Antibiotic and Heavy Metal Co-Resistant Strain Isolated from Enrichment Culture of Marine Sediments, with Potential for Environmental Bioremediation Applications
Abstract
:1. Introduction
2. Results and Discussion
2.1. Analysis of the Community Structure of the Isolated Strains
2.2. Biochemical and Phenotypic Properties of Strain ZC255
2.3. Phylogenetic Analysis of the Strain
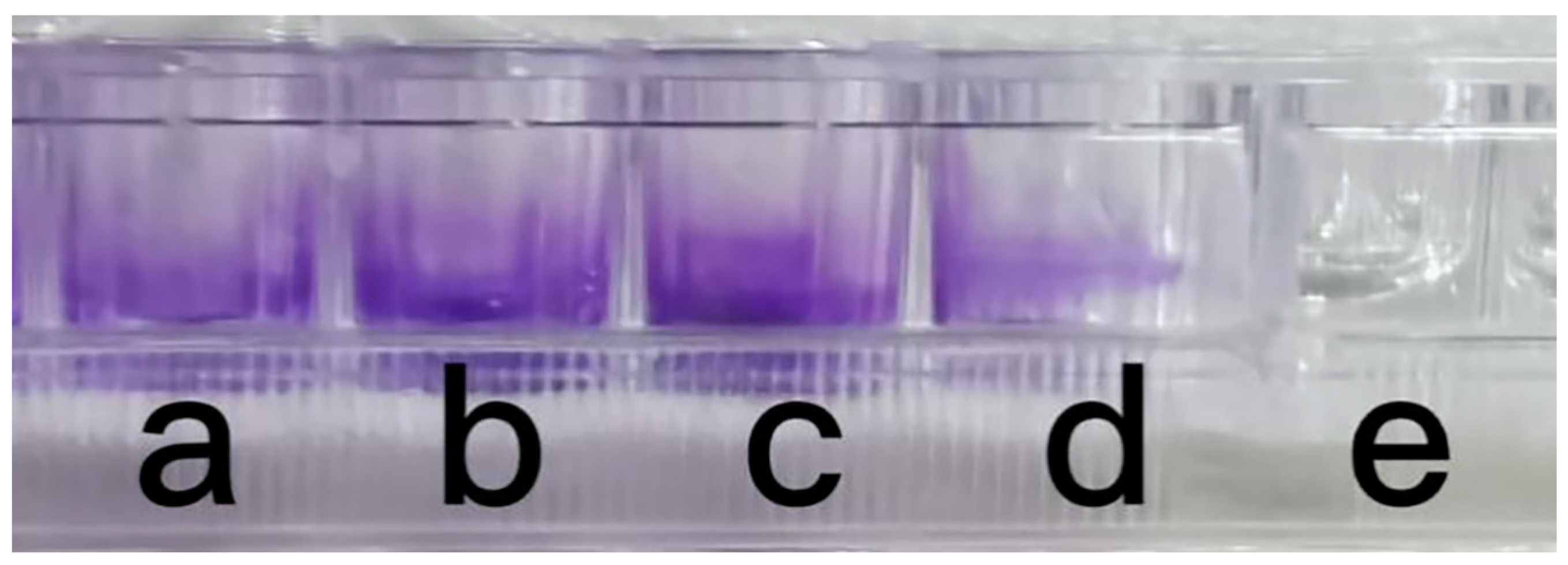
2.4. Biofilm Formation Assay
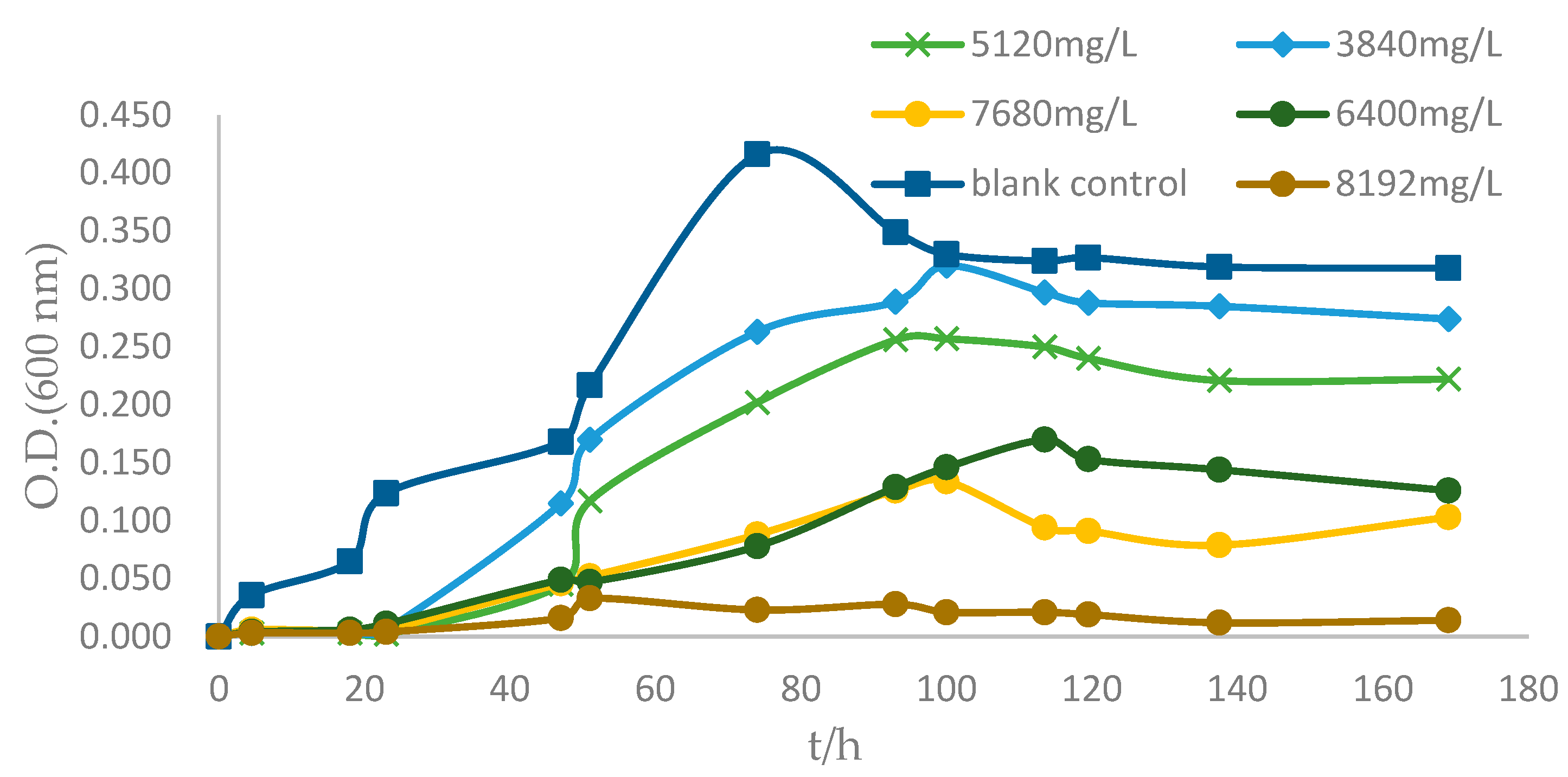
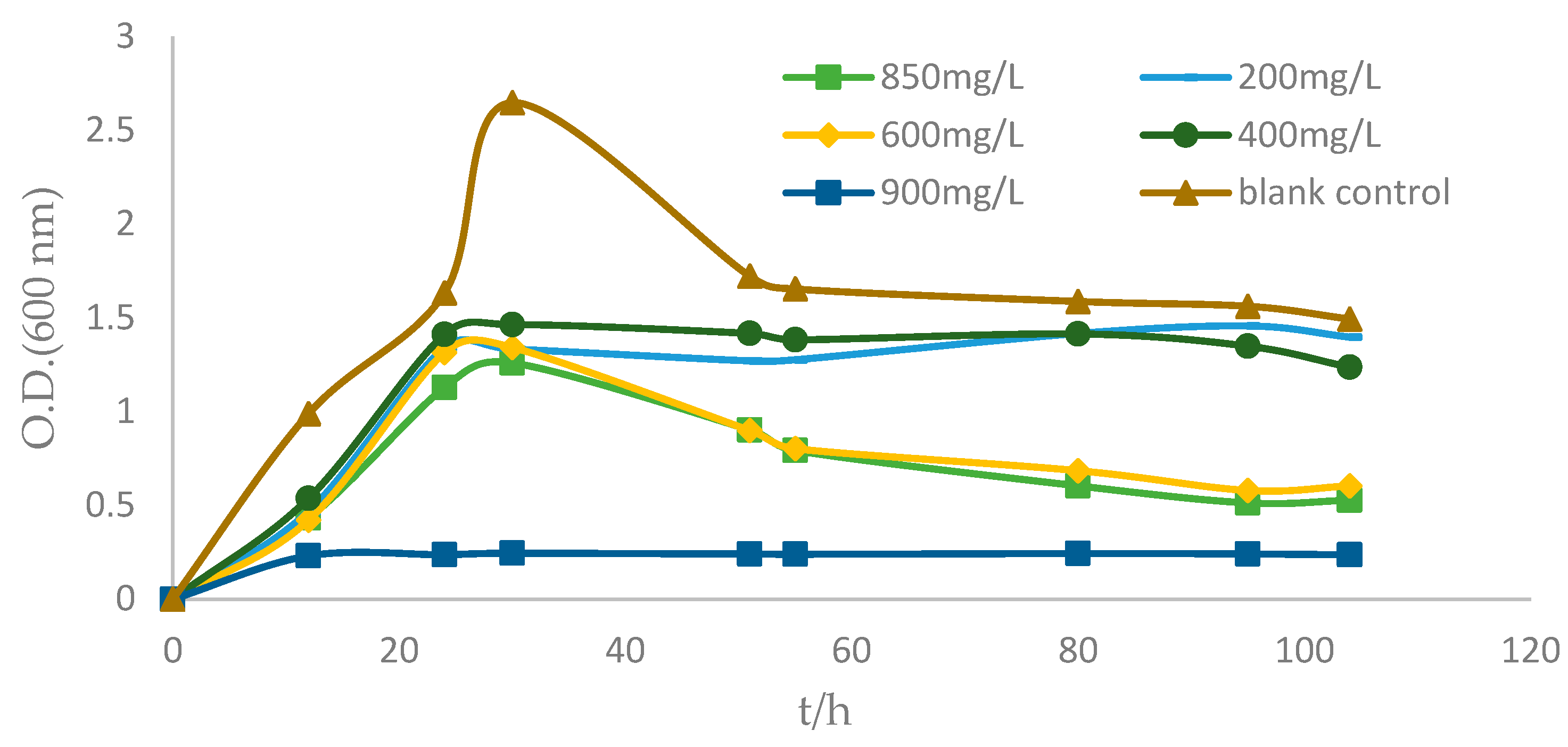
2.5. Drug Resistance of Strain ZC255
2.6. Evaluation of the Bioremoval Capacity of Strain ZC255
3. Materials and Methods
3.1. Sampling and Enrichment
3.2. Bacterial Isolation and Identification
3.3. Bacterial Resistance Assay against Antibiotics and Heavy Metals
3.4. Characterization of a Novel Resistant Strain
3.4.1. Morphological, Physiological and Biochemical Characteristics
3.4.2. Phylogenetic Analysis
3.4.3. Biofilm Formation Assay
3.4.4. Quantifying the Resistance to Different Drugs
3.4.5. Bioremoval Experiment
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, C.; Zou, X.; Feng, Z.; Hao, Z.; Gao, J. Distribution and transport of heavy metals in estuarine–inner shelf regions of the East China Sea. Sci. Total Environ. 2018, 644, 298–305. [Google Scholar] [CrossRef]
- Ojuederie, O.B.; Babalola, O.O. Microbial and plant-assisted bioremediation of heavy metal polluted environments: A review. Int. J. Environ. Res. Public Health 2017, 14, 1504. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Ji, C.; Li, F.; Wu, H. Bioaccumulation and human health implications of trace metals in oysters from coastal areas of China. Mar. Environ. Res. 2023, 184, 105872. [Google Scholar] [CrossRef]
- Lin, A.Y.; Yu, T.; Lin, C. Pharmaceutical contamination in residential, industrial, and agricultural waste streams: Risk to aqueous environments in Taiwan. Chemosphere 2008, 74, 131–141. [Google Scholar] [CrossRef]
- Fraser, M.; Surette, C.; Vaillancourt, C. Fish and seafood availability in markets in the Baie des Chaleurs Region, New Brunswick, Canada: A heavy metal contamination baseline study. Environ. Sci. Pollut. Res. Int. 2013, 20, 761–770. [Google Scholar] [CrossRef]
- Liu, X.; Steele, J.C.; Meng, X. Usage, residue, and human health risk of antibiotics in Chinese aquaculture: A review. Environ. Pollut. 2017, 223, 161–169. [Google Scholar] [CrossRef]
- Heuer, O.E.; Kruse, H.; Grave, K.; Collignon, P.; Karunasagar, I.; Angulo, F.J. Human health consequences of use of antimicrobial agents in aquaculture. Clin. Infect. Dis. 2009, 49, 1248–1253. [Google Scholar] [CrossRef]
- Okeke, E.S.; Chukwudozie, K.I.; Nyaruaba, R.; Ita, R.E.; Oladipo, A.; Ejeromedoghene, O.; Atakpa, E.O.; Agu, C.V.; Okoye, C.O. Antibiotic resistance in aquaculture and aquatic organisms: A review of current nanotechnology applications for sustainable management. Environ. Sci. Pollut. Res. Int. 2022, 29, 69241–69274. [Google Scholar] [CrossRef]
- Lei, K.; Lai, H. Effects of sunlight, microbial activity, and temperature on the declines of antibiotic lincomycin in freshwater and saline aquaculture pond waters and sediments. Environ. Sci. Pollut. Res. Int. 2019, 26, 33988–33994. [Google Scholar] [CrossRef] [PubMed]
- Cheung, K.C.; Leung, H.M.; Kong, K.Y.; Wong, M.H. Residual levels of ddts and pahs in freshwater and marine fish from Hong Kong markets and their health risk assessment. Chemosphere 2007, 66, 460–468. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Li, Y.; Qi, Z.; Yue, Y.; Min, M.; Peng, S.; Shi, Z.; Gao, Y. Diverse and abundant antibiotic resistance genes from mariculture sites of China’s coastline. Sci. Total Environ. 2018, 630, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Soni, K.; Jyoti, K.; Chandra, H.; Chandra, R. Bacterial antibiotic resistance in municipal wastewater treatment plant; Mechanism and its impacts on human health and economy. Bioresour. Technol. Rep. 2022, 19, 101080. [Google Scholar] [CrossRef]
- Zhou, L.; Li, S.; Li, F. Damage and elimination of soil and water antibiotic and heavy metal pollution caused by livestock husbandry. Environ. Res. 2022, 215, 114188. [Google Scholar] [CrossRef]
- Wang, M.; Yu, Y.; Ren, Y.; Wang, J.; Chen, H. Effect of antibiotic and/or heavy metal on nitrogen cycle of sediment-water interface in aquaculture system: Implications from sea cucumber culture. Environ. Pollut. 2023, 325, 121453. [Google Scholar] [CrossRef]
- Du, J.; Zhao, H.; Liu, S.; Xie, H.; Wang, Y.; Chen, J. Antibiotics in the coastal water of the south yellow sea in China: Occurrence, distribution and ecological risks. Sci. Total Environ. 2017, 595, 521–527. [Google Scholar] [CrossRef]
- Sui, Q.; Zhao, W.; Cao, X.; Lu, S.; Qiu, Z.; Gu, X.; Yu, G. Pharmaceuticals and personal care products in the leachates from a typical landfill reservoir of municipal solid waste in Shanghai, China: Occurrence and removal by a full-scale membrane bioreactor. J. Hazard. Mater. 2017, 323, 99–108. [Google Scholar] [CrossRef]
- Peng, Q.; Song, J.; Li, X.; Yuan, H.; Li, N.; Duan, L.; Zhang, Q.; Liang, X. Biogeochemical characteristics and ecological risk assessment of pharmaceutically active compounds (phacs) in the surface seawaters of Jiaozhou Bay, North China. Environ. Pollut. 2019, 255, 113247. [Google Scholar] [CrossRef] [PubMed]
- Mehrtens, A.; Licha, T.; Burke, V. Occurrence, effects and behaviour of the antibiotic lincomycin in the agricultural and aquatic environment—A review. Sci. Total Environ. 2021, 778, 146306. [Google Scholar] [CrossRef] [PubMed]
- Bertelkamp, C.; Reungoat, J.; Cornelissen, E.R.; Singhal, N.; Reynisson, J.; Cabo, A.J.; van der Hoek, J.P.; Verliefde, A.R.D. Sorption and biodegradation of organic micropollutants during river bank filtration: A laboratory column study. Water Res. 2014, 52, 231–241. [Google Scholar] [CrossRef]
- Bergmann, T.; Richardson, T.L.; Paerl, H.W.; Pinckney, J.L.; Schofield, O. Synergy of light and nutrients on the photosynthetic efficiency of phytoplankton populations from the Neuse River estuary, North Carolina. J. Plankton Res. 2002, 24, 923–933. [Google Scholar] [CrossRef]
- Andrade, L.R.; Farina, M.; Amado Filho, G.M. Effects of copper on Enteromorpha flexuosa (chlorophyta) in vitro. Ecotoxicol. Environ. Saf. 2004, 58, 117–125. [Google Scholar] [CrossRef]
- Mirzaei, M.; Azini, M.R.; Aminrad, T. Seasonal variation of heavy metal in seawater, sediment and hooded oyster, saccostreacucullata, in Iranian southern waters (Chabahar Coast). Res. Mar. Sci. 2016, 1, 3–12. [Google Scholar]
- Jardine, J.; Mavumengwana, V.; Ubomba-Jaswa, E. Antibiotic resistance and heavy metal tolerance in cultured bacteria from hot springs as indicators of environmental intrinsic resistance and tolerance levels. Environ. Pollut. 2019, 249, 696–702. [Google Scholar] [CrossRef]
- Li, J.; Li, W.; Liu, K.; Guo, Y.; Ding, C.; Han, J.; Li, P. Global review of macrolide antibiotics in the aquatic environment: Sources, occurrence, fate, ecotoxicity, and risk assessment. J. Hazard. Mater. 2022, 439, 129628. [Google Scholar] [CrossRef]
- Rakib, M.R.J.; Rahman, M.A.; Onyena, A.P.; Kumar, R.; Sarker, A.; Hossain, M.B.; Islam, A.R.M.T.; Islam, M.S.; Rahman, M.M.; Idris, Y.N.J.A.; et al. A comprehensive review of heavy metal pollution in the coastal areas of Bangladesh: Abundance, bioaccumulation, health implications, and challenges. Environ. Sci. Pollut. Res. 2022, 45, 67532–67558. [Google Scholar] [CrossRef]
- Zhao, M.; Xu, Y.; Zhang, C.; Rong, H.; Zeng, G. New trends in removing heavy metals from wastewater. Appl. Microbiol. Biotechnol. 2016, 100, 6509–6518. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, L.; Tian, W.; Ma, L. A review of the distribution of antibiotics in water in different regions of China and current antibiotic degradation pathways. Front. Environ. Sci. 2021, 9, 692298. [Google Scholar] [CrossRef]
- Kumarasamy, K.K.; Toleman, M.A.; Walsh, T.R.; Bagaria, J.; Butt, F.; Balakrishnan, R.; Chaudhary, U.; Doumith, M.; Giske, C.G.; Irfan, S.; et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect. Dis. 2010, 10, 597–602. [Google Scholar] [CrossRef]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Alequis Pavón, D.R.V.J.; Manzano, C.; Lopez-Joven, C.; Reyes-Cerpa, S.; Navarrete, P.; Pavez, L.; García, K. The high risk of bivalve farming in coastal areas with heavy metal pollution and antibiotic-resistant bacteria: A Chilean perspective. Front. Cell. Infect. Microbiol. 2022, 12, 377. [Google Scholar]
- von Wintersdorff, C.J.H.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef]
- Kamala-Kannan, S.; Lee, K.J. Metal tolerance and antibiotic resistance of bacillus species isolated from Sunchon Bay sediments, South Korea. Biotechnology 2008, 7, 149–152. [Google Scholar] [CrossRef]
- Nithya, C.; Pandian, S.K. Isolation of heterotrophic bacteria from Palk Bay sediments showing heavy metal tolerance and antibiotic production. Microbiol. Res. 2010, 7, 578–593. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Kim, I.; Kang, K.H.; Oh, T.; Park, Y. Bacillus marisflavi sp. Nov. and bacillus aquimaris sp. Nov., Isolated from sea water of a tidal flat of the Yellow Sea in Korea. Int. J. Syst. Evol. Microbiol. 2003, 53, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Haruko Noguchi, M.U.O.S.; Komagata, L.K.N. Bacillus vietnamensis sp. Nov., A moderately halotolerant, aerobic, endospore-forming bacterium isolated from Vietnamese fish sauce. Int. J. Syst. Evol. Microbiol. 2004, 54, 2117–2120. [Google Scholar] [CrossRef]
- Dastager, S.G.; Mawlankar, R.; Tang, S.-K.; Srinivasan, K.; Ramana, V.V.; Shouche, Y.S. Bacillus enclensis sp. Nov., Isolated from sediment sample. Antonie Van Leeuwenhoek 2013, 105, 199–206. [Google Scholar] [CrossRef]
- Navarro-Torre, S.; Lorena, C.; José, M.I.A.M. Rossellomorea arthrocnemi sp. Nov., A novel plant growthpromoting bacterium used in heavy metal polluted soils as a phytoremediation tool. Int. J. Syst. Evol. Microbiol. 2021, 71, 005015. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.W.; Kwon, S.; Kim, S.; Kim, S.Y.; Kim, J.J.; Lee, J.S.; Oh, M.; Kim, A.; Chung, K.S. Bacillus oryzaecorticis sp. Nov., A moderately halophilic bacterium isolated from rice husks. Int. J. Syst. Evol. Microbiol. 2014, 64, 2786–2791. [Google Scholar] [CrossRef]
- Gupta, R.S.; Patel, S.; Saini, N.; Chen, S. Robust demarcation of 17 distinct bacillus species clades, proposed as novel bacillaceae genera, by phylogenomics and comparative genomic analyses: Description of Robertmurraya kyonggiensis sp. Nov. And proposal for an emended genus bacillus limiting it only to the members of the subtilis and cereus clades of species. Int. J. Syst. Evol. Microbiol. 2020, 70, 5753. [Google Scholar]
- Fu, H.; Chen, F.; Liu, W.; Kong, W.; Wang, C.; Fang, X.; Ye, J. Adding nutrients to the biocontrol strain jk-sh007 promotes biofilm formation and improves resistance to stress. AMB Express 2020, 10, 32. [Google Scholar] [CrossRef]
- Fernandez, M.; Paulucci, N.S.; Reynoso, E.; Morales, G.M.; Agostini, E.; González, P.S. Morphological and structural response of Bacillus sp. Sfc 500-1e after cr(vi) and phenol treatment. J. Basic Microbiol. 2020, 8, 679–690. [Google Scholar] [CrossRef]
- Harrison, J.J.; Ceri, H.; Yerly, J.; Rabiei, M.; Hu, Y.; Martinuzzi, R.; Turner, R.J. Metal ions may suppress or enhance cellular differentiation in candida albicans and candida tropicalis biofilms. Appl. Environ. Microbiol. 2007, 73, 4940–4949. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Xu, Y.; Qiao, Y.; Zhang, Z.; Liang, J.; Peng, Y.; Liao, J.; Qiao, Y.; Shang, C.; Guo, Z.; et al. A novel yeast strain Geotrichum sp. Cs-67 capable of accumulating heavy metal ions. Ecotoxicol. Environ. Saf. 2022, 236, 113497. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Song, W.; Zhang, X.; Chen, M.; Li, J.; Yang, X.; Zhang, L. Potential application of novel cadmium-tolerant bacteria in bioremediation of cd-contaminated soil. Ecotoxicol. Environ. Saf. 2023, 255, 114766. [Google Scholar] [CrossRef]
- Singh, P.; Raghukumar, C.; Verma, A.K.; Meena, R.M. Differentially expressed genes under simulated deep-sea conditions in the psychrotolerant yeast Cryptococcus sp. Niocc#py13. Extremophiles 2012, 16, 777–785. [Google Scholar]
- Catania, S.; Bottinelli, M.; Fincato, A.; Gastaldelli, M.; Barberio, A.; Gobbo, F.; Vicenzoni, G. Evaluation of minimum inhibitory concentrations for 154 mycoplasma synoviae isolates from Italy collected during 2012–2017. PLoS ONE 2019, 14, e224903. [Google Scholar] [CrossRef]
- Andrzejak, T.; Raje, H.; Lafleur, G.; Willis, J.; Boopathy, R. Water quality and antibiotic resistance in the recreational waters. Bioresour. Technol. 2023, 370, 128546. [Google Scholar] [CrossRef] [PubMed]
- Iskandar, N.L.; Zainudin, N.A.I.M.; Tan, S.G. Tolerance and biosorption of copper (cu) and lead (pb) by filamentous fungi isolated from a freshwater ecosystem. J. Environ. Sci. 2011, 23, 824–830. [Google Scholar] [CrossRef]
- Wong, C.; Tan, L.T.; Mujahid, A.; Lihan, S.; Wee, J.L.S.; Ting, L.F.; Müller, M. Biosorption of copper by endophytic fungi isolated from nepenthes ampullaria. Lett. Appl. Microbiol. 2018, 4, 384–391. [Google Scholar] [CrossRef]
- Wang, M.; Cai, C.; Zhang, B.; Liu, H. Characterization and mechanism analysis of lincomycin biodegradation with Clostridium sp. Strain lcm-b isolated from lincomycin mycelial residue (lmr). Chemosphere 2018, 193, 611–617. [Google Scholar] [CrossRef]
- Apreja, M.; Sharma, A.; Balda, S.; Kataria, K.; Capalash, N.; Sharma, P. Antibiotic residues in environment: Antimicrobial resistance development, ecological risks, and bioremediation. Environ. Sci. Pollut. Res. Int. 2021, 2022, 3355–3371. [Google Scholar]
- Wróbel, M.; Sliwakowski, W.; Kowalczyk, P.; Kramkowski, K.; Dobrzyński, J. Bioremediation of heavy metals by the genus bacillus. Int. J. Environ. Res. Public Health 2023, 6, 4964. [Google Scholar]
- Ye, M.; Chen, G.; Du, Z. Effects of antibiotics on the bacterial community, metabolic functions and antibiotic resistance genes in mariculture sediments during enrichment culturing. J. Mar. Sci. Eng. 2020, 8, 604. [Google Scholar]
- Kang, C.; Shin, Y.; Yu, H.; Kim, S.; So, J. Antibiotic and heavy-metal resistance of vibrio parahaemolyticus isolated from oysters in Korea. Mar. Pollut. Bull. 2018, 135, 69–74. [Google Scholar]
- Jordan, E.M.; Thompson, F.L.; Zhang, X.; Li, Y.; Vancanneyt, M.; Kroppenstedt, R.M.; Priest, F.G.; Austin, B. Sneathiella chinensis gen. Nov., Sp. Nov., A novel marine alphaproteobacterium isolated from coastal sediment in Qingdao, China. Int. J. Syst. Evol. Microbiol. 2007, 57 Pt 1, 114. [Google Scholar] [PubMed]
- Yoon, S.; Ha, S.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing ezbiocloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Altuğ, G.; Çardak, M.; Türetken, P.S.Ç.; Kalkan, S.; Gürün, S. Antibiotic and heavy metal resistant bacteria isolated from aegean sea water and sediment in Güllük Bay, Turkey: Quantifying the resistance of identified bacteria species with potential for environmental remediation applications. Johns Matthey Technol. Rev. 2020, 64, 507–525. [Google Scholar]
- Yamina, B.; Tahar, B.; Laure, F.M. Isolation and screening of heavy metal resistant bacteria from wastewater: A study of heavy metal co-resistance and antibiotics resistance. Water Sci. Technol. 2012, 10, 2041–2048. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.Z.; Cai, M.Y. Determination of Biochemical Characteristics. In Manual for the Systematic Identification of General Bacteria, 14th ed.; Science Press: Beijing, China, 2001. [Google Scholar]
- Edgar, R.C. Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 19, 1792–1797. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 4, 783–791. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7, Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 7, 1870–1874. [Google Scholar]
- Maity, S.; Sarkar, D.; Poddar, K.; Patil, P.; Sarkar, A. Biofilm-mediated heavy metal removal from aqueous system by multi-metal-resistant bacterial strain Bacillus sp. Gh-s29. Appl. Biochem. Biotechnol. 2023, 195, 4832–4850. [Google Scholar] [PubMed]
- Cai, X.; Zheng, X.; Zhang, D.; Iqbal, W.; Liu, C.; Yang, B.; Zhao, X.; Lu, X.; Mao, Y. Microbial characterization of heavy metal resistant bacterial strains isolated from an electroplating wastewater treatment plant. Ecotoxicol. Environ. Saf. 2019, 181, 472–480. [Google Scholar]
- Mihhalevski, A.; Sarand, I.; Viiard, E.; Salumets, A.; Paalme, T. Growth characterization of individual rye sourdough bacteria by isothermal microcalorimetry. J. Appl. Microbiol. 2011, 110, 529–540. [Google Scholar] [PubMed]
- Pennekamp, A.; Punter, V.; Zbinden, R. Disk diffusion, agar dilution and the e-test for susceptibility testing of corynebacterium jeikeium. Clin. Microbiol. Infect. 1996, 2, 209–213. [Google Scholar] [PubMed]
- Liaqat, I.; Muhammad, N.; Ara, C.; Hanif, U.; Andleeb, S.; Arshad, M.; Aftab, M.N.; Raza, C.; Mubin, M. Bioremediation of heavy metals polluted environment and decolourization of black liquor using microbial biofilms. Mol. Biol. Rep. 2023, 50, 3985–3997. [Google Scholar] [PubMed]
- Di, Z.; Chaoyang, L.; Mengxi, Z.; Yunlin, Z.; Zhenggang, X.; Guiyan, Y. Curvularia coatesiae xk8, a potential bioadsorbent material for adsorbing cd(ii) and sb(iii) compound pollution: Characteristics and effects. Front. Microbiol. 2022, 12, 816312. [Google Scholar] [PubMed]


| Heavy Metals | MIC (mg/L) | MTC (mg/L) |
|---|---|---|
| Zn (II) | 600 | 2660 |
| Cu (II) | 1600 | 7680 |
| Ni (II) | 400 | 1210 |
| Cr (III) | 320 | 820 |
| Cd (II) | 100 | 400 |
| Pb (II) | 440 | 1640 |
| Mn (II) | 10 | 20 |
| Antibiotics | Test Result a |
|---|---|
| Lincomycin | R |
| Carbenicillin | S |
| Vancomycin | S |
| Norfloxacin | R |
| Kanamycin | R |
| Ofloxacin | R |
| Ampicillin | S |
| Penicillin | S |
| Polymyxin B | R |
| Ceftriaxone | R |
| Erythromycin | R |
| Chloramphenicol | S |
| Streptomycin | R |
| Clarithromycin | S |
| Rifampin | I |
| Clarithromycin | I |
| Neomycin | R |
| Tobramycin | R |
| Tetracycline | R |
| Gentamycin | R |
| Metal Ion | Initial Concentration (mg/L) | Final Concentration (mg/L) | Bioremoval Percentage | Bioremoval Capacity (mg/g Biomass) |
|---|---|---|---|---|
| Cu (II) | 1200 | 1000 | 16.67% | 350.2 ± 2.6 |
| 3420 | 3234 | 5.43% | 651.4 ± 1.7 |
| Antibiotics | Initial Concentration (mg/L) | Final Concentration (mg/L) | Bioremoval Percentage | Bioremoval Capacity (mg/g Biomass) |
|---|---|---|---|---|
| Lincomycin | 424 | 385 | 9.19% | 32.5 ± 0.4 |
| 199 | 187 | 6.03% | 8.1 ± 0.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, H.-S.; Liang, X.; Liu, J.-C.; Zhong, H.-Y.; Yang, Y.-H.; Guan, W.-P.; Du, Z.-J.; Ye, M.-Q. Antibiotic and Heavy Metal Co-Resistant Strain Isolated from Enrichment Culture of Marine Sediments, with Potential for Environmental Bioremediation Applications. Antibiotics 2023, 12, 1379. https://doi.org/10.3390/antibiotics12091379
Zhu H-S, Liang X, Liu J-C, Zhong H-Y, Yang Y-H, Guan W-P, Du Z-J, Ye M-Q. Antibiotic and Heavy Metal Co-Resistant Strain Isolated from Enrichment Culture of Marine Sediments, with Potential for Environmental Bioremediation Applications. Antibiotics. 2023; 12(9):1379. https://doi.org/10.3390/antibiotics12091379
Chicago/Turabian StyleZhu, Han-Sheng, Xiao Liang, Jun-Cheng Liu, Han-Yang Zhong, Yuan-Hang Yang, Wen-Peng Guan, Zong-Jun Du, and Meng-Qi Ye. 2023. "Antibiotic and Heavy Metal Co-Resistant Strain Isolated from Enrichment Culture of Marine Sediments, with Potential for Environmental Bioremediation Applications" Antibiotics 12, no. 9: 1379. https://doi.org/10.3390/antibiotics12091379






