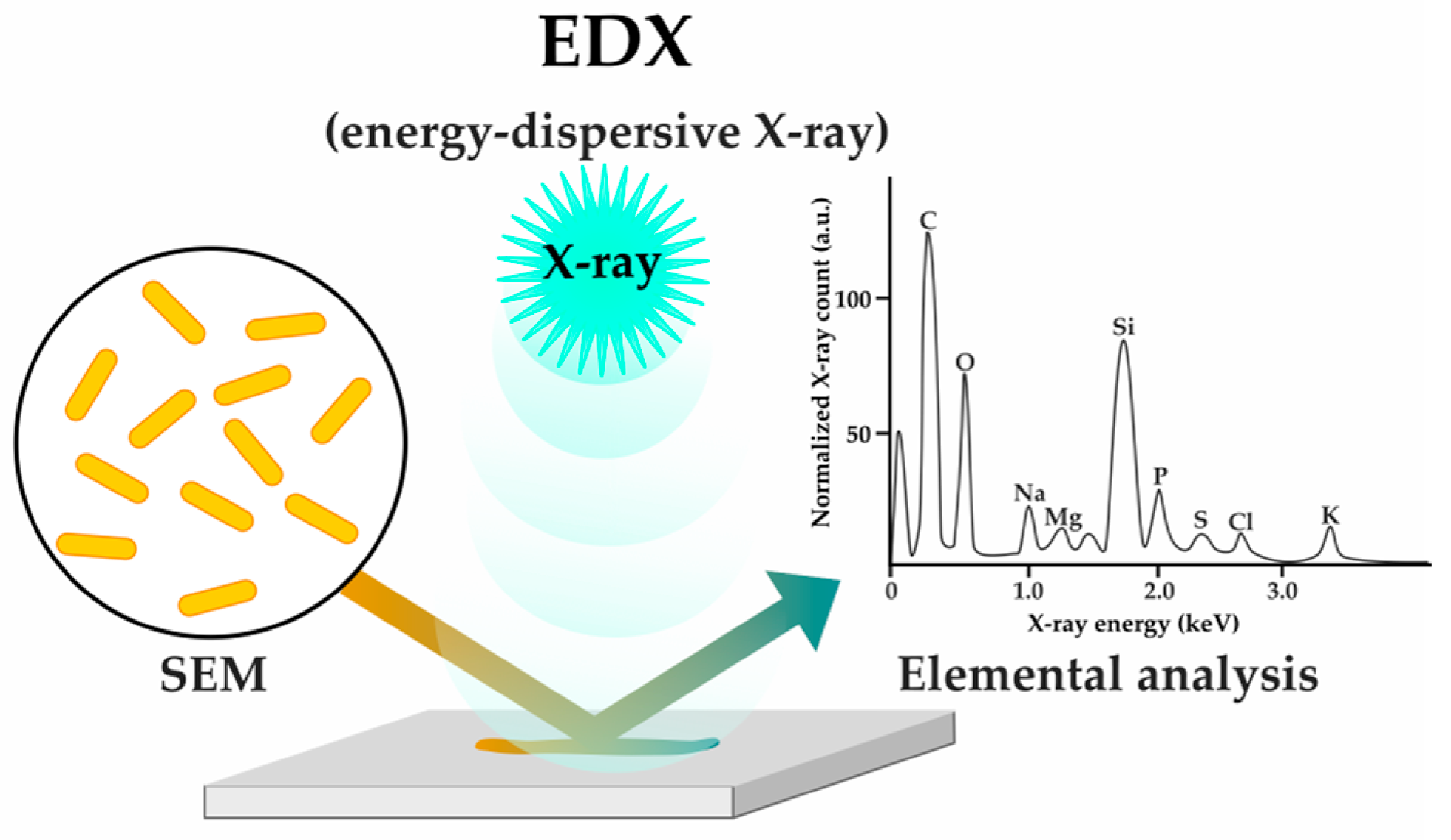
| X-ray analysis (EDX) | [53] | |||
| Mechanism-specific methods | ||||
| Reporter strains | Sensitive detection of artificial phenotypic alteration | Narrow spectrum of applicability, each mechanism requires the development of the specific reporter strain | Development of reporter strains with wide applications | [60,61,62] |
| Reporter strain for citizen science application | [63] | |||
| Membrane-targeting studies | Informative for membrane studies, otherwise difficult to approach | Molecular mode of action can be elucidated only by combination of multiple techniques | FRET-based aggregation probes | [65] |
| High-throughput assays | [22,66] | |||
| Novel dye development | [67,68,69,71,72,73,74,75] | |||
| Peptidiglycan targeting | Deep insight into cell wall-associated MoA | Narrow spectrum of applicability | Reporter strains | [77,78] |
| Biosensors | [79] | |||
| Protein target identification | Improved accuracy compared with phenotyping methods | Narrow spectrum of applicability, verification by classic approaches required | MS-assisted phenotyping | [89] |
| Biosensors | [90] | |||
| Method | Advantages | Disadvantages | Development Points | References |
|---|---|---|---|---|
| Phenotyping | Classical approach, providing information on both bacterial growth and susceptibility | Application in medicine requires more rapid and cost-effective methods | Ion-selective sensors | [124] |
| Colorimetric and photothermal assays | [125,126] | |||
| SERS | [120,127,128] | |||
| Miniaturization in microfluidic platforms | [133,134,135,136,138,140,141,142] | |||
| Inactivating enzymes detection | Sensitive and rapid assay for inactivating enzyme detection | Narrow spectrum of applicability, mostly used for β-lactamase detection | Carbapenemase-selective probes | [146,147,148,149,150,151] |
| Efflux/influx probes | Well-established method, based on fluorescent probes | Classical approach is suitable only for population-level studies | Single-cell studies | [155] |
| Porine modelling | [159] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baranova, A.A.; Tyurin, A.P.; Korshun, V.A.; Alferova, V.A. Sensing of Antibiotic–Bacteria Interactions. Antibiotics 2023, 12, 1340. https://doi.org/10.3390/antibiotics12081340
Baranova AA, Tyurin AP, Korshun VA, Alferova VA. Sensing of Antibiotic–Bacteria Interactions. Antibiotics. 2023; 12(8):1340. https://doi.org/10.3390/antibiotics12081340
Chicago/Turabian StyleBaranova, Anna A., Anton P. Tyurin, Vladimir A. Korshun, and Vera A. Alferova. 2023. "Sensing of Antibiotic–Bacteria Interactions" Antibiotics 12, no. 8: 1340. https://doi.org/10.3390/antibiotics12081340
APA StyleBaranova, A. A., Tyurin, A. P., Korshun, V. A., & Alferova, V. A. (2023). Sensing of Antibiotic–Bacteria Interactions. Antibiotics, 12(8), 1340. https://doi.org/10.3390/antibiotics12081340






