Detection, Identification and Diagnostic Characterization of the Staphylococcal Small Colony-Variant (SCV) Phenotype
Abstract
1. Introduction
2. Why Should We Specifically Look for Staphylococcal SCVs When Appropriate?
3. When Should We Specifically Consider Staphylococcal SCVs in Diagnostics?
4. What Should Be Noted in the Preanalytics of SCV Detection?
5. What Should Be Considered during Cultivation of Staphylococcal SCVs?
6. How Can the Species of SCVs Be Determined?
7. How Can the SCV Phenotype Be Objectified and Characterized?
8. Is It Necessary to Test SCVs Separately for Resistance Determination and How Can This Be Performed?
9. How Should an SCV Finding Be Reported, Commented on and Managed?
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Proctor, R.A.; von Eiff, C.; Kahl, B.C.; Becker, K.; McNamara, P.; Herrmann, M.; Peters, G. Small colony variants: A pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 2006, 4, 295–305. [Google Scholar] [CrossRef]
- Kahl, B.C.; Becker, K.; Löffler, B. Clinical Significance and Pathogenesis of Staphylococcal Small Colony Variants in Persistent Infections. Clin. Microbiol. Rev. 2016, 29, 401–427. [Google Scholar] [CrossRef]
- Jiang, B.; You, B.; Tan, L.; Yu, S.; Li, H.; Bai, G.; Li, S.; Rao, X.; Xie, Z.; Shi, X.; et al. Clinical Staphylococcus argenteus Develops to Small Colony Variants to Promote Persistent Infection. Front. Microbiol. 2018, 9, 1347. [Google Scholar] [CrossRef]
- von Eiff, C.; Vaudaux, P.; Kahl, B.C.; Lew, D.; Emler, S.; Schmidt, A.; Peters, G.; Proctor, R.A. Bloodstream infections caused by small-colony variants of coagulase-negative staphylococci following pacemaker implantation. Clin. Infect. Dis. 1999, 29, 932–934. [Google Scholar] [CrossRef]
- Baddour, L.M.; Barker, L.P.; Christensen, G.D.; Parisi, J.T.; Simpson, W.A. Phenotypic variation of Staphylococcus epidermidis in infection of transvenous endocardial pacemaker electrodes. J. Clin. Microbiol. 1990, 28, 676–679. [Google Scholar] [CrossRef]
- Bogut, A.; Niedźwiadek, J.; Kozioł-Montewka, M.; Strzelec-Nowak, D.; Blacha, J.; Mazurkiewicz, T.; Marczyński, W.; Plewik, D. Characterization of Staphylococcus epidermidis and Staphyloccocus warneri small-colony variants associated with prosthetic-joint infections. J. Med. Microbiol. 2014, 63, 176–185. [Google Scholar] [CrossRef][Green Version]
- Seifert, H.; Oltmanns, D.; Becker, K.; Wisplinghoff, H.; von Eiff, C. Staphylococcus lugdunensis pacemaker-related infection. Emerg. Infect. Dis. 2005, 11, 1283–1286. [Google Scholar] [CrossRef] [PubMed]
- Argemi, X.; Dahyot, S.; Lebeurre, J.; Hansmann, Y.; Ronde Oustau, C.; Prévost, G. Staphylococcus lugdunensis small colony variant conversion resulting in chronic prosthetic joint infection. Med. Mal. Infect. 2017, 47, 498–501. [Google Scholar] [CrossRef] [PubMed]
- Savini, V.; Carretto, E.; Polilli, E.; Marrollo, R.; Santarone, S.; Fazii, P.; D’Antonio, D.; Rossano, A.; Perreten, V. Small colony variant of methicillin-resistant Staphylococcus pseudintermedius ST71 presenting as a sticky phenotype. J. Clin. Microbiol. 2014, 52, 1225–1227. [Google Scholar] [CrossRef] [PubMed]
- Hale, J.H. Studies on Staphylococcus mutation; characteristics of the “G” (gonidial) variant and factors concerned in its production. Br. J. Exp. Pathol. 1947, 28, 202–210. [Google Scholar]
- Goudie, J.G.; Goudie, R.B. Recurrent infections by a stable dwarf-colony variant of Staphylococcus aureus. J. Clin. Pathol. 1955, 8, 284–287. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Swingle, E.L. Studies on Small Colony Variants of Staphylococcus aureus. J. Bacteriol. 1935, 29, 467–489. [Google Scholar] [CrossRef]
- Tuchscherr, L.; Heitmann, V.; Hussain, M.; Viemann, D.; Roth, J.; von Eiff, C.; Peters, G.; Becker, K.; Löffler, B. Staphylococcus aureus small-colony variants are adapted phenotypes for intracellular persistence. J. Infect. Dis. 2010, 202, 1031–1040. [Google Scholar] [CrossRef]
- Tuchscherr, L.; Medina, E.; Hussain, M.; Volker, W.; Heitmann, V.; Niemann, S.; Holzinger, D.; Roth, J.; Proctor, R.A.; Becker, K.; et al. Staphylococcus aureus phenotype switching: An effective bacterial strategy to escape host immune response and establish a chronic infection. EMBO Mol. Med. 2011, 3, 129–141. [Google Scholar] [CrossRef]
- McNamara, P.J.; Proctor, R.A. Staphylococcus aureus small colony variants, electron transport and persistent infections. Int. J. Antimicrob. Agents 2000, 14, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, I.; Kriegeskorte, A.; Fischer, A.; Deiwick, S.; Theimann, N.; Proctor, R.A.; Peters, G.; Herrmann, M.; Kahl, B.C. In vivo mutations of thymidylate synthase (encoded by thyA) are responsible for thymidine dependency in clinical small-colony variants of Staphylococcus aureus. J. Bacteriol. 2008, 190, 834–842. [Google Scholar] [CrossRef] [PubMed]
- Schleimer, N.; Kaspar, U.; Drescher, M.; Seggewiß, J.; von Eiff, C.; Proctor, R.A.; Peters, G.; Kriegeskorte, A.; Becker, K. The energy-coupling factor transporter module EcfAA’T, a novel candidate for the genetic basis of fatty acid-auxotrophic small-colony variants of Staphylococcus aureus. Front. Microbiol. 2018, 9, 1863. [Google Scholar] [CrossRef]
- Tuchscherr, L.; Löffler, B. Staphylococcus aureus dynamically adapts global regulators and virulence factor expression in the course from acute to chronic infection. Curr. Genet. 2016, 62, 15–17. [Google Scholar] [CrossRef]
- Proctor, R.A.; Peters, G. Small colony variants in staphylococcal infections: Diagnostic and therapeutic implications. Clin. Infect. Dis. 1998, 27, 419–422. [Google Scholar] [CrossRef]
- Proctor, R.A.; Balwit, J.M.; Vesga, O. Variant subpopulations of Staphylococcus aureus as cause of persistent and recurrent infections. Infect. Agents Dis. 1994, 3, 302–312. [Google Scholar]
- Garcia, L.G.; Lemaire, S.; Kahl, B.C.; Becker, K.; Proctor, R.A.; Denis, O.; Tulkens, P.M.; Van Bambeke, F. Antibiotic activity against small-colony variants of Staphylococcus aureus: Review of in vitro, animal and clinical data. J. Antimicrob. Chemother. 2013, 68, 1455–1464. [Google Scholar] [CrossRef]
- Seaman, P.F.; Ochs, D.; Day, M.J. Small-colony variants: A novel mechanism for triclosan resistance in methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 2007, 59, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Kriegeskorte, A.; König, S.; Sander, G.; Pirkl, A.; Mahabir, E.; Proctor, R.A.; von Eiff, C.; Peters, G.; Becker, K. Small colony variants of Staphylococcus aureus reveal distinct protein profiles. Proteomics 2011, 11, 2476–2490. [Google Scholar] [CrossRef]
- Vesga, O.; Groeschel, M.C.; Otten, M.F.; Brar, D.W.; Vann, J.M.; Proctor, R.A. Staphylococcus aureus small colony variants are induced by the endothelial cell intracellular milieu. J. Infect. Dis. 1996, 173, 739–742. [Google Scholar] [CrossRef] [PubMed]
- von Eiff, C.; Bettin, D.; Proctor, R.A.; Rolauffs, B.; Lindner, N.; Winkelmann, W.; Peters, G. Recovery of small colony variants of Staphylococcus aureus following gentamicin bead placement for osteomyelitis. Clin. Infect. Dis. 1997, 25, 1250–1251. [Google Scholar] [CrossRef]
- Rolauffs, B.; Bernhardt, T.M.; von Eiff, C.; Hart, M.L.; Bettin, D. Osteopetrosis, femoral fracture, and chronic osteomyelitis caused by Staphylococcus aureus small colony variants (SCV) treated by girdlestone resection—6-year follow-up. Arch. Orthop. Trauma Surg. 2002, 122, 547–550. [Google Scholar] [CrossRef]
- Proctor, R.A.; van Langevelde, P.; Kristjansson, M.; Maslow, J.N.; Arbeit, R.D. Persistent and relapsing infections associated with small-colony variants of Staphylococcus aureus. Clin. Infect. Dis. 1995, 20, 95–102. [Google Scholar] [CrossRef]
- von Eiff, C.; Becker, K. Small-colony variants (SCVs) of staphylococci: A role in foreign body-associated infections. Int. J. Artif. Organs 2007, 30, 778–785. [Google Scholar] [CrossRef]
- von Eiff, C.; Becker, K.; Metze, D.; Lubritz, G.; Hockmann, J.; Schwarz, T.; Peters, G. Intracellular persistence of Staphylococcus aureus small-colony variants within keratinocytes: A cause for antibiotic treatment failure in a patient with darier’s disease. Clin. Infect. Dis. 2001, 32, 1643–1647. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.T.; Tsai, J.C.; Yamamoto, T.; Chen, H.J.; Hung, W.C.; Hsueh, P.R.; Teng, L.J. Emergence of a small colony variant of vancomycin-intermediate Staphylococcus aureus in a patient with septic arthritis during long-term treatment with daptomycin. J. Antimicrob. Chemother. 2016, 71, 1807–1814. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, X.C.; Nielsen, F.T.; Kurtzhals, J.A.; Moser, C.; Boye, K.; Christensen, J.J.; Johansen, U.R.; Westh, H. Management of recurrent pacemaker-related bacteraemia with small colony variant Staphylococcus aureus in a haemodialysis patient. BMJ Case Rep. 2009, 2009, bcr0520091910. [Google Scholar] [CrossRef]
- Sendi, P.; Rohrbach, M.; Graber, P.; Frei, R.; Ochsner, P.E.; Zimmerli, W. Staphylococcus aureus small colony variants in prosthetic joint infection. Clin. Infect. Dis. 2006, 43, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Mashayamombe, M.; Walsh, T.P.; Kuang, B.K.P.; Pena, G.N.; Vreugde, S.; Cooksley, C.; Carda-Diéguez, M.; Mira, A.; Jesudason, D.; et al. The bacteriology of diabetic foot ulcers and infections and incidence of Staphylococcus aureus Small Colony Variants. J. Med. Microbiol. 2023, 72, 001716. [Google Scholar] [CrossRef] [PubMed]
- Kahl, B.; Herrmann, M.; Everding, A.S.; Koch, H.G.; Becker, K.; Harms, E.; Proctor, R.A.; Peters, G. Persistent infection with small colony variant strains of Staphylococcus aureus in patients with cystic fibrosis. J. Infect. Dis. 1998, 177, 1023–1029. [Google Scholar] [CrossRef]
- Branger, C.; Gardye, C.; Lambert-Zechovsky, N. Persistence of Staphylococcus aureus strains among cystic fibrosis patients over extended periods of time. J. Med. Microbiol. 1996, 45, 294–301. [Google Scholar] [CrossRef]
- Hirschhausen, N.; Block, D.; Bianconi, I.; Bragonzi, A.; Birtel, J.; Lee, J.C.; Dübbers, A.; Küster, P.; Kahl, J.; Peters, G.; et al. Extended Staphylococcus aureus persistence in cystic fibrosis is associated with bacterial adaptation. Int. J. Med. Microbiol. 2013, 303, 685–692. [Google Scholar] [CrossRef] [PubMed]
- Ong, T.; Ramsey, B.W. Cystic Fibrosis: A Review. JAMA 2023, 329, 1859–1871. [Google Scholar] [CrossRef] [PubMed]
- Cramer, N.; Wiehlmann, L.; Ciofu, O.; Tamm, S.; Hoiby, N.; Tümmler, B. Molecular epidemiology of chronic Pseudomonas aeruginosa airway infections in cystic fibrosis. PLoS ONE 2012, 7, e50731. [Google Scholar] [CrossRef] [PubMed]
- Hogardt, M.; Heesemann, J. Microevolution of Pseudomonas aeruginosa to a chronic pathogen of the cystic fibrosis lung. Between Pathog. Commensalism 2013, 358, 91–118. [Google Scholar] [CrossRef]
- Kahl, B.C.; Duebbers, A.; Lubritz, G.; Haeberle, J.; Koch, H.G.; Ritzerfeld, B.; Reilly, M.; Harms, E.; Proctor, R.A.; Herrmann, M.; et al. Population dynamics of persistent Staphylococcus aureus isolated from the airways of cystic fibrosis patients during a 6-year prospective study. J. Clin. Microbiol. 2003, 41, 4424–4427. [Google Scholar] [CrossRef]
- Besier, S.; Smaczny, C.; von Mallinckrodt, C.; Krahl, A.; Ackermann, H.; Brade, V.; Wichelhaus, T.A. Prevalence and clinical significance of Staphylococcus aureus small-colony variants in cystic fibrosis lung disease. J. Clin. Microbiol. 2007, 45, 168–172. [Google Scholar] [CrossRef]
- Wolter, D.J.; Emerson, J.C.; McNamara, S.; Buccat, A.M.; Qin, X.; Cochrane, E.; Houston, L.S.; Rogers, G.B.; Marsh, P.; Prehar, K.; et al. Staphylococcus aureus small-colony variants are independently associated with worse lung disease in children with cystic fibrosis. Clin. Infect. Dis. 2013, 57, 384–391. [Google Scholar] [CrossRef]
- Becker, K.; Laham, N.A.; Fegeler, W.; Proctor, R.A.; Peters, G.; von Eiff, C. Fourier-transform infrared spectroscopic analysis is a powerful tool for studying the dynamic changes in Staphylococcus aureus small-colony variants. J. Clin. Microbiol. 2006, 44, 3274–3278. [Google Scholar] [CrossRef]
- Schleifer, K.H.; Bell, J.A. Family VIII. Staphylococcaceae fam. nov. In Bergey’s Manual of Systematic Bacteriology, Second Edition, Volume Three, The Firmicutes; De Vos, P., Garrity, G.M., Jones, D., Krieg, N.R., Ludwig, W., Rainey, F.A., Schleifer, K.H., Whitman, W.B., Eds.; Springer: Dordrecht, The Netherlands; Berlin/Heidelberg, Germany; London, UK; New York, NY, USA, 2009; pp. 392–433. [Google Scholar]
- von Eiff, C.; Heilmann, C.; Proctor, R.A.; Woltz, C.; Peters, G.; Götz, F. A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J. Bacteriol. 1997, 179, 4706–4712. [Google Scholar] [CrossRef]
- Kriegeskorte, A.; Grubmuller, S.; Huber, C.; Kahl, B.C.; von Eiff, C.; Proctor, R.A.; Peters, G.; Eisenreich, W.; Becker, K. Staphylococcus aureus small colony variants show common metabolic features in central metabolism irrespective of the underlying auxotrophism. Front. Cell. Infect. Microbiol. 2014, 4, 141. [Google Scholar] [CrossRef]
- Lenhard, J.R.; von Eiff, C.; Hong, I.S.; Holden, P.N.; Bear, M.D.; Suen, A.; Bulman, Z.P.; Tsuji, B.T. Evolution of Staphylococcus aureus under vancomycin selective pressure: The role of the small-colony variant phenotype. Antimicrob. Agents Chemother. 2015, 59, 1347–1351. [Google Scholar] [CrossRef]
- van de Rijn, I.; Kessler, R.E. Growth characteristics of group A streptococci in a new chemically defined medium. Infect. Immun. 1980, 27, 444–448. [Google Scholar] [CrossRef]
- Kipp, F.; Kahl, B.C.; Becker, K.; Baron, E.J.; Proctor, R.A.; Peters, G.; von Eiff, C. Evaluation of two chromogenic agar media for recovery and identification of Staphylococcus aureus small-colony variants. J. Clin. Microbiol. 2005, 43, 1956–1959. [Google Scholar] [CrossRef]
- Kloos, W.E.; Schleifer, K.H. Simplified scheme for routine identification of human Staphylococcus species. J. Clin. Microbiol. 1975, 1, 82–88. [Google Scholar] [CrossRef]
- Mellmann, A.; Becker, K.; von Eiff, C.; Keckevoet, U.; Schumann, P.; Harmsen, D. Sequencing and staphylococci identification. Emerg. Infect. Dis. 2006, 12, 333–336. [Google Scholar] [CrossRef]
- Ota, Y.; Matsumoto, T.; Sugano, M.; Honda, T. Identification of Clinical Thymidine-Dependent Small-Colony Variants of Staphylococcus aureus by Using Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry. Jpn. J. Clin. Pathol. 2015, 63, 683–687. [Google Scholar]
- Gröbner, S.; Beck, J.; Schaller, M.; Autenrieth, I.B.; Schulte, B. Characterization of an Enterococcus faecium small-colony variant isolated from blood culture. Int. J. Med. Microbiol. 2012, 302, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Hoshi, S.; Niwa, T.; Ariyoshi, E.; Matsumoto, T. A case of acute bacterial cystitis caused by carbon dioxide-dependent Escherichia coli with a deletion mutation in the can. J. Infect. Chemother. 2023, 29, 803–805. [Google Scholar] [CrossRef]
- Frickmann, H.; Zautner, A.E.; Moter, A.; Kikhney, J.; Hagen, R.M.; Stender, H.; Poppert, S. Fluorescence in situ hybridization (FISH) in the microbiological diagnostic routine laboratory: A review. Crit. Rev. Microbiol. 2017, 43, 263–293. [Google Scholar] [CrossRef]
- Kipp, F.; Ziebuhr, W.; Becker, K.; Krimmer, V.; Höß, N.; Peters, G.; Von Eiff, C. Detection of Staphylococcus aureus by 16S rRNA directed in situ hybridisation in a patient with a brain abscess caused by small colony variants. J. Clin. Pathol. 2003, 74, 1000–1002. [Google Scholar]
- Goh, S.H.; Santucci, Z.; Kloos, W.E.; Faltyn, M.; George, C.G.; Driedger, D.; Hemmingsen, S.M. Identification of Staphylococcus species and subspecies by the chaperonin 60 gene identification method and reverse checkerboard hybridization. J. Clin. Microbiol. 1997, 35, 3116–3121. [Google Scholar] [CrossRef]
- Martineau, F.; Picard, F.J.; Ke, D.; Paradis, S.; Roy, P.H.; Ouellette, M.; Bergeron, M.G. Development of a PCR assay for identification of staphylococci at genus and species levels. J. Clin. Microbiol. 2001, 39, 2541–2547. [Google Scholar] [CrossRef]
- Poyart, C.; Quesne, G.; Boumaila, C.; Trieu-Cuot, P. Rapid and accurate species-level identification of coagulase-negative staphylococci by using the sodA gene as a target. J. Clin. Microbiol. 2001, 39, 4296–4301. [Google Scholar] [CrossRef]
- Pizauro, L.J.L.; de Almeida, C.C.; Soltes, G.A.; Slavic, D.; Rossi-Junior, O.D.; de Ávila, F.A.; Zafalon, L.F.; MacInnes, J.I. Species level identification of coagulase negative Staphylococcus spp. from buffalo using matrix-assisted laser desorption ionization-time of flight mass spectrometry and cydB real-time quantitative PCR. Vet. Microbiol. 2017, 204, 8–14. [Google Scholar] [CrossRef]
- Acar, J.F.; Goldstein, F.W.; Lagrange, P. Human infections caused by thiamine- or menadione-requiring Staphylococcus aureus. J. Clin. Microbiol. 1978, 8, 142–147. [Google Scholar] [CrossRef]
- Devriese, L.A. Hemin-dependent mutants isolated from methicillin-resistant Staphylococcus aureus strains. Antonie Van Leeuwenhoek 1973, 39, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Slifkin, M.; Merkow, L.P.; Kreuzberger, S.A.; Engwall, C.; Pardo, M. Characterization of CO2 dependent microcolony variants of Staphylococcus aureus. Am. J. Clin. Pathol. 1971, 56, 584–592. [Google Scholar] [CrossRef] [PubMed]
- Zeroual, W.; Choisy, C.; Doglia, S.M.; Bobichon, H.; Angiboust, J.F.; Manfait, M. Monitoring of bacterial growth and structural analysis as probed by FT-IR spectroscopy. Biochim. Biophys. Acta 1994, 1222, 171–178. [Google Scholar] [CrossRef]
- Helm, D.; Labischinski, H.; Schallehn, G.; Naumann, D. Classification and identification of bacteria by Fourier-transform infrared spectroscopy. Microbiology 1991, 137, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Seggewiß, J.; Becker, K.; Kotte, O.; Eisenacher, M.; Yazdi, M.R.; Fischer, A.; McNamara, P.; Al Laham, N.; Proctor, R.; Peters, G.; et al. Reporter metabolite analysis of transcriptional profiles of a Staphylococcus aureus strain with normal phenotype and its isogenic hemB mutant displaying the small-colony-variant phenotype. J. Bacteriol. 2006, 188, 7765–7777. [Google Scholar] [CrossRef]
- Schaaff, F.; Bierbaum, G.; Baumert, N.; Bartmann, P.; Sahl, H.G. Mutations are involved in emergence of aminoglycoside-induced small colony variants of Staphylococcus aureus. Int. J. Med. Microbiol. 2003, 293, 427–435. [Google Scholar] [CrossRef]
- Köser, C.U.; Holden, M.T.; Ellington, M.J.; Cartwright, E.J.; Brown, N.M.; Ogilvy-Stuart, A.L.; Hsu, L.Y.; Chewapreecha, C.; Croucher, N.J.; Harris, S.R.; et al. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N. Engl. J. Med. 2012, 366, 2267–2275. [Google Scholar] [CrossRef]
- Lannergård, J.; von Eiff, C.; Sander, G.; Cordes, T.; Seggewiß, J.; Peters, G.; Proctor, R.A.; Becker, K.; Hughes, D. Identification of the genetic basis for clinical menadione-auxotrophic small-colony variant isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 2008, 52, 4017–4022. [Google Scholar] [CrossRef]
- Clements, M.O.; Watson, S.P.; Poole, R.K.; Foster, S.J. CtaA of Staphylococcus aureus is required for starvation survival, recovery, and cytochrome biosynthesis. J. Bacteriol. 1999, 181, 501–507. [Google Scholar] [CrossRef]
- Besier, S.; Ludwig, A.; Ohlsen, K.; Brade, V.; Wichelhaus, T.A. Molecular analysis of the thymidine-auxotrophic small colony variant phenotype of Staphylococcus aureus. Int. J. Med. Microbiol. 2007, 297, 217–225. [Google Scholar] [CrossRef]
- Kriegeskorte, A.; Block, D.; Drescher, M.; Windmüller, N.; Mellmann, A.; Baum, C.; Neumann, C.; Lorè, N.I.; Bragonzi, A.; Liebau, E.; et al. Inactivation of thyA in Staphylococcus aureus attenuates virulence and has a strong impact on metabolism and virulence gene expression. MBio 2014, 5, e01447-14. [Google Scholar] [CrossRef]
- Abu-Qatouseh, L.F.; Chinni, S.V.; Seggewiß, J.; Proctor, R.A.; Brosius, J.; Rozhdestvensky, T.S.; Peters, G.; von Eiff, C.; Becker, K. Identification of differentially expressed small non-protein-coding RNAs in Staphylococcus aureus displaying both the normal and the small-colony variant phenotype. J. Mol. Med. 2010, 88, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Chua, K.; Davies, J.K.; Newton, H.J.; Seemann, T.; Harrison, P.F.; Holmes, N.E.; Rhee, H.W.; Hong, J.I.; Hartland, E.L.; et al. Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog. 2010, 6, e1000944. [Google Scholar] [CrossRef] [PubMed]
- Gómez-González, C.; Acosta, J.; Villa, J.; Barrado, L.; Sanz, F.; Orellana, M.Á.; Otero, J.R.; Chaves, F. Clinical and molecular characteristics of infections with CO2-dependent small-colony variants of Staphylococcus aureus. J. Clin. Microbiol. 2010, 48, 2878–2884. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Painter, K.L.; Strange, E.; Parkhill, J.; Bamford, K.B.; Armstrong-James, D.; Edwards, A.M. Staphylococcus aureus adapts to oxidative stress by producing H2O2-resistant small-colony variants via the SOS response. Infect. Immun. 2015, 83, 1830–1844. [Google Scholar] [CrossRef] [PubMed]
- Bazaid, A.S.; Forbes, S.; Humphreys, G.J.; Ledder, R.G.; O’Cualain, R.; McBain, A.J. Fatty Acid Supplementation Reverses the Small Colony Variant Phenotype in Triclosan-Adapted Staphylococcus aureus: Genetic, Proteomic and Phenotypic Analyses. Sci. Rep. 2018, 8, 3876. [Google Scholar] [CrossRef]
- Enright, M.C.; Day, N.P.; Davies, C.E.; Peacock, S.J.; Spratt, B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000, 38, 1008–1015. [Google Scholar] [CrossRef]
- Frenay, H.M.; Bunschoten, A.E.; Schouls, L.M.; van Leeuwen, W.J.; Vandenbroucke-Grauls, C.M.; Verhoef, J.; Mooi, F.R. Molecular typing of methicillin-resistant Staphylococcus aureus on the basis of protein A gene polymorphism. Eur. J. Clin. Microbiol. Infect. Dis. 1996, 15, 60–64. [Google Scholar] [CrossRef]
- Shopsin, B.; Gomez, M.; Montgomery, S.O.; Smith, D.H.; Waddington, M.; Dodge, D.E.; Bost, D.A.; Riehman, M.; Naidich, S.; Kreiswirth, B.N. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 1999, 37, 3556–3563. [Google Scholar] [CrossRef]
- Harmsen, D.; Claus, H.; Witte, W.; Rothgänger, J.; Claus, H.; Turnwald, D.; Vogel, U. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 2003, 41, 5442–5448. [Google Scholar] [CrossRef]
- Vergison, A.; Denis, O.; Deplano, A.; Casimir, G.; Claeys, G.; DeBaets, F.; DeBoeck, K.; Douat, N.; Franckx, H.; Gigi, J.; et al. National survey of molecular epidemiology of Staphylococcus aureus colonization in Belgian cystic fibrosis patients. J. Antimicrob. Chemother. 2007, 59, 893–899. [Google Scholar] [CrossRef] [PubMed]
- Baumert, N.; von Eiff, C.; Schaaff, F.; Peters, G.; Proctor, R.A.; Sahl, H.G. Physiology and antibiotic susceptibility of Staphylococcus aureus small colony variants. Microb. Drug Resist. 2002, 8, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Kahl, B.C.; Belling, G.; Becker, P.; Chatterjee, I.; Wardecki, K.; Hilgert, K.; Cheung, A.L.; Peters, G.; Herrmann, M. Thymidine-dependent Staphylococcus aureus small-colony variants are associated with extensive alterations in regulator and virulence gene expression profiles. Infect. Immun. 2005, 73, 4119–4126. [Google Scholar] [CrossRef] [PubMed]
- Schnitzer, R.J.; Camagni, L.J.; Buck, M. Resistance of small colony variants (G-forms) of a Staphylococcus toward the bacteriostatic activity of penicillin. Proc. Soc. Exp. Biol. Med. 1943, 53, 75–78. [Google Scholar] [CrossRef]
- Bulger, R.J. In vitro studies on highly resistant small colony variants of Staphylococcus aureus resistant to methicillin. J. Infect. Dis. 1969, 120, 491–494. [Google Scholar] [CrossRef]
- von Eiff, C.; Friedrich, A.W.; Becker, K.; Peters, G. Comparative in vitro activity of ceftobiprole against staphylococci displaying normal and small-colony variant phenotypes. Antimicrob. Agents Chemother. 2005, 49, 4372–4374. [Google Scholar] [CrossRef]
- Garcia, L.G.; Lemaire, S.; Kahl, B.C.; Becker, K.; Proctor, R.A.; Tulkens, P.M.; Van Bambeke, F. Intracellular forms of menadione-dependent small-colony variants of methicillin-resistant Staphylococcus aureus are hypersusceptible to β-lactams in a THP-1 cell model due to cooperation between vacuolar acidic pH and oxidant species. J. Antimicrob. Chemother. 2012, 67, 2873–2881. [Google Scholar] [CrossRef]
- Kipp, F.; Becker, K.; Peters, G.; von Eiff, C. Evaluation of different methods to detect methicillin resistance in small-colony variants of Staphylococcus aureus. J. Clin. Microbiol. 2004, 42, 1277–1279. [Google Scholar] [CrossRef]
- Nguyen, H.A.; Denis, O.; Vergison, A.; Theunis, A.; Tulkens, P.M.; Struelens, M.J.; Van Bambeke, F. Intracellular activity of antibiotics in a model of human THP-1 macrophages infected by a Staphylococcus aureus small-colony variant strain isolated from a cystic fibrosis patient: Pharmacodynamic evaluation and comparison with isogenic normal-phenotype and revertant strains. Antimicrob. Agents Chemother. 2009, 53, 1434–1442. [Google Scholar] [CrossRef]
- Barcia-Macay, M.; Seral, C.; Mingeot-Leclercq, M.P.; Tulkens, P.M.; Van Bambeke, F. Pharmacodynamic evaluation of the intracellular activities of antibiotics against Staphylococcus aureus in a model of THP-1 macrophages. Antimicrob. Agents Chemother. 2006, 50, 841–851. [Google Scholar] [CrossRef]
- Dik, J.W.; Poelman, R.; Friedrich, A.W.; Panday, P.N.; Lo-Ten-Foe, J.R.; van Assen, S.; van Gemert-Pijnen, J.E.; Niesters, H.G.; Hendrix, R.; Sinha, B. An integrated stewardship model: Antimicrobial, infection prevention and diagnostic (AID). Future Microbiol. 2016, 11, 93–102. [Google Scholar] [CrossRef] [PubMed]
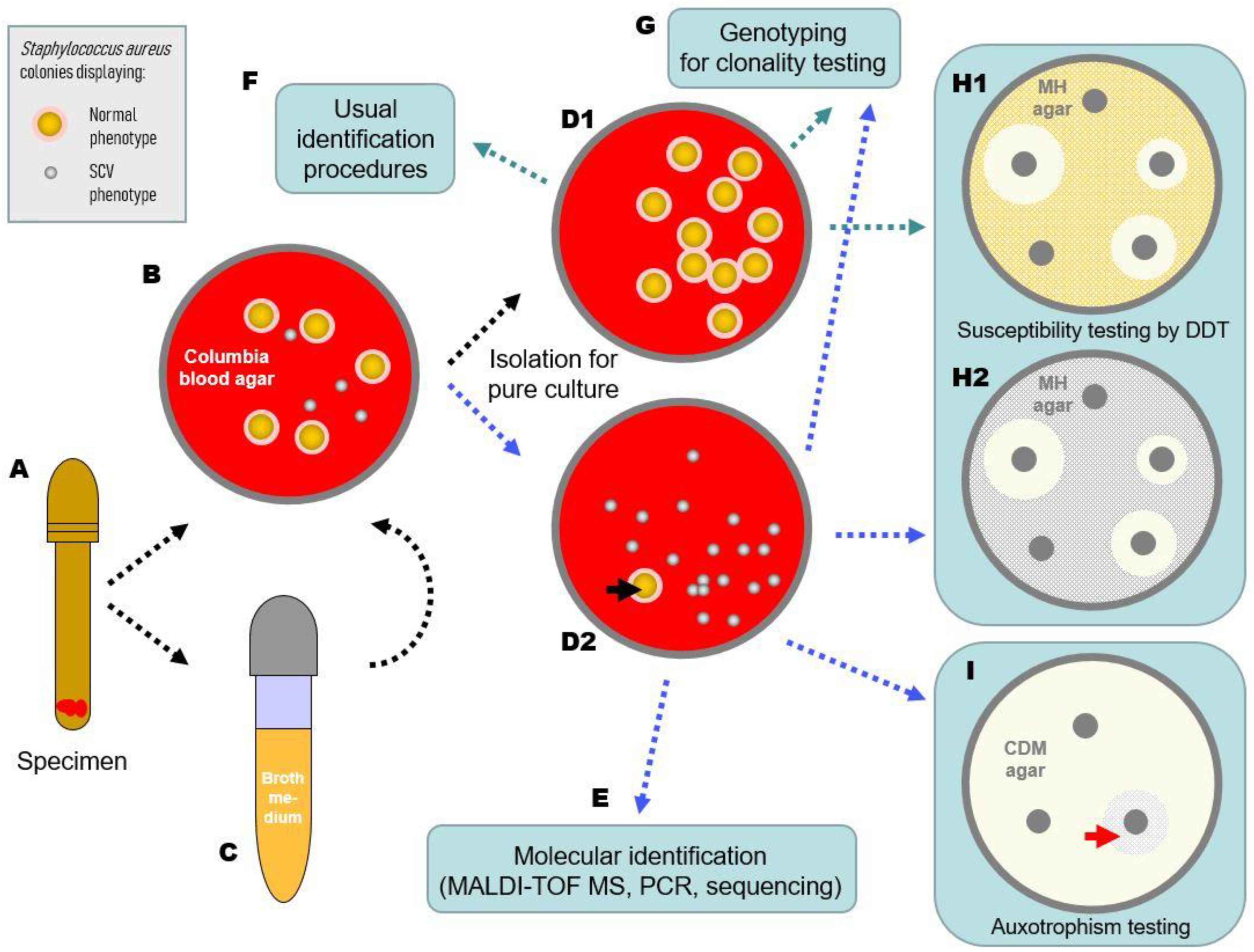

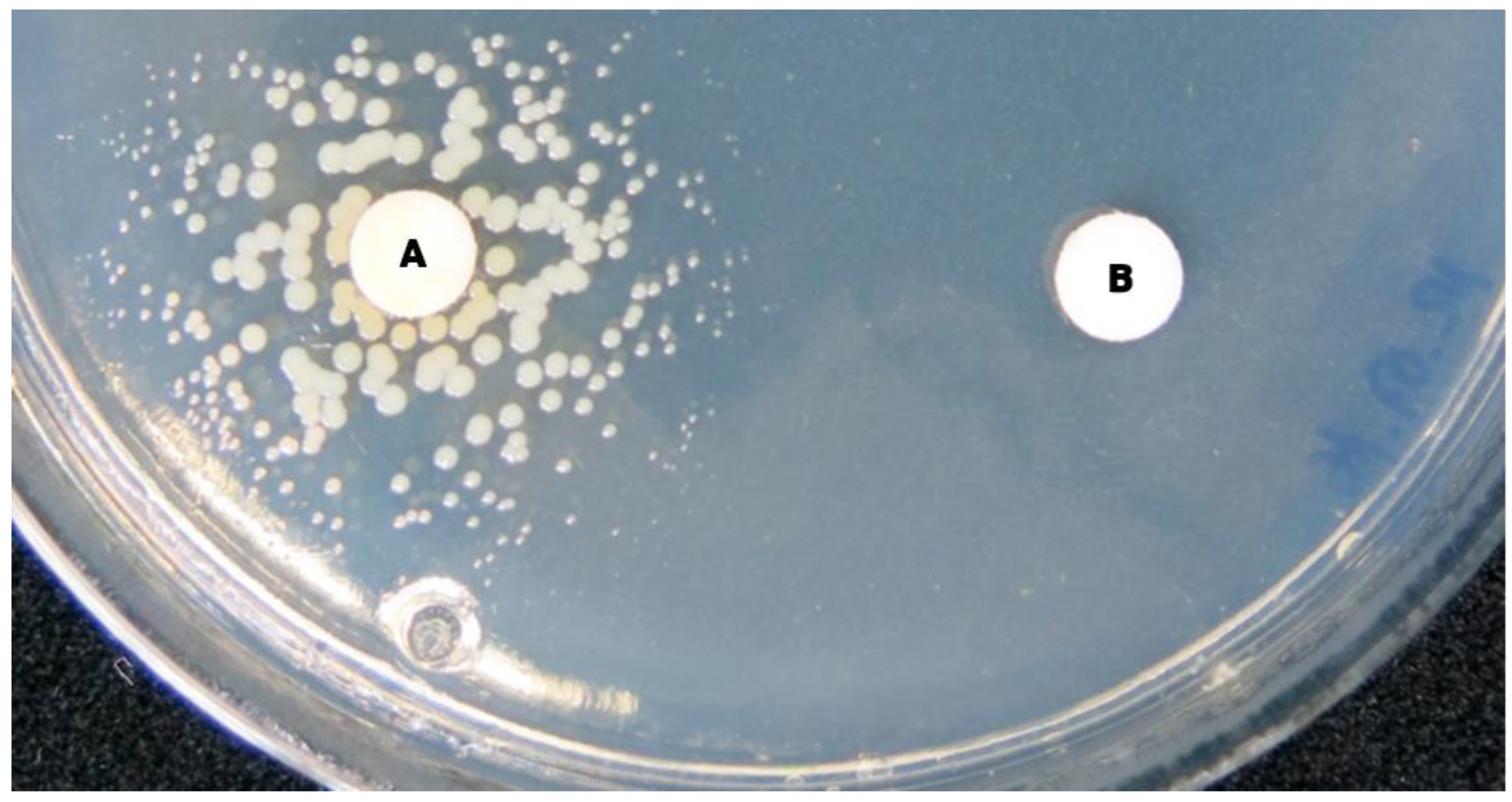
| Colony Morphological Characteristics * | SCV Phenotype ** | Normal Phenotype *** |
|---|---|---|
| Colony appearance | Pinpoint or “fried-egg”-like | Smooth, raised and glistening colonies with entire margins |
| Size after 24 h of incubation | Invisible or tiny (ca. 1/10 of the normal phenotype) | 1–3 mm or larger |
| Pigmentation | Not present or reduced | Unpigmented or coloured, varying from grey to yellow–orange |
| Hemolysis | Not present or reduced | Weak to strong zone of β-hemolysis (in particular, S. aureus and S. haemolyticus) or without hemolysis (most of the CoNS species) |
| Feature * | SCV Phenotype | Normal Phenotype (NP) |
|---|---|---|
| Growth rate | Reduced ** (approx. 48 to >72 h) | Normal (6 to 24 h) |
| Auxotrophism | Present (different types; most frequent hemin-, menadione-, thymidine-dependent SCVs, less often dependency on fatty acids, CO2 and other factors) | Absent |
| Fermentation and other biochemical reactions | (Strongly) delayed to absent | Normal (“overnight”) |
| Coagulase activity *** | (Strongly) delayed | (2) 4 to 24 h |
| Catalase activity | (Strongly) delayed | Rapid reaction |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Becker, K. Detection, Identification and Diagnostic Characterization of the Staphylococcal Small Colony-Variant (SCV) Phenotype. Antibiotics 2023, 12, 1446. https://doi.org/10.3390/antibiotics12091446
Becker K. Detection, Identification and Diagnostic Characterization of the Staphylococcal Small Colony-Variant (SCV) Phenotype. Antibiotics. 2023; 12(9):1446. https://doi.org/10.3390/antibiotics12091446
Chicago/Turabian StyleBecker, Karsten. 2023. "Detection, Identification and Diagnostic Characterization of the Staphylococcal Small Colony-Variant (SCV) Phenotype" Antibiotics 12, no. 9: 1446. https://doi.org/10.3390/antibiotics12091446
APA StyleBecker, K. (2023). Detection, Identification and Diagnostic Characterization of the Staphylococcal Small Colony-Variant (SCV) Phenotype. Antibiotics, 12(9), 1446. https://doi.org/10.3390/antibiotics12091446






