Antiviral Activity of Anthranilamide Peptidomimetics against Herpes Simplex Virus 1 and a Coronavirus
Abstract
:1. Introduction
2. Results and Discussion
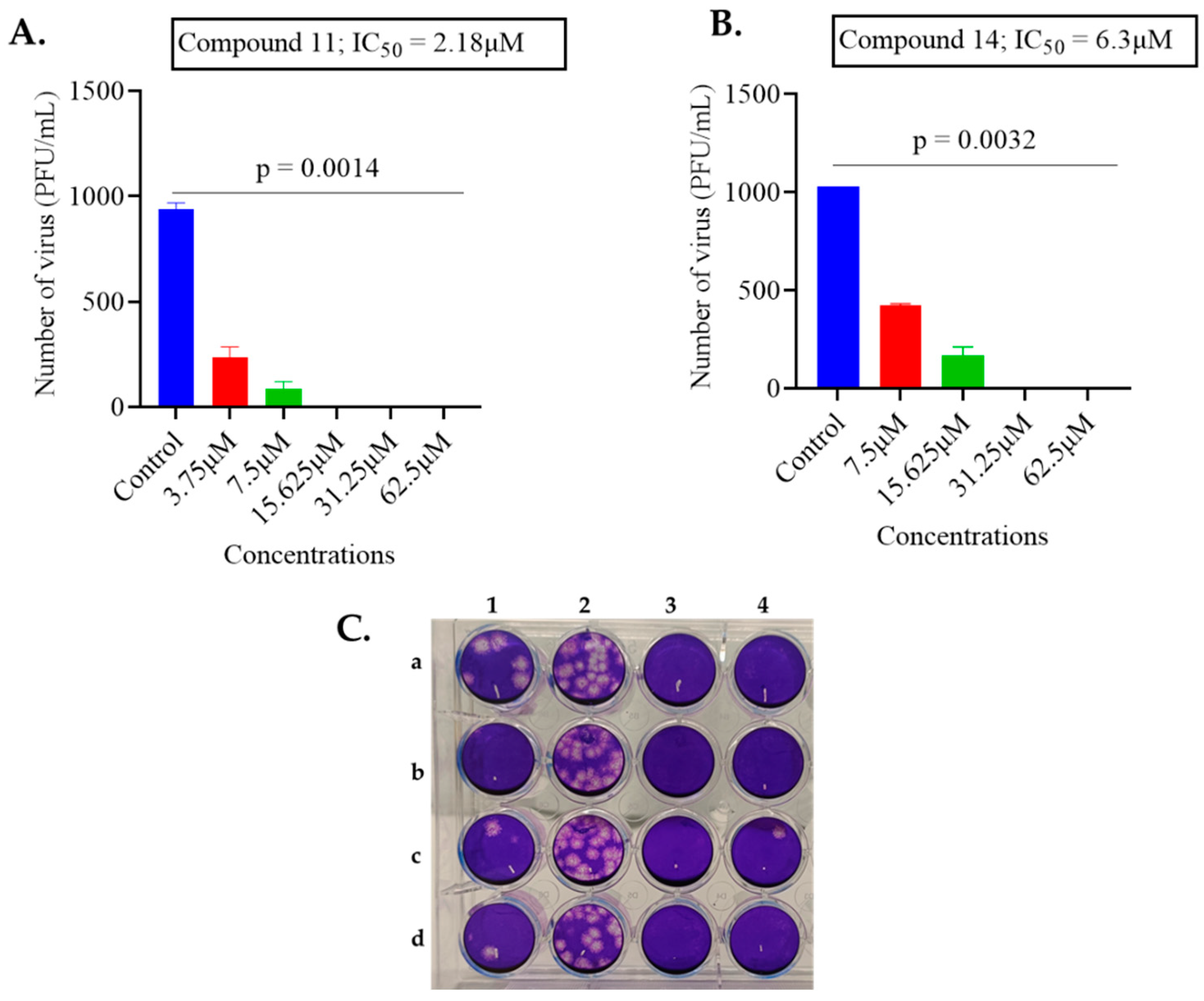
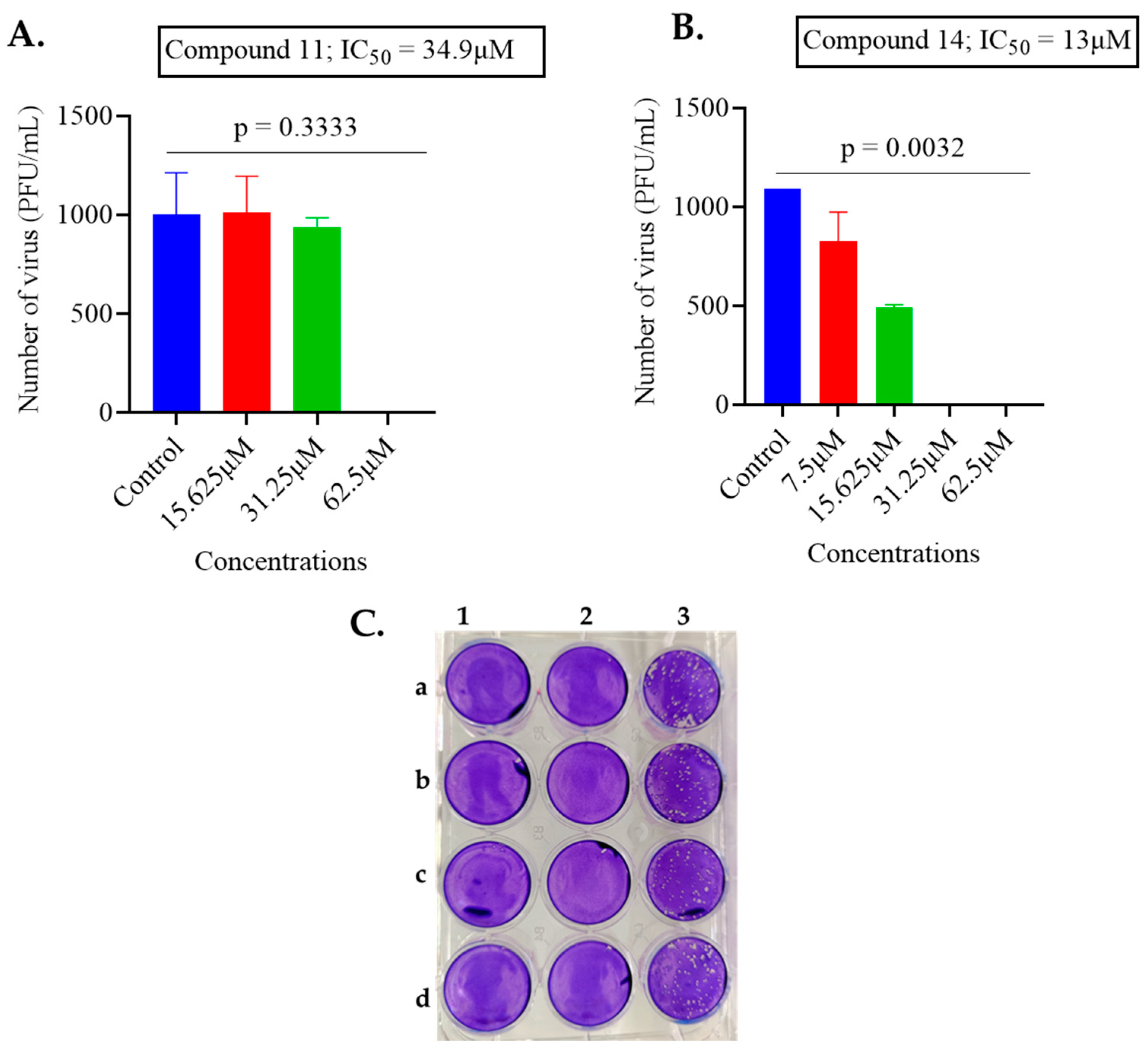
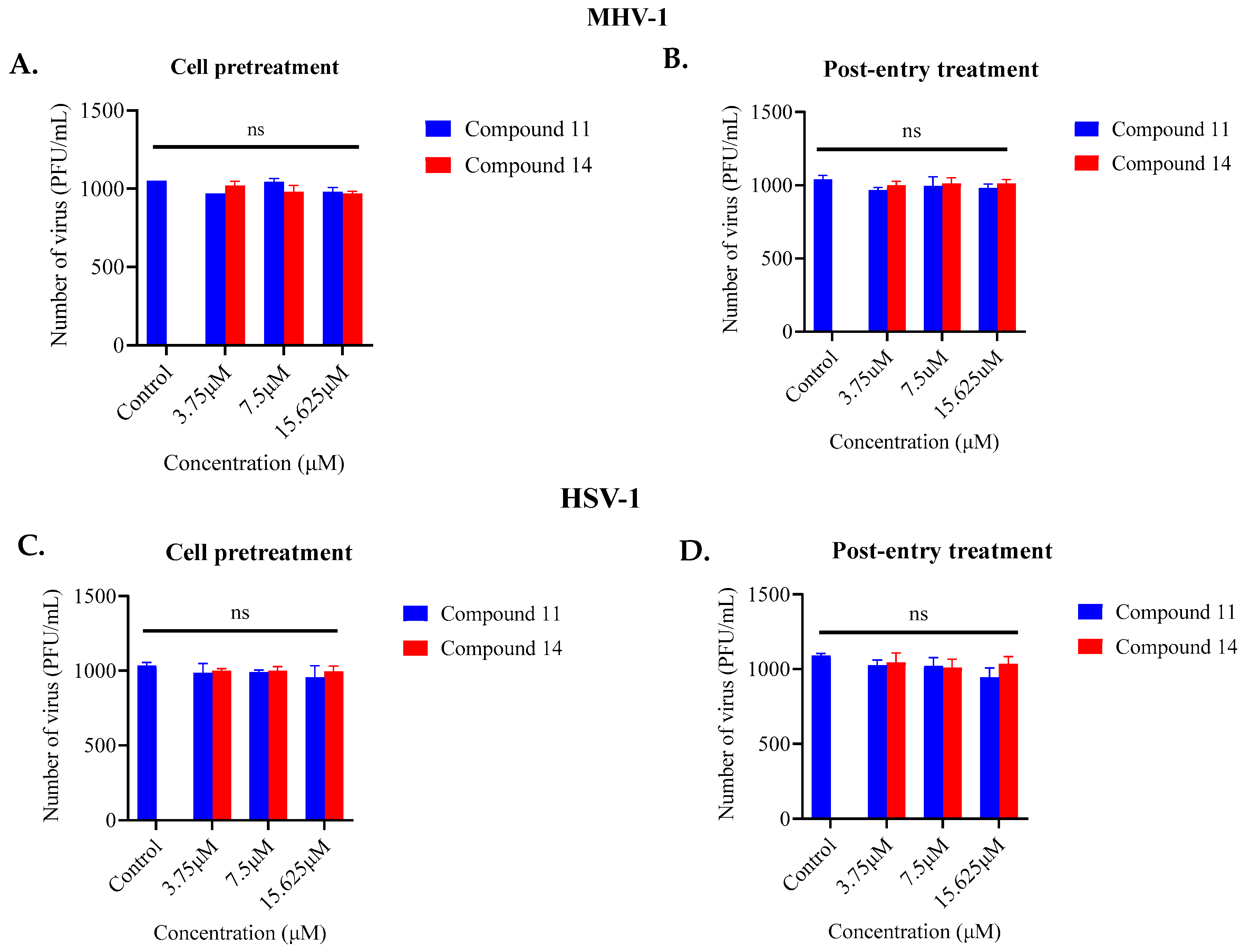
2.1. Screening of Anthranilamide Peptidomimetics against MHV-1 and HSV-1
2.2. TEM Images
2.3. Cytotoxic Activities of Active Mimetics
3. Materials and Methods
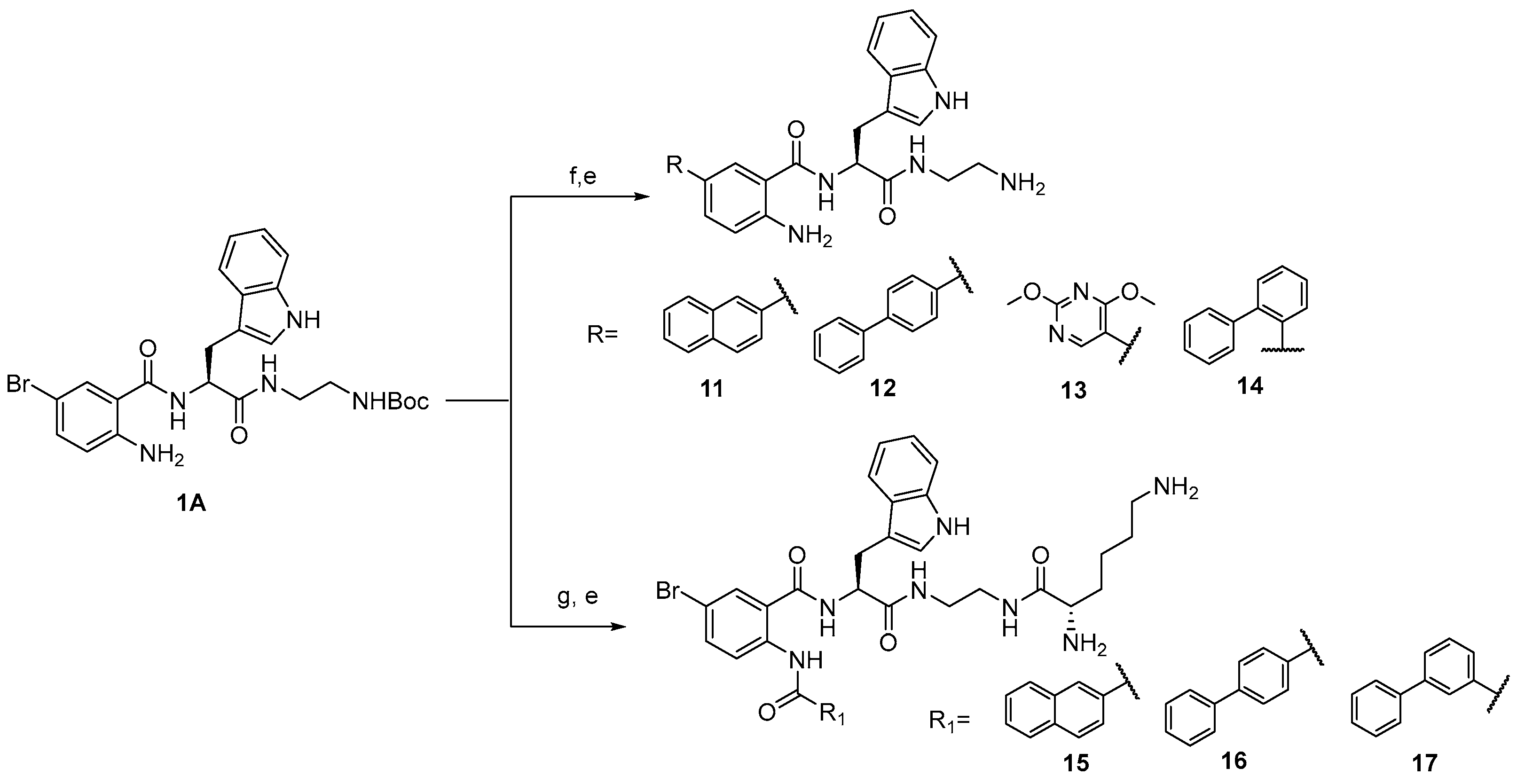
3.1. Chemistry
3.2. Analytical Data
3.3. Virus and Cell Culture
3.4. Antiviral Testing
3.5. Cytotoxicity Assay
3.6. Transmission Electron Microscopy
3.7. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, A.; Morling, J. Living with Endemic Covid-19. Public Health 2022, 205, 26. [Google Scholar] [CrossRef] [PubMed]
- Javanian, M.; Barary, M.; Ghebrehewet, S.; Koppolu, V.; Vasigala, V.; Ebrahimpour, S. A Brief Review of Influenza Virus Infection. J. Med. Virol. 2021, 93, 4638–4646. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Viejo-Borbolla, A. Pathogenesis and Virulence of Herpes Simplex Virus. Virulence 2021, 12, 2670–2702. [Google Scholar] [CrossRef] [PubMed]
- Gershon, A.A.; Breuer, J.; Cohen, J.I.; Cohrs, R.J.; Gershon, M.D.; Gilden, D.; Grose, C.; Hambleton, S.; Kennedy, P.G.; Oxman, M.N. Varicella Zoster Virus Infection. Nat. Rev. Dis. Primers 2015, 1, 15016. [Google Scholar] [CrossRef]
- Robilotti, E.; Deresinski, S.; Pinsky, B.A. Norovirus. Clin. Microbiol. Rev. 2015, 28, 134–164. [Google Scholar] [CrossRef]
- Li, X.; Chang, J.; Chen, S.; Wang, L.; Yau, T.O.; Zhao, Q.; Hong, Z.; Ruan, J.; Duan, G.; Gao, S. Genomic Feature Analysis of Betacoronavirus Provides Insights into Sars and Covid-19 Pandemics. Front. Microbiol. 2021, 12, 614494. [Google Scholar] [CrossRef]
- Bender, S.J.; Weiss, S.R. Pathogenesis of Murine Coronavirus in the Central Nervous System. J. Neuroimmune Pharmacol. 2010, 5, 336–354. [Google Scholar] [CrossRef]
- Featherstone, A.B.; Brown, A.C.; Chitlapilly Dass, S. Murine Hepatitis Virus, a Biosafety Level 2 Model for SARS-CoV-2, Can Remain Viable on Meat and Meat Packaging Materials for at Least 48 Hours. Microbiol. Spectr. 2022, 10, e0186222. [Google Scholar] [CrossRef]
- Casanova, L.M.; Jeon, S.; Rutala, W.A.; Weber, D.J.; Sobsey, M.D. Effects of Air Temperature and Relative Humidity on Coronavirus Survival on Surfaces. Appl. Environ. Microbiol. 2010, 76, 2712–2717. [Google Scholar] [CrossRef]
- Dellanno, C.; Vega, Q.; Boesenberg, D. The Antiviral Action of Common Household Disinfectants and Antiseptics against Murine Hepatitis Virus, a Potential Surrogate for Sars Coronavirus. Am. J. Infect. Control 2009, 37, 649–652. [Google Scholar] [CrossRef]
- Yasir, M.; Kumar Vijay, A.; Willcox, M. Antiviral Effect of Multipurpose Contact Lens Disinfecting Solutions against Coronavirus. Cont. Lens Anterior Eye 2021, 45, 101513. [Google Scholar] [CrossRef] [PubMed]
- Agostini, M.L.; Andres, E.L.; Sims, A.C.; Graham, R.L.; Sheahan, T.P.; Lu, X.; Smith, E.C.; Case, J.B.; Feng, J.Y.; Jordan, R.; et al. Coronavirus Susceptibility to the Antiviral Remdesivir (Gs-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. mBio 2018, 9, e00221-18. [Google Scholar] [CrossRef] [PubMed]
- Burrer, R.; Neuman, B.W.; Ting, J.P.; Stein, D.A.; Moulton, H.M.; Iversen, P.L.; Kuhn, P.; Buchmeier, M.J. Antiviral Effects of Antisense Morpholino Oligomers in Murine Coronavirus Infection Models. J. Virol. 2007, 81, 5637–5648. [Google Scholar] [CrossRef]
- Agostini, M.L.; Pruijssers, A.J.; Chappell, J.D.; Gribble, J.; Lu, X.; Andres, E.L.; Bluemling, G.R.; Lockwood, M.A.; Sheahan, T.P.; Sims, A.C.; et al. Small-Molecule Antiviral Β-D-N(4)-Hydroxycytidine Inhibits a Proofreading-Intact Coronavirus with a High Genetic Barrier to Resistance. J. Virol. 2019, 93, e01348-19. [Google Scholar] [CrossRef]
- Zhou, Q.; Luo, Y.; Zhu, Y.; Chen, Q.; Qiu, J.; Cong, F.; Li, Y.; Zhang, X. Nonsteroidal Anti-Inflammatory Drugs (Nsaids) and Nucleotide Analog Gs-441524 Conjugates with Potent in Vivo Efficacy against Coronaviruses. Eur. J. Med. Chem. 2023, 249, 115113. [Google Scholar] [CrossRef] [PubMed]
- Tu, E.C.; Hsu, W.L.; Tzen, J.T.C. Strictinin, a Major Ingredient in Yunnan Kucha Tea Possessing Inhibitory Activity on the Infection of Mouse Hepatitis Virus to Mouse L Cells. Molecules 2023, 28, 1080. [Google Scholar] [CrossRef] [PubMed]
- Graff Reis, J.; Dai Prá, I.; Michelon, W.; Viancelli, A.; Piedrahita Marquez, D.G.; Schmitz, C.; Maraschin, M.; Moura, S.; Thaís Silva, I.; de Oliveira Costa, G.; et al. Characterization of Planktochlorella Nurekis Extracts and Virucidal Activity against a Coronavirus Model, the Murine Coronavirus 3. Int. J. Environ. Res. Public Health 2022, 19, 15823. [Google Scholar] [CrossRef]
- Sainz, B., Jr.; Mossel, E.C.; Gallaher, W.R.; Wimley, W.C.; Peters, C.J.; Wilson, R.B.; Garry, R.F. Inhibition of Severe Acute Respiratory Syndrome-Associated Coronavirus (Sars-Cov) Infectivity by Peptides Analogous to the Viral Spike Protein. Virus Res. 2006, 120, 146–155. [Google Scholar] [CrossRef]
- Kim, H.Y.; Eo, E.Y.; Park, H.; Kim, Y.C.; Park, S.; Shin, H.J.; Kim, K. Medicinal Herbal Extracts of Sophorae Radix, Acanthopanacis Cortex, Sanguisorbae Radix and Torilis Fructus Inhibit Coronavirus Replication in Vitro. Antivir. Ther. 2010, 15, 697–709. [Google Scholar] [CrossRef]
- Parums, D.V. Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Patients. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2022, 28, e935952-1–e935952-4. [Google Scholar] [CrossRef]
- Whitley, R.J.; Roizman, B. Herpes Simplex Virus Infections. Lancet 2001, 357, 1513–1518. [Google Scholar] [CrossRef] [PubMed]
- Looker, K.J.; Magaret, A.S.; May, M.T.; Turner, K.M.; Vickerman, P.; Gottlieb, S.L.; Newman, L.M. Global and Regional Estimates of Prevalent and Incident Herpes Simplex Virus Type 1 Infections in 2012. PLoS ONE 2015, 10, e0140765. [Google Scholar] [CrossRef] [PubMed]
- Kłysik, K.; Pietraszek, A.; Karewicz, A.; Nowakowska, M. Acyclovir in the Treatment of Herpes Viruses—A Review. Curr. Med. Chem. 2020, 27, 4118–4137. [Google Scholar] [CrossRef] [PubMed]
- Schalkwijk, H.H.; Snoeck, R.; Andrei, G. Acyclovir Resistance in Herpes Simplex Viruses: Prevalence and Therapeutic Alternatives. Biochem. Pharmacol. 2022, 206, 115322. [Google Scholar] [CrossRef]
- Kuo, J.Y.; Yeh, C.S.; Wang, S.M.; Chen, S.H.; Wang, J.R.; Chen, T.Y.; Tsai, H.P. Acyclovir-Resistant Hsv-1 Isolates among Immunocompromised Patients in Southern Taiwan: Low Prevalence and Novel Mutations. J. Med. Virol. 2023, 95, e28985. [Google Scholar] [CrossRef]
- Mousavi Maleki, M.S.; Sardari, S.; Ghandehari Alavijeh, A.; Madanchi, H. Recent Patents and Fda-Approved Drugs Based on Antiviral Peptides and Other Peptide-Related Antivirals. Int. J. Pept. Res. Ther. 2023, 29, 5. [Google Scholar] [CrossRef]
- Urmi, U.L.; Vijay, A.K.; Kuppusamy, R.; Islam, S.; Willcox, M.D.P. A Review of the Antiviral Activity of Cationic Antimicrobial Peptides. Peptides 2023, 166, 171024. [Google Scholar] [CrossRef]
- Chowdhury, A.S.; Reehl, S.M.; Kehn-Hall, K.; Bishop, B.; Webb-Robertson, B.M. Better Understanding and Prediction of Antiviral Peptides through Primary and Secondary Structure Feature Importance. Sci. Rep. 2020, 10, 19260. [Google Scholar] [CrossRef]
- Bradshaw, J. Cationic Antimicrobial Peptides: Issues for Potential Clinical Use. BioDrugs 2003, 17, 233–240. [Google Scholar] [CrossRef]
- Gurwitz, D. Peptide Mimetics: Fast-Forward Look. Drug Dev. Res. 2017, 78, 231–235. [Google Scholar] [CrossRef]
- Li Petri, G.; Di Martino, S.; De Rosa, M. Peptidomimetics: An Overview of Recent Medicinal Chemistry Efforts toward the Discovery of Novel Small Molecule Inhibitors. J. Med. Chem. 2022, 65, 7438–7475. [Google Scholar] [CrossRef]
- Ding, D.; Xu, S.; da Silva-Júnior, E.F.; Liu, X.; Zhan, P. Medicinal Chemistry Insights into Antiviral Peptidomimetics. Drug Discov. Today 2023, 28, 103468. [Google Scholar] [CrossRef]
- Lenci, E.; Trabocchi, A. Peptidomimetic Toolbox for Drug Discovery. Chem. Soc. Rev. 2020, 49, 3262–3277. [Google Scholar] [CrossRef] [PubMed]
- Nitsche, C.; Zhang, L.; Weigel, L.F.; Schilz, J.; Graf, D.; Bartenschlager, R.; Hilgenfeld, R.; Klein, C.D. Peptide–Boronic Acid Inhibitors of Flaviviral Proteases: Medicinal Chemistry and Structural Biology. J. Med. Chem. 2017, 60, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Osswald, H.L.; Prato, G. Recent Progress in the Development of Hiv-1 Protease Inhibitors for the Treatment of Hiv/Aids. J. Med. Chem. 2016, 59, 5172–5208. [Google Scholar] [CrossRef] [PubMed]
- Su, B.; Yao, C.; Zhao, Q.-X.; Cai, W.-P.; Wang, M.; Lu, H.-Z.; Mu, T.-T.; Chen, Y.-Y.; Liu, L.; Wang, H. Long-Acting Hiv Fusion Inhibitor Albuvirtide Combined with Ritonavir-Boosted Lopinavir for Hiv-1-Infected Patients after Failing the First-Line Antiretroviral Therapy: 48-Week Randomized, Controlled, Phase 3 Non-Inferiority Talent Study. J. Infect. 2022, 85, 334–363. [Google Scholar] [CrossRef]
- Awahara, C.; Oku, D.; Furuta, S.; Kobayashi, K.; Teruya, K.; Akaji, K.; Hattori, Y. The Effects of Side-Chain Configurations of a Retro–Inverso-Type Inhibitor on the Human T-Cell Leukemia Virus (Htlv)-1 Protease. Molecules 2022, 27, 1646. [Google Scholar] [CrossRef]
- Rosenquist, Å.; Samuelsson, B.; Johansson, P.-O.; Cummings, M.D.; Lenz, O.; Raboisson, P.; Simmen, K.; Vendeville, S.; de Kock, H.; Nilsson, M. Discovery and Development of Simeprevir (Tmc435), a Hcv Ns3/4a Protease Inhibitor. J. Med. Chem. 2014, 57, 1673–1693. [Google Scholar] [CrossRef]
- Diamond, G.; Molchanova, N.; Herlan, C.; Fortkort, J.A.; Lin, J.S.; Figgins, E.; Bopp, N.; Ryan, L.K.; Chung, D.; Adcock, R.S.; et al. Potent Antiviral Activity against Hsv-1 and SARS-CoV-2 by Antimicrobial Peptoids. Pharmaceuticals 2021, 14, 304. [Google Scholar] [CrossRef]
- Kuppusamy, R.; Yasir, M.; Yu, T.T.; Voli, F.; Vittorio, O.; Miller, M.J.; Lewis, P.; Black, D.S.; Willcox, M.; Kumar, N. Tuning the Anthranilamide Peptidomimetic Design to Selectively Target Planktonic Bacteria and Biofilm. Antibiotics 2023, 12, 585. [Google Scholar] [CrossRef]
- Barraza, S.J.; Delekta, P.C.; Sindac, J.A.; Dobry, C.J.; Xiang, J.; Keep, R.F.; Miller, D.J.; Larsen, S.D. Discovery of Anthranilamides as a Novel Class of Inhibitors of Neurotropic Alphavirus Replication. Bioorg. Med. Chem. 2015, 23, 1569–1587. [Google Scholar] [CrossRef] [PubMed]
- Chianese, A.; Zannella, C.; Monti, A.; De Filippis, A.; Doti, N.; Franci, G.; Galdiero, M. The Broad-Spectrum Antiviral Potential of the Amphibian Peptide Ar-23. Int. J. Mol. Sci. 2022, 23, 883. [Google Scholar] [CrossRef] [PubMed]
- Zannella, C.; Chianese, A.; Palomba, L.; Marcocci, M.E.; Bellavita, R.; Merlino, F.; Grieco, P.; Folliero, V.; De Filippis, A.; Mangoni, M.; et al. Broad-Spectrum Antiviral Activity of the Amphibian Antimicrobial Peptide Temporin L and Its Analogs. Int. J. Mol. Sci. 2022, 23, 2060. [Google Scholar] [CrossRef]
- Tooze, S.A.; Tooze, J.; Warren, G. Site of Addition of N-Acetyl-Galactosamine to the E1 Glycoprotein of Mouse Hepatitis Virus-A59. J. Cell Biol. 1988, 106, 1475–1487. [Google Scholar] [CrossRef]
- Perrier, A.; Bonnin, A.; Desmarets, L.; Danneels, A.; Goffard, A.; Rouille, Y.; Dubuisson, J.; Belouzard, S. The C-Terminal Domain of the Mers Coronavirus M Protein Contains a Trans-Golgi Network Localization Signal. J. Biol. Chem. 2019, 294, 14406–14421. [Google Scholar] [CrossRef] [PubMed]
- McBride, C.E.; Li, J.; Machamer, C.E. The Cytoplasmic Tail of the Severe Acute Respiratory Syndrome Coronavirus Spike Protein Contains a Novel Endoplasmic Reticulum Retrieval Signal That Binds Copi and Promotes Interaction with Membrane Protein. J. Virol. 2007, 81, 2418–2428. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Wilson, D.W. Hsv-1 Cytoplasmic Envelopment and Egress. Int. J. Mol. Sci. 2020, 21, 5969. [Google Scholar] [CrossRef]
- Ivanova, P.T.; Myers, D.S.; Milne, S.B.; McClaren, J.L.; Thomas, P.G.; Brown, H.A. Lipid Composition of the Viral Envelope of Three Strains of Influenza Virus Not All Viruses Are Created Equal. ACS Infect. Dis. 2015, 1, 435–442. [Google Scholar] [CrossRef]
- Chazal, N.; Gerlier, D. Virus Entry, Assembly, Budding, and Membrane Rafts. Microbiol. Mol. Biol. Rev. 2003, 67, 226–237. [Google Scholar] [CrossRef]
- Kirui, J.; Abidine, Y.; Lenman, A.; Islam, K.; Gwon, Y.-D.; Lasswitz, L.; Evander, M.; Bally, M.; Gerold, G. The Phosphatidylserine Receptor Tim-1 Enhances Authentic Chikungunya Virus Cell Entry. Cells 2021, 10, 1828. [Google Scholar] [CrossRef]
- Tate, P.M.; Mastrodomenico, V.; Cunha, C.; McClure, J.; Barron, A.E.; Diamond, G.; Mounce, B.C.; Kirshenbaum, K. Peptidomimetic Oligomers Targeting Membrane Phosphatidylserine Exhibit Broad Antiviral Activity. ACS Infect. Dis. 2023, 9, 1508–1522. [Google Scholar] [CrossRef] [PubMed]
- Lorin, C.; Saidi, H.; Belaid, A.; Zairi, A.; Baleux, F.; Hocini, H.; Bélec, L.; Hani, K.; Tangy, F. The Antimicrobial Peptide Dermaseptin S4 Inhibits Hiv-1 Infectivity in Vitro. Virology 2005, 334, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Kuppusamy, R.; Yasir, M.; Yee, E.; Willcox, M.; Black, D.S.; Kumar, N. Guanidine Functionalized Anthranilamides as Effective Antibacterials with Biofilm Disruption Activity. Org. Biomol. Chem. 2018, 16, 5871–5888. [Google Scholar] [CrossRef] [PubMed]
- Savoia, D.; Donalisio, M.; Civra, A.; Salvadori, S.; Guerrini, R. In Vitro Activity of Dermaseptin S1 Derivatives against Genital Pathogens. Apmis 2010, 118, 674–680. [Google Scholar] [CrossRef]
- Tang, W.H.; Wang, C.F.; Liao, Y.D. Fetal Bovine Serum Albumin Inhibits Antimicrobial Peptide Activity and Binds Drug Only in Complex with A1-Antitrypsin. Sci. Rep. 2021, 11, 1267. [Google Scholar] [CrossRef]
- Zeng, Z.; Zhang, Q.; Hong, W.; Xie, Y.; Liu, Y.; Li, W.; Wu, Y.; Cao, Z. A Scorpion Defensin Bmkdfsin4 Inhibits Hepatitis B Virus Replication in Vitro. Toxins 2016, 8, 124. [Google Scholar] [CrossRef]
- Xiao, C.; Chen, X.; Xie, Q.; Li, G.; Xiao, H.; Song, J.; Han, H. Virus Identification in Electron Microscopy Images by Residual Mixed Attention Network. Comput. Methods Programs Biomed. 2021, 198, 105766. [Google Scholar] [CrossRef]
- Zannella, C.; Chianese, A.; Annunziata, G.; Ambrosino, A.; De Filippis, A.; Tenore, G.C.; Novellino, E.; Stornaiuolo, M.; Galdiero, M. Antiherpetic Activity of Taurisolo(®), a Grape Pomace Polyphenolic Extract. Microorganisms 2023, 11, 1346. [Google Scholar] [CrossRef]
- Liu, P.; Zhong, L.; Xiao, J.; Hu, Y.; Liu, T.; Ren, Z.; Wang, Y.; Zheng, K. Ethanol Extract from Artemisia Argyi Leaves Inhibits Hsv-1 Infection by Destroying the Viral Envelope. Virol. J. 2023, 20, 8. [Google Scholar] [CrossRef]



| Compound Number | MHV-1 IC50 (µM) | HSV-1 IC50 (µM) |
|---|---|---|
| 1 | >62.5 | >62.5 |
| 2 | >62.5 | >62.5 |
| 3 | >62.5 | >62.5 |
| 4 | >62.5 | >62.5 |
| 5 | >62.5 | >62.5 |
| 6 | >62.5 | >62.5 |
| 7 | >62.5 | 32.7 |
| 8 | >62.5 | >62.5 |
| 9 | >62.5 | 14.8 |
| 10 | >62.5 | >62.5 |
| 11 | 2.38 | 34.9 |
| 12 | >62.5 | >62.5 |
| 13 | >62.5 | >62.5 |
| 14 | 6.3 | 13 |
| 15 | 11.09 | >62.5 |
| 16 | 22.5 | >62.5 |
| 17 | 11.8 | >62.5 |
| Compound Number | MHV-1 (A9 Cells) | HSV-1 (Vero Cells) | ||||
|---|---|---|---|---|---|---|
| IC50 (µM) | CC50 (µM) | Therapeutic Index | IC50 (µM) | CC50 (µM) | Therapeutic Index | |
| 7 | >62.5 | - | - | 32.76 | 127 | 3.87 |
| 9 | >62.5 | - | - | 14.88 | 45.02 | 3.03 |
| 11 | 2.18 | 20.52 | 9.41 | 34.9 | 16.69 | 0.48 |
| 14 | 6.3 | 10.20 | 1.6 | 13 | 15.18 | 1.16 |
| 15 | 11.09 | 62.68 | 5.65 | >62.5 | - | - |
| 16 | 22.56 | 81.64 | 3.62 | >62.5 | - | - |
| 17 | 11.79 | 114.5 | 9.71 | >62.5 | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urmi, U.L.; Attard, S.; Vijay, A.K.; Willcox, M.D.P.; Kumar, N.; Islam, S.; Kuppusamy, R. Antiviral Activity of Anthranilamide Peptidomimetics against Herpes Simplex Virus 1 and a Coronavirus. Antibiotics 2023, 12, 1436. https://doi.org/10.3390/antibiotics12091436
Urmi UL, Attard S, Vijay AK, Willcox MDP, Kumar N, Islam S, Kuppusamy R. Antiviral Activity of Anthranilamide Peptidomimetics against Herpes Simplex Virus 1 and a Coronavirus. Antibiotics. 2023; 12(9):1436. https://doi.org/10.3390/antibiotics12091436
Chicago/Turabian StyleUrmi, Umme Laila, Samuel Attard, Ajay Kumar Vijay, Mark D. P. Willcox, Naresh Kumar, Salequl Islam, and Rajesh Kuppusamy. 2023. "Antiviral Activity of Anthranilamide Peptidomimetics against Herpes Simplex Virus 1 and a Coronavirus" Antibiotics 12, no. 9: 1436. https://doi.org/10.3390/antibiotics12091436
APA StyleUrmi, U. L., Attard, S., Vijay, A. K., Willcox, M. D. P., Kumar, N., Islam, S., & Kuppusamy, R. (2023). Antiviral Activity of Anthranilamide Peptidomimetics against Herpes Simplex Virus 1 and a Coronavirus. Antibiotics, 12(9), 1436. https://doi.org/10.3390/antibiotics12091436









