Multi-Omics Analysis Reveals the Gut Microbiota Characteristics of Diarrheal Piglets Treated with Gentamicin
Abstract
:1. Introduction
2. Results
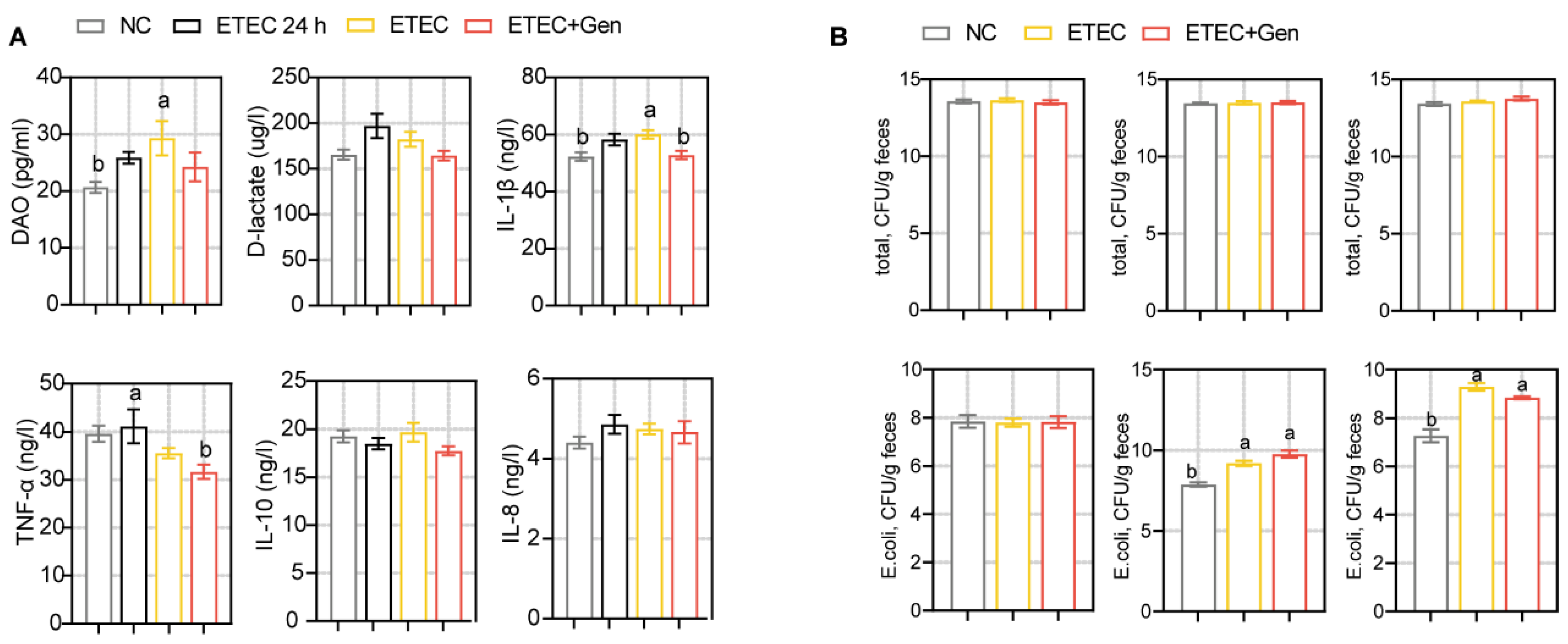
2.1. Effects of Gentamicin on ETEC Infection in Piglets
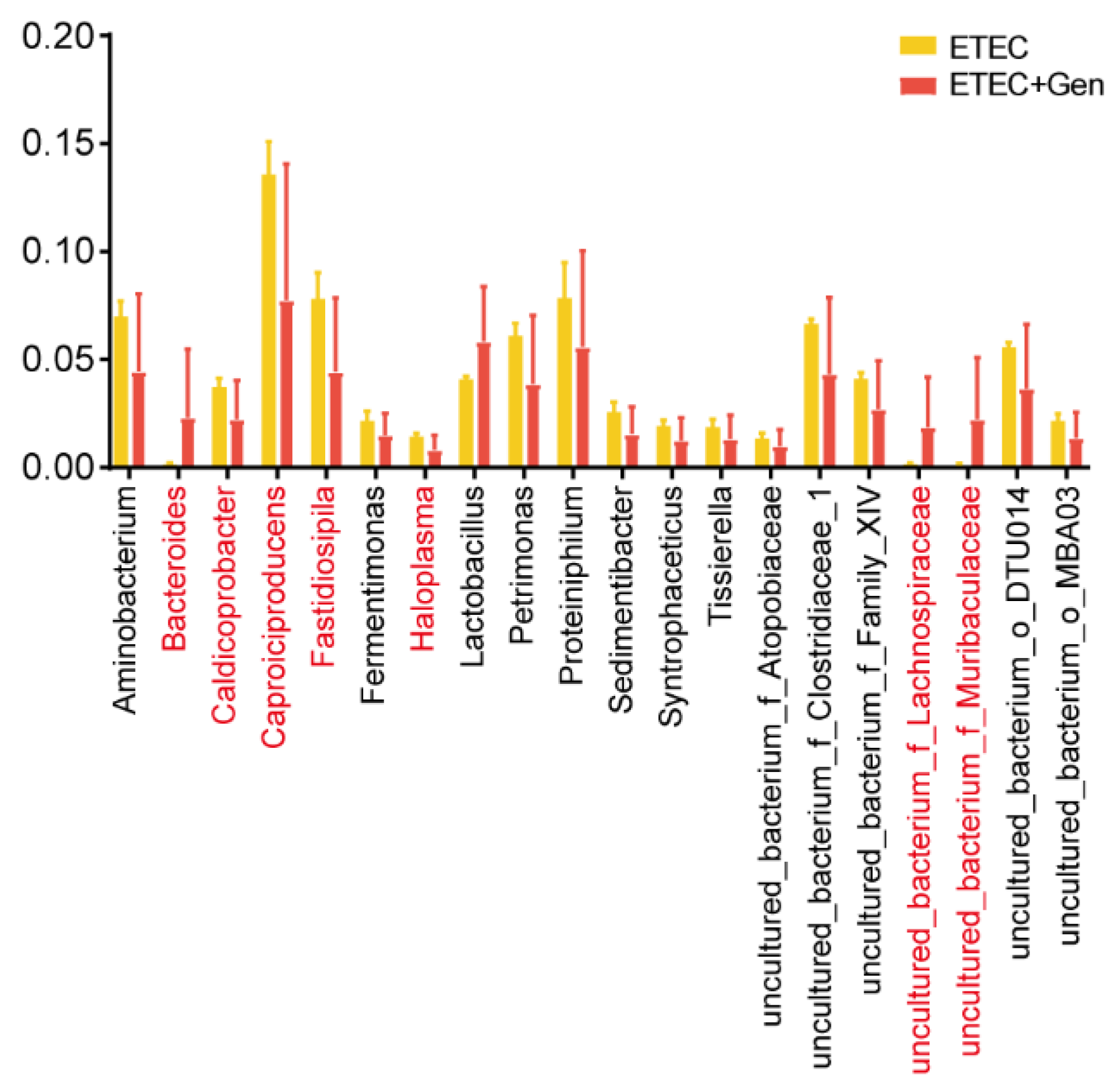
2.2. Diversity Analysis of Intestinal Microbiota
2.3. Gentamicin Treatment Altered Metabolite Levels
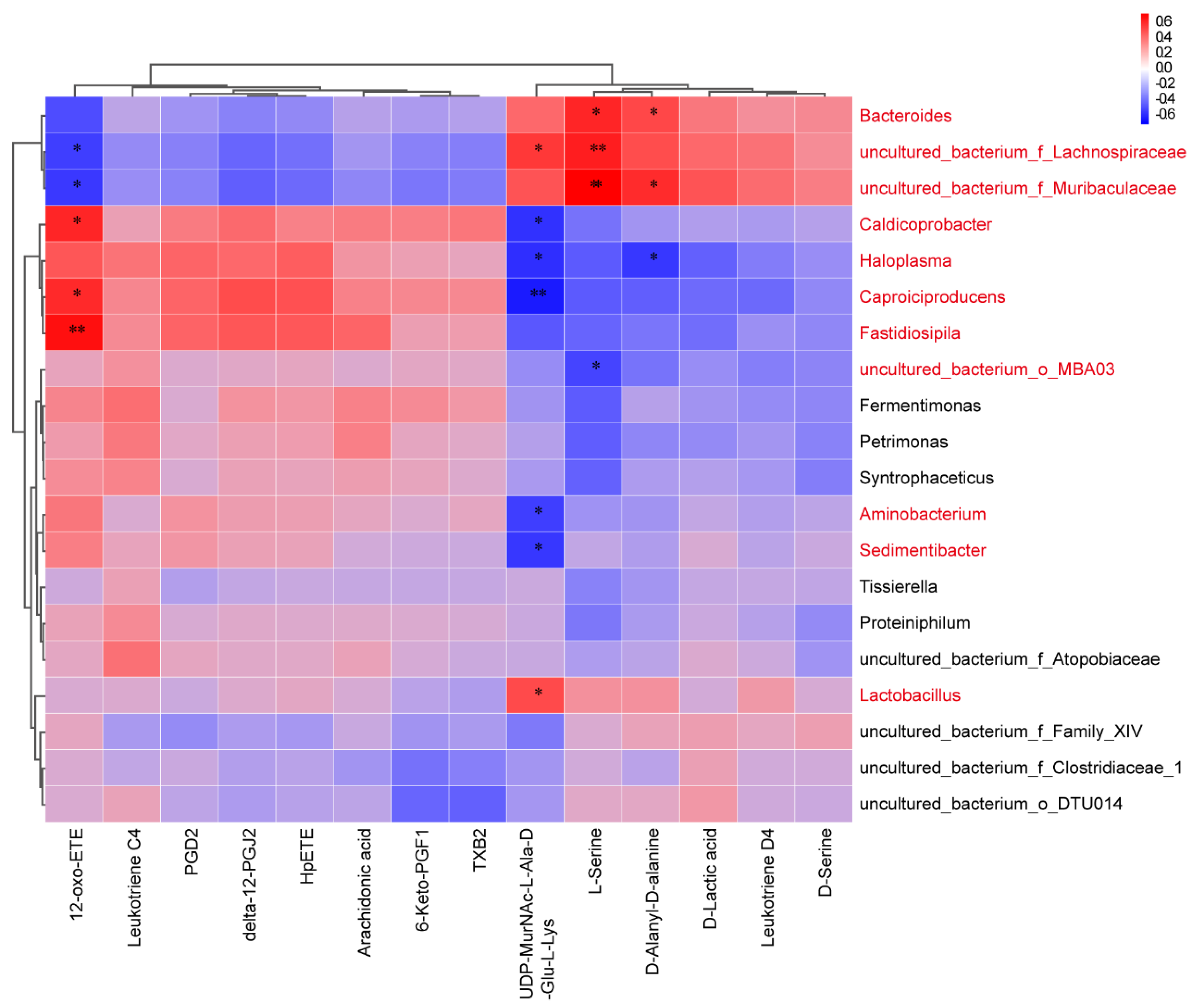
2.4. Correlation Analysis between Specific Metabolites and Microbiota
3. Discussion
4. Materials and Methods
4.1. Piglet Selection
4.2. Bacterial Strain and Culture Conditions
4.3. Experimental Design for Piglets
4.4. ELISA
4.5. Fecal Indexes
4.6. Histology
4.7. Microbiota Composition Assessed via 16S rRNA Sequencing Analysis
4.8. LC-MS/MS Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Blaser, M.J. Antibiotic use and its consequences for the normal microbiome. Science 2016, 352, 544–545. [Google Scholar] [CrossRef]
- Yao, J.; Carter, R.A.; Vuagniaux, G.; Barbier, M.; Rosch, J.W.; Rock, C.O. A Pathogen-Selective Antibiotic Minimizes Disturbance to the Microbiome. Antimicrob. Agents Chemother. 2016, 60, 4264–4273. [Google Scholar] [CrossRef]
- Brüssow, H. Growth promotion and gut microbiota: Insights from antibiotic use. Environ. Microbiol. 2015, 17, 2216–2227. [Google Scholar] [CrossRef]
- Korpela, K.; Salonen, A.; Virta, L.J.; Kekkonen, R.A.; Forslund, K.; Bork, P.; de Vos, W.M. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat. Commun. 2016, 7, 10410. [Google Scholar] [CrossRef]
- Pérez-Cobas, A.E.; Gosalbes, M.J.; Friedrichs, A.; Knecht, H.; Artacho, A.; Eismann, K.; Otto, W.; Rojo, D.; Bargiela, R.; von Bergen, M.; et al. Gut microbiota disturbance during antibiotic therapy: A multi-omic approach. Gut 2013, 62, 1591–1601. [Google Scholar] [CrossRef]
- He, Y.; Wen, Q.; Yao, F.; Xu, D.; Huang, Y.; Wang, J. Gut-lung axis: The microbial contributions and clinical implications. Crit. Rev. Microbiol. 2017, 43, 81–95. [Google Scholar] [CrossRef]
- Sittipo, P.; Shim, J.W.; Lee, Y.K. Microbial Metabolites Determine Host Health and the Status of Some Diseases. Int. J. Mol. Sci. 2019, 20, 5296. [Google Scholar] [CrossRef] [PubMed]
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.; Sandhu, K.V.; Bastiaanssen, T.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The Microbiota-Gut-Brain Axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef] [PubMed]
- Greenhalgh, K.; Meyer, K.M.; Aagaard, K.M.; Wilmes, P. The human gut microbiome in health: Establishment and resilience of microbiota over a lifetime. Environ. Microbiol. 2016, 18, 2103–2116. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Government of the United Kingdom: London, UK, 2016.
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance. Available online: https://www.who.int/publications/i/item/9789241564748 (accessed on 24 May 2023).
- Sánchez, V.A.; Lavilla, L.L.; Benomar, N.; Gálvez, A.; Pérez, P.R.; Abriouel, H. Phenotypic and molecular antibiotic resistance profile of Enterococcus faecalis and Enterococcus faecium isolated from different traditional fermented foods. Foodborne Pathog. Dis. 2013, 10, 143–149. [Google Scholar] [CrossRef]
- Nieto-Arribas, P.; Seseña, S.; Poveda, J.M.; Chicón, R.; Cabezas, L.; Palop, L. Enterococcus populations in artisanal Manchego cheese: Biodiversity, technological and safety aspects. Food Microbiol. 2011, 28, 891–899. [Google Scholar] [CrossRef]
- Marshall, B.M.; Levy, S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [PubMed]
- Dibner, J.J.; Richards, J.D. Antibiotic growth promoters in agriculture: History and mode of action. Poult. Sci. 2005, 84, 634–643. [Google Scholar]
- Andersson, D.I.; Hughes, D. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 2014, 12, 465–478. [Google Scholar]
- Dewey, C.; Cox, B.D.; Straw, B.; Bush, E.; Hurd, S. Use of antimicrobials in swine feeds in the United States. J. Swine Health Prod. 1999, 7, 19–25. [Google Scholar]
- European, F.S.A.; European, C.F.D.P. European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from animals and food in the European Union in 2009. EFSA J. 2011, 9, 2154. [Google Scholar]
- Roubos-van, D.H.P.; Litjens, R.; Oudshoorn, A.K.; Resink, J.W.; Smits, C. New perspectives to the enterotoxigenic E. coli F4 porcine infection model: Susceptibility genotypes in relation to performance, diarrhoea and bacterial shedding. Vet. Microbiol. 2017, 202, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Madec, F.; Bridoux, N.; Bounaix, S.; Cariolet, R.; Duval-Iflah, Y.; Hampson, D.J.; Jestin, A. Experimental models of porcine post-weaning colibacillosis and their relationship to post-weaning diarrhoea and digestive disorders as encountered in the field. Vet. Microbiol. 2000, 72, 295–310. [Google Scholar] [CrossRef]
- Fairbrother, J.M.; Nadeau, E.; Gyles, C.L. Escherichia coli in postweaning diarrhea in pigs: An update on bacterial types, pathogenesis, and prevention strategies. Anim. Health Res. Rev. 2005, 6, 17–39. [Google Scholar] [CrossRef]
- Nagy, B.; Fekete, P.Z. Enterotoxigenic Escherichia coli (ETEC) in farm animals. Vet. Res. 1999, 30, 259–284. [Google Scholar]
- Moya, A.; Ferrer, M. Functional Redundancy-Induced Stability of Gut Microbiota Subjected to Disturbance. Trends Microbiol. 2016, 24, 402–413. [Google Scholar] [PubMed]
- Kim, K.; Song, M.; Liu, Y.; Ji, P. Enterotoxigenic Escherichia coli infection of weaned pigs: Intestinal challenges and nutritional intervention to enhance disease resistance. Front. Immunol. 2022, 13, 885253. [Google Scholar] [PubMed]
- Uddin, M.B.; Alam, M.N.; Hasan, M.; Hossain, S.; Debnath, M.; Begum, R.; Samad, M.A.; Hoque, S.F.; Chowdhury, M.; Rahman, M.M.; et al. Molecular Detection of Colistin Resistance mcr-1 Gene in Multidrug-Resistant Escherichia coli Isolated from Chicken. Antibiotics 2022, 11, 97. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, Z.; Aslam, B.; Zahoor, M.A.; Siddique, A.; Rafique, A.; Aslam, R.; Qamar, M.U.; Ali, S.; Mubeen, M. Frequency of Extended Spectrum Beta Lactamase Producing Escherichia coli in Fresh and Frozen Meat. Pak. Vet. J. 2020, 41, 102–106. [Google Scholar] [CrossRef]
- Jensen, G.M.; Frydendahl, K.; Svendsen, O.; Jørgensen, C.B.; Cirera, S.; Fredholm, M.; Nielsen, J.P.; Møller, K. Experimental infection with Escherichia coli O149:F4ac in weaned piglets. Vet. Microbiol. 2006, 115, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Trevisi, P.; Colombo, M.; Priori, D.; Fontanesi, L.; Galimberti, G.; Calò, G.; Motta, V.; Latorre, R.; Fanelli, F.; Mezzullo, M.; et al. Comparison of three patterns of feed supplementation with live Saccharomyces cerevisiae yeast on postweaning diarrhea, health status, and blood metabolic profile of susceptible weaning pigs orally challenged with Escherichia coli F4ac. J. Anim. Sci. 2015, 93, 2225–2233. [Google Scholar] [CrossRef]
- Kim, K.; He, Y.; Jinno, C.; Kovanda, L.; Li, X.; Song, M.; Liu, Y. Trace amounts of antibiotic exacerbated diarrhea and systemic inflammation of weaned pigs infected with a pathogenic Escherichia coli. J. Anim. Sci. 2021, 99, skab073. [Google Scholar] [CrossRef]
- Kim, K.; He, Y.; Jinno, C.; Kovanda, L.; Li, X.; Bravo, D.; Cox, E.; Liu, Y. Supplementation of oligosaccharide-based polymer enhanced growth and disease resistance of weaned pigs by modulating intestinal integrity and systemic immunity. J. Anim. Sci. Biotechnol. 2022, 13, 10. [Google Scholar]
- Becattini, S.; Taur, Y.; Pamer, E.G. Antibiotic-Induced Changes in the Intestinal Microbiota and Disease. Trends Mol. Med. 2016, 22, 458–478. [Google Scholar]
- Dubourg, G.; Lagier, J.C.; Robert, C.; Armougom, F.; Hugon, P.; Metidji, S.; Dione, N.; Dangui, N.P.; Pfleiderer, A.; Abrahao, J.; et al. Culturomics and pyrosequencing evidence of the reduction in gut microbiota diversity in patients with broad-spectrum antibiotics. Int. J. Antimicrob. Agents 2014, 44, 117–124. [Google Scholar] [CrossRef]
- Hill, D.A.; Hoffmann, C.; Abt, M.C.; Du, Y.; Kobuley, D.; Kirn, T.J.; Bushman, F.D.; Artis, D. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010, 3, 148–158. [Google Scholar] [PubMed]
- Naeem, S.; Kawabata, Z.; Loreau, M. Transcending boundaries in biodiversity research. Trends Ecol. Evol. 1998, 13, 134–135. [Google Scholar] [CrossRef] [PubMed]
- Konopka, A. What is microbial community ecology? ISME J. 2009, 3, 1223–1230. [Google Scholar]
- Ben-Amor, K.; Heilig, H.; Smidt, H.; Vaughan, E.E.; Abee, T.; de Vos, W.M. Genetic diversity of viable, injured, and dead fecal bacteria assessed by fluorescence-activated cell sorting and 16S rRNA gene analysis. Appl. Environ. Microb. 2005, 71, 4679–4689. [Google Scholar]
- Potgieter, M.; Bester, J.; Kell, D.B.; Pretorius, E. The dormant blood microbiome in chronic, inflammatory diseases. FEMS Microbiol. Rev. 2015, 39, 567–591. [Google Scholar] [PubMed]
- Maurice, C.F.; Haiser, H.J.; Turnbaugh, P.J. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 2013, 152, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Peris-Bondia, F.; Latorre, A.; Artacho, A.; Moya, A.; D’Auria, G. The active human gut microbiota differs from the total microbiota. PLoS ONE 2011, 6, e22448. [Google Scholar]
- Ferrer, M.; Méndez-García, C.; Rojo, D.; Barbas, C.; Moya, A. Antibiotic use and microbiome function. Biochem. Pharmacol. 2017, 134, 114–126. [Google Scholar]
- Wang, B.; Wu, L.; Chen, J.; Dong, L.; Chen, C.; Wen, Z.; Hu, J.; Fleming, I.; Wang, D.W. Metabolism pathways of arachidonic acids: Mechanisms and potential therapeutic targets. Signal Transduct. Tar. 2021, 6, 94. [Google Scholar]
- Suzuki, S.; Horinouchi, T.; Furusawa, C. Prediction of antibiotic resistance by gene expression profiles. Nat. Commun. 2014, 5, 5792. [Google Scholar]
- Frese, S.A.; Parker, K.; Calvert, C.C.; Mills, D.A. Diet shapes the gut microbiome of pigs during nursing and weaning. Microbiome 2015, 3, 28. [Google Scholar] [PubMed]
- Mach, N.; Berri, M.; Estellé, J.; Levenez, F.; Lemonnier, G.; Denis, C.; Leplat, J.J.; Chevaleyre, C.; Billon, Y.; Doré, J.; et al. Early-life establishment of the swine gut microbiome and impact on host phenotypes. Environ. Microbiol. Rep. 2015, 7, 554–569. [Google Scholar]
- Chen, L.; Xu, Y.; Chen, X.; Fang, C.; Zhao, L.; Chen, F. The Maturing Development of Gut Microbiota in Commercial Piglets during the Weaning Transition. Front. Microbiol. 2017, 8, 1688. [Google Scholar]
- Gresse, R.; Chaucheyras-Durand, F.; Fleury, M.A.; Van de Wiele, T.; Forano, E.; Blanquet-Diot, S. Gut Microbiota Dysbiosis in Postweaning Piglets: Understanding the Keys to Health. Trends Microbiol. 2017, 25, 851–873. [Google Scholar] [PubMed]
- Zhao, J.; Liu, P.; Wu, Y.; Guo, P.; Liu, L.; Ma, N.; Levesque, C.; Chen, Y.; Zhao, J.; Zhang, J.; et al. Dietary Fiber Increases Butyrate-Producing Bacteria and Improves the Growth Performance of Weaned Piglets. J. Agric. Food Chem. 2018, 66, 7995–8004. [Google Scholar] [CrossRef]
- Saladrigas-García, M.; D’Angelo, M.; Ko, H.L.; Nolis, P.; Ramayo-Caldas, Y.; Folch, J.M.; Llonch, P.; Solà-Oriol, D.; Pérez, J.F.; Martín-Orúe, S.M. Understanding host-microbiota interactions in the commercial piglet around weaning. Sci. Rep. 2021, 11, 23488. [Google Scholar]
- Hiraishi, K.; Zhao, F.; Kurahara, L.H.; Li, X.; Yamashita, T.; Hashimoto, T.; Matsuda, Y.; Sun, Z.; Zhang, H.; Hirano, K. Lactulose Modulates the Structure of Gut Microbiota and Alleviates Colitis-Associated Tumorigenesis. Nutrients 2022, 14, 649. [Google Scholar]
- Pereira, F.C.; Wasmund, K.; Cobankovic, I.; Jehmlich, N.; Herbold, C.W.; Lee, K.S.; Sziranyi, B.; Vesely, C.; Decker, T.; Stocker, R.; et al. Rational design of a microbial consortium of mucosal sugar utilizers reduces Clostridiodes difficile colonization. Nat. Commun. 2020, 11, 5104. [Google Scholar]
- Chevalier, C.; Kieser, S.; Çolakoğlu, M.; Hadadi, N.; Brun, J.; Rigo, D.; Suárez-Zamorano, N.; Spiljar, M.; Fabbiano, S.; Busse, B.; et al. Warmth Prevents Bone Loss Through the Gut Microbiota. Cell Metab. 2020, 32, 575–590. [Google Scholar]
- National Research Council. Nutrient Requirements of Swine, 11th ed.; The National Academies Press: Washington, DC, USA, 2012. [Google Scholar]
- Wang, L.; Martínez, V.; Kimura, H.; Taché, Y. 5-Hydroxytryptophan activates colonic myenteric neurons and propulsive motor function through 5-HT4 receptors in conscious mice. Am. J. Physiol.-Gastrointest. Liver Physiol. 2007, 292, G419–G428. [Google Scholar] [CrossRef]
- Tsukahara, T.; Nakanishi, N.; Nakayama, K.; Matsubara, N.; Ushida, K. Experimental infection of enterotoxemic Escherichia coli associated with porcine edema disease and its pathologic characteristics in the intestine. J. Vet. Med. Sci. 2005, 67, 1167–1171. [Google Scholar]
- Han, G.Q.; Xiang, Z.T.; Yu, B.; Chen, D.W.; Qi, H.W.; Mao, X.B.; Chen, H.; Mao, Q.; Huang, Z.Q. Effects of different starch sources on Bacillus spp. in intestinal tract and expression of intestinal development related genes of weanling piglets. Mol. Biol. Rep. 2012, 39, 1869–1876. [Google Scholar] [PubMed]
- Hong, C.; Si, Y.; Xing, Y.; Li, Y. Illumina MiSeq sequencing investigation on the contrasting soil bacterial community structures in different iron mining areas. Environ. Sci. Pollut. Res. 2015, 22, 10788–10799. [Google Scholar]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shang, L.; Yang, F.; Wei, Y.; Dai, Z.; Chen, Q.; Zeng, X.; Qiao, S.; Yu, H. Multi-Omics Analysis Reveals the Gut Microbiota Characteristics of Diarrheal Piglets Treated with Gentamicin. Antibiotics 2023, 12, 1349. https://doi.org/10.3390/antibiotics12091349
Shang L, Yang F, Wei Y, Dai Z, Chen Q, Zeng X, Qiao S, Yu H. Multi-Omics Analysis Reveals the Gut Microbiota Characteristics of Diarrheal Piglets Treated with Gentamicin. Antibiotics. 2023; 12(9):1349. https://doi.org/10.3390/antibiotics12091349
Chicago/Turabian StyleShang, Lijun, Fengjuan Yang, Yushu Wei, Ziqi Dai, Qingyun Chen, Xiangfang Zeng, Shiyan Qiao, and Haitao Yu. 2023. "Multi-Omics Analysis Reveals the Gut Microbiota Characteristics of Diarrheal Piglets Treated with Gentamicin" Antibiotics 12, no. 9: 1349. https://doi.org/10.3390/antibiotics12091349
APA StyleShang, L., Yang, F., Wei, Y., Dai, Z., Chen, Q., Zeng, X., Qiao, S., & Yu, H. (2023). Multi-Omics Analysis Reveals the Gut Microbiota Characteristics of Diarrheal Piglets Treated with Gentamicin. Antibiotics, 12(9), 1349. https://doi.org/10.3390/antibiotics12091349




