Dilution Reduces Sample Matrix Effects for Rapid, Direct, and Miniaturised Phenotypic Antibiotic Susceptibility Tests for Bovine Mastitis
Abstract
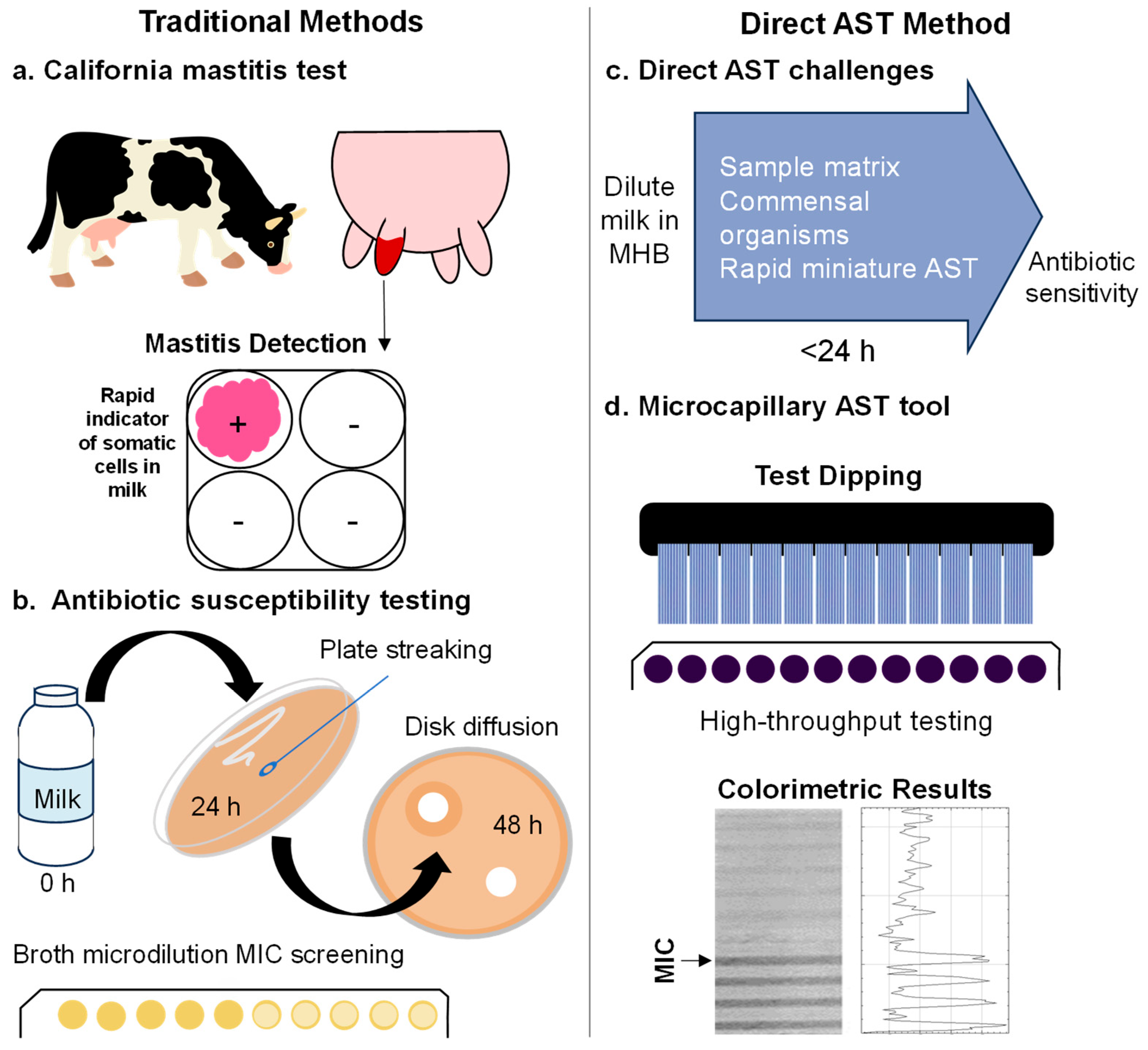
:1. Introduction
2. Results
2.1. Outlining Challenges with Direct Rapid Field Antibiotic Susceptibility Testing
2.2. Colourimetric Microfluidic Bacterial Growth Detection in the Presence of Milk
2.3. Impact of Milk Sample Matrix on Antibiotic Minimum Inhibitory Concentration Measurement
2.4. Exploring Whether Commensal Organisms in Milk Might Affect Direct Microfluidic AST
3. Discussion
4. Materials and Methods
4.1. Bacterial Isolates and Reagents
4.2. Preparation of Antibiotic Microcapillary Dip-Stick Test Strips
4.3. Microcapillary Antibiotic Susceptibility Test
4.4. Total Plate Count and Bacterial Identification of Fresh Milk Samples from Healthy Cows
4.5. MIC Determination in Simulated Mastitis Samples with Mixed Bacterial Populations
4.6. Imaging and Image Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharma, C.; Rokana, N.; Chandra, M.; Singh, B.P.; Gulhane, R.D.; Gill, J.P.S.; Ray, P.; Puniya, A.K.; Panwar, H. Antimicrobial Resistance: Its Surveillance, Impact, and Alternative Management Strategies in Dairy Animals. Front. Vet. Sci. 2017, 4, 237. [Google Scholar] [CrossRef] [PubMed]
- Woolhouse, M.; Ward, M.; van Bunnik, B.; Farrar, J. Antimicrobial resistance in humans, livestock and the wider environment. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140083. [Google Scholar] [CrossRef] [PubMed]
- Call, D.R.; Davis, M.A.; Sawant, A.A. Antimicrobial resistance in beef and dairy cattle production. Anim. Health Res. Rev. 2008, 9, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Scott, H.M.; Acuff, G.; Bergeron, G.; Bourassa, M.W.; Gill, J.; Graham, D.W.; Kahn, L.H.; Morley, P.S.; Salois, M.J.; Simjee, S.; et al. Critically important antibiotics: Criteria and approaches for measuring and reducing their use in food animal agriculture. Ann. N. Y. Acad. Sci. 2019, 1441, 8–16. [Google Scholar] [CrossRef]
- OIE. OIE List of Antimicrobials of Veterinary Importance; World Organization for Animal Health: Paris, France, 2019; pp. 1–9. [Google Scholar]
- Rainard, P.; Foucras, G.; Fitzgerald, J.R.; Watts, J.L.; Koop, G.; Middleton, J.R. Knowledge gaps and research priorities in Staphylococcus aureus mastitis control. Transbound. Emerg Dis. 2018, 65 (Suppl. 1), 149–165. [Google Scholar] [CrossRef]
- Rainard, P. Mammary microbiota of dairy ruminants: Fact or fiction? Vet. Res. 2017, 48, 25. [Google Scholar] [CrossRef]
- Oikonomou, G.; Machado, V.S.; Santisteban, C.; Schukken, Y.H.; Bicalho, R.C. Microbial diversity of bovine mastitic milk as described by pyrosequencing of metagenomic 16s rDNA. PLoS ONE 2012, 7, e47671. [Google Scholar] [CrossRef]
- Taponen, S.; McGuinness, D.; Hiitio, H.; Simojoki, H.; Zadoks, R.; Pyorala, S. Bovine milk microbiome: A more complex issue than expected. Vet. Res. 2019, 50, 44. [Google Scholar] [CrossRef]
- Addis, M.F.; Tanca, A.; Uzzau, S.; Oikonomou, G.; Bicalho, R.C.; Moroni, P. The bovine milk microbiota: Insights and perspectives from -omics studies. Mol. Biosyst. 2016, 12, 2359–2372. [Google Scholar] [CrossRef]
- Bronzo, V.; Lopreiato, V.; Riva, F.; Amadori, M.; Curone, G.; Addis, M.F.; Cremonesi, P.; Moroni, P.; Trevisi, E.; Castiglioni, B. The Role of Innate Immune Response and Microbiome in Resilience of Dairy Cattle to Disease: The Mastitis Model. Animals 2020, 10, 1397. [Google Scholar] [CrossRef]
- Dong, L.; Meng, L.; Liu, H.; Wu, H.; Schroyen, M.; Zheng, N.; Wang, J. Effect of Cephalosporin Treatment on the Microbiota and Antibiotic Resistance Genes in Feces of Dairy Cows with Clinical Mastitis. Antibiotics 2022, 11, 117. [Google Scholar] [CrossRef] [PubMed]
- Levison, L.J.; Miller-Cushon, E.K.; Tucker, A.L.; Bergeron, R.; Leslie, K.E.; Barkema, H.W.; DeVries, T.J. Incidence rate of pathogen-specific clinical mastitis on conventional and organic Canadian dairy farms. J. Dairy Sci. 2016, 99, 1341–1350. [Google Scholar] [CrossRef] [PubMed]
- Oikonomou, G.; Bicalho, M.L.; Meira, E.; Rossi, R.E.; Foditsch, C.; Machado, V.S.; Teixeira, A.G.; Santisteban, C.; Schukken, Y.H.; Bicalho, R.C. Microbiota of cow’s milk; distinguishing healthy, sub-clinically and clinically diseased quarters. PLoS ONE 2014, 9, e85904. [Google Scholar] [CrossRef]
- Kuehn, J.S.; Gorden, P.J.; Munro, D.; Rong, R.; Dong, Q.; Plummer, P.J.; Wang, C.; Phillips, G.J. Bacterial community profiling of milk samples as a means to understand culture-negative bovine clinical mastitis. PLoS ONE 2013, 8, e61959. [Google Scholar] [CrossRef]
- Machado, N.A.F.; Da Costa, L.B.S.; Barbosa-Filho, J.A.D.; De Oliveira, K.P.L.; De Sampaio, L.C.; Peixoto, M.S.M.; Damasceno, F.A. Using infrared thermography to detect subclinical mastitis in dairy cows in compost barn systems. J. Therm. Biol. 2021, 97, 102881. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kang, X.; He, Z.; Feng, Y.; Liu, G. Accurate detection of dairy cow mastitis with deep learning technology: A new and comprehensive detection method based on infrared thermal images. Animal 2022, 16, 100646. [Google Scholar] [CrossRef]
- Morgans, L. Developing targeted antimicrobial treatment protocols for mastitis. Vet. Rec. 2020, 187, 398–400. [Google Scholar] [CrossRef]
- Leimbach, S.; Kromker, V. Laboratory evaluation of a novel rapid tube test system for differentiation of mastitis-causing pathogen groups. J. Dairy Sci. 2018, 101, 6357–6365. [Google Scholar] [CrossRef]
- Jones, G.; Bork, O.; Ferguson, S.A.; Bates, A. Comparison of an on-farm point-of-care diagnostic with conventional culture in analysing bovine mastitis samples. J. Dairy Res. 2019, 86, 222–225. [Google Scholar] [CrossRef]
- Malcata, F.B.; Pepler, P.T.; O’Reilly, E.L.; Brady, N.; Eckersall, P.D.; Zadoks, R.N.; Viora, L. Point-of-care tests for bovine clinical mastitis: What do we have and what do we need? J. Dairy Res. 2020, 87, 60–66. [Google Scholar] [CrossRef]
- Griffioen, K.; Hop, G.E.; Holstege, M.M.C.; Velthuis, A.G.J.; Lam, T.; 1Health4Food–Dutch Mastitis Diagnostics Consortium. Dutch dairy farmers’ need for microbiological mastitis diagnostics. J. Dairy Sci. 2016, 99, 5551–5561. [Google Scholar] [CrossRef] [PubMed]
- Reis, N.M.; Pivetal, J.; Loo-Zazueta, A.L.; Barros, J.M.; Edwards, A.D. Lab on a stick: Multi-analyte cellular assays in a microfluidic dipstick. Lab Chip 2016, 16, 2891–2899. [Google Scholar] [CrossRef]
- Rodoplu, D.; Chang, C.S.; Kao, C.Y.; Hsu, C.H. A simple magnetic-assisted microfluidic method for rapid detection and phenotypic characterization of ultralow concentrations of bacteria. Talanta 2021, 230, 122291. [Google Scholar] [CrossRef]
- Donmez, S.I.; Needs, S.H.; Osborn, H.M.I.; Edwards, A.D. Label-free smartphone quantitation of bacteria by darkfield imaging of light scattering in fluoropolymer micro capillary film allows portable detection of bacteriophage lysis. Sens. Actuators B Chem. 2020, 323, 128645. [Google Scholar] [CrossRef]
- Huang, X.; Xu, D.; Chen, J.; Liu, J.; Li, Y.; Song, J.; Ma, X.; Guo, J. Smartphone-based analytical biosensors. Analyst 2018, 143, 5339–5351. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Caen, O.; Vrignon, J.; Zonta, E.; El Harrak, Z.; Nizard, P.; Baret, J.C.; Taly, V. Author Correction: High throughput single cell counting in droplet-based microfluidics. Sci. Rep. 2020, 10, 10061. [Google Scholar] [CrossRef] [PubMed]
- Needs, S.H.; Diep, T.T.; Bull, S.P.; Lindley-Decaire, A.; Ray, P.; Edwards, A.D. Exploiting open source 3D printer architecture for laboratory robotics to automate high-throughput time-lapse imaging for analytical microbiology. PLoS ONE 2019, 14, e0224878. [Google Scholar] [CrossRef]
- Chakraborty, S.; Dhama, K.; Tiwari, R.; Iqbal Yatoo, M.; Khurana, S.K.; Khandia, R.; Munjal, A.; Munuswamy, P.; Kumar, M.A.; Singh, M.; et al. Technological interventions and advances in the diagnosis of intramammary infections in animals with emphasis on bovine population—A review. Vet. Q. 2019, 39, 76–94. [Google Scholar] [CrossRef]
- Berlanda, S.F.; Breitfeld, M.; Dietsche, C.L.; Dittrich, P.S. Recent Advances in Microfluidic Technology for Bioanalysis and Diagnostics. Anal. Chem. 2021, 93, 311–331. [Google Scholar] [CrossRef]
- Needs, S.H.; Saiprom, N.; Rafaque, Z.; Imtiaz, W.; Chantratita, N.; Runcharoen, C.; Thammachote, J.; Anun, S.; Peacock, S.J.; Ray, P.; et al. Miniaturised broth microdilution for simplified antibiotic susceptibility testing of Gram negative clinical isolates using microcapillary devices. Analyst 2022, 147, 3558–3569. [Google Scholar] [CrossRef]
- Reis, N.M.; Needs, S.H.; Jegouic, S.M.; Gill, K.K.; Sirivisoot, S.; Howard, S.; Kempe, J.; Bola, S.; Al-Hakeem, K.; Jones, I.M.; et al. Gravity-Driven Microfluidic Siphons: Fluidic Characterization and Application to Quantitative Immunoassays. ACS Sens. 2021, 6, 4338–4348. [Google Scholar] [CrossRef]
- Long, M.M.; Diep, T.T.; Needs, S.H.; Ross, M.J.; Edwards, A.D. PiRamid: A compact Raspberry Pi imaging box to automate small-scale time-lapse digital analysis, suitable for laboratory and field use. HardwareX 2022, 12, e00377. [Google Scholar] [CrossRef] [PubMed]
- Needs, S.H.; Osborn, H.M.I.; Edwards, A.D. Counting bacteria in microfluidic devices: Smartphone compatible ‘dip-and-test’ viable cell quantitation using resazurin amplified detection in microliter capillary arrays. J. Microbiol. Methods 2021, 187, 106199. [Google Scholar] [CrossRef]
- Nagasawa, Y.; Kiku, Y.; Sugawara, K.; Yabusaki, T.; Oono, K.; Fujii, K.; Suzuki, T.; Maehana, K.; Hayashi, T. The bacterial load in milk is associated with clinical severity in cases of bovine coliform mastitis. J. Vet. Med. Sci. 2019, 81, 107–112. [Google Scholar] [CrossRef]
- Needs, S.H.; Dönmez, S.İ.; Edwards, A.D. Direct microfluidic antibiotic resistance testing in urine with smartphone capture: Significant variation in sample matrix interference between individual human urine samples. RSC Adv. 2021, 11, 38258–38263. [Google Scholar] [CrossRef]
- Alakomi, H.L.; Skytta, E.; Saarela, M.; Mattila-Sandholm, T.; Latva-Kala, K.; Helander, I.M. Lactic acid permeabilizes gram-negative bacteria by disrupting the outer membrane. Appl. Environ. Microbiol. 2000, 66, 2001–2005. [Google Scholar] [CrossRef]
- European Commission. Regulation (EC) No. 853/2004 of the European Parliament and of the Council of 29 April 2004 Laying Down Specific Hygiene Rules for Food of Animal Origin; European Commission: Brussels, Belgium, 2021. [Google Scholar]
- Conucil of the European Union. Council Directive 92/46/EC Laying Down the Health Rules for the Production and Placing on the Market of Raw Milk, Heat Treated Milk and Milk-Based Products; Official Journal of the European Union: Luxembourg, Luxembourg, 1992; pp. 1–31.
- Quigley, L.; O’Sullivan, O.; Stanton, C.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. The complex microbiota of raw milk. FEMS Microbiol. Rev. 2013, 37, 664–698. [Google Scholar] [CrossRef] [PubMed]
- Martin, N.H.; Torres-Frenzel, P.; Wiedmann, M. Invited review: Controlling dairy product spoilage to reduce food loss and waste. J. Dairy Sci. 2021, 104, 1251–1261. [Google Scholar] [CrossRef] [PubMed]
- Asif, M.; Awan, F.R.; Khan, Q.M.; Ngamsom, B.; Pamme, N. Paper-based analytical devices for colorimetric detection of S. aureus and E. coli and their antibiotic resistant strains in milk. Analyst 2020, 145, 7320–7329. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Xiang, W.; Wang, Q.; Yin, J.; Tian, T.; Yang, Q.; Zhang, M.; Ge, G.; Li, J.; Diao, N.; et al. Prevalence and risk factors of Klebsiella spp. in milk samples from dairy cows with mastitis—A global systematic review. Front. Vet. Sci. 2023, 10, 1143257. [Google Scholar] [CrossRef]
- Park, H.R.; Hong, M.K.; Hwang, S.Y.; Park, Y.K.; Kwon, K.H.; Yoon, J.W.; Shin, S.; Kim, J.H.; Park, Y.H. Characterisation of Pseudomonas aeruginosa related to bovine mastitis. Acta Vet. Hung. 2014, 62, 1–12. [Google Scholar] [CrossRef]
- Kawai, K.; Shinozuka, Y.; Uchida, I.; Hirose, K.; Mitamura, T.; Watanabe, A.; Kuruhara, K.; Yuasa, R.; Sato, R.; Onda, K.; et al. Control of Pseudomonas mastitis on a large dairy farm by using slightly acidic electrolyzed water. Anim. Sci. J. 2017, 88, 1601–1605. [Google Scholar] [CrossRef]
- Schauer, B.; Wald, R.; Urbantke, V.; Loncaric, I.; Baumgartner, M. Tracing Mastitis Pathogens-Epidemiological Investigations of a Pseudomonas aeruginosa Mastitis Outbreak in an Austrian Dairy Herd. Animals 2021, 11, 279. [Google Scholar] [CrossRef] [PubMed]
- Hoque, M.N.; Istiaq, A.; Clement, R.A.; Gibson, K.M.; Saha, O.; Islam, O.K.; Abir, R.A.; Sultana, M.; Siddiki, A.Z.; Crandall, K.A.; et al. Insights Into the Resistome of Bovine Clinical Mastitis Microbiome, a Key Factor in Disease Complication. Front. Microbiol. 2020, 11, 860. [Google Scholar] [CrossRef] [PubMed]
- Burian, A.; Erdogan, Z.; Jandrisits, C.; Zeitlinger, M. Impact of pH on activity of trimethoprim, fosfomycin, amikacin, colistin and ertapenem in human urine. Pharmacology 2012, 90, 281–287. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Long, M.M.; Needs, S.H.; Edwards, A.D. Dilution Reduces Sample Matrix Effects for Rapid, Direct, and Miniaturised Phenotypic Antibiotic Susceptibility Tests for Bovine Mastitis. Antibiotics 2023, 12, 1363. https://doi.org/10.3390/antibiotics12091363
Long MM, Needs SH, Edwards AD. Dilution Reduces Sample Matrix Effects for Rapid, Direct, and Miniaturised Phenotypic Antibiotic Susceptibility Tests for Bovine Mastitis. Antibiotics. 2023; 12(9):1363. https://doi.org/10.3390/antibiotics12091363
Chicago/Turabian StyleLong, Matthew Michael, Sarah Helen Needs, and Alexander Daniel Edwards. 2023. "Dilution Reduces Sample Matrix Effects for Rapid, Direct, and Miniaturised Phenotypic Antibiotic Susceptibility Tests for Bovine Mastitis" Antibiotics 12, no. 9: 1363. https://doi.org/10.3390/antibiotics12091363
APA StyleLong, M. M., Needs, S. H., & Edwards, A. D. (2023). Dilution Reduces Sample Matrix Effects for Rapid, Direct, and Miniaturised Phenotypic Antibiotic Susceptibility Tests for Bovine Mastitis. Antibiotics, 12(9), 1363. https://doi.org/10.3390/antibiotics12091363







